Abstract
Adult stem cells sustain tissue maintenance and regeneration throughout the lifetime of an animal1,2. These cells often reside in specific signalling niches that orchestrate the stem cell’s balancing act between quiescence and cell-cycle re-entry based on the demand for tissue regeneration2–4. How stem cells maintain their capacity to replenish themselves after tissue regeneration is poorly understood. Here we use RNA-interference-based loss-of-function screening as a powerful approach to uncover transcriptional regulators that govern the self-renewal capacity and regenerative potential of stem cells. Hair follicle stem cells provide an ideal model. These cells have been purified and characterized from their native niche in vivo and, in contrast to their rapidly dividing progeny, they can be maintained and passaged long-term in vitro5–7. Focusing on the nuclear proteins and/or transcription factors that are enriched in stem cells compared with their progeny5,6, we screened ~2,000 short hairpin RNAs for their effect on long-term, but not short-term, stem cell self-renewal in vitro. To address the physiological relevance of our findings, we selected one candidate that was uncovered in the screen: TBX1. This transcription factor is expressed in many tissues but has not been studied in the context of stem cell biology. By conditionally ablating Tbx1 in vivo, we showed that during homeostasis, tissue regeneration occurs normally but is markedly delayed. We then devised an in vivo assay for stem cell replenishment and found that when challenged with repetitive rounds of regeneration, the Tbx1-deficient stem cell niche becomes progressively depleted. Addressing the mechanism of TBX1 action, we discovered that TBX1 acts as an intrinsic rheostat of BMP signalling: it is a gatekeeper that governs the transition between stem cell quiescence and proliferation in hair follicles. Our results validate the RNA interference screen and underscore its power in unearthing new molecules that govern stem cell self-renewal and tissue-regenerative potential.
Stem cell self-renewal is the process by which stem cells proliferate and generate more stem cells. This process requires control of the cell cycle and maintenance of the undifferentiated state. Embryonic stem cells are refractory to most proliferation checkpoints4, and they typically promote self-renewal by suppressing differentiation8. By contrast, the few established regulators of self-renewal in adult stem cells function by regulating cell-cycle progression9–12. A prerequisite to unlocking the key to regenerative medicine is to dissect the complex mechanisms governing stem cell self-renewal in adult tissues.
With their enormous capacity for tissue regeneration, hair follicles offer an ideal system to explore these mechanisms. Hair follicle stem cells (HF-SCs) become activated early in each hair growth cycle, when a few of these cells exit their niche (called the bulge) to generate a new hair follicle. The differentiation of stem cells into lineage-restricted progeny probably requires micro-environmental stimuli that are not present in the stem cell niche, because this process happens gradually along the follicle outer root sheath13–15. The stepwise process culminates at the base of the mature follicle, where committed transit-amplifying matrix cells differentiate into the six lineages that are involved in hair production.
Following their activation, stem cells within the bulge and its vicinity (the upper outer root sheath, which becomes the bulge in the next cycle) briefly self-renew, replenishing the expended stem cells and ensuring long-term tissue regeneration13–15. Niche HF-SCs also proliferate following injury and repair wounds13–15. Another feature that distinguishes HF-SCs from their committed progeny is their ability to be propagated for at least five passages in vitro, reflecting their capacity for long-term proliferative potential6.
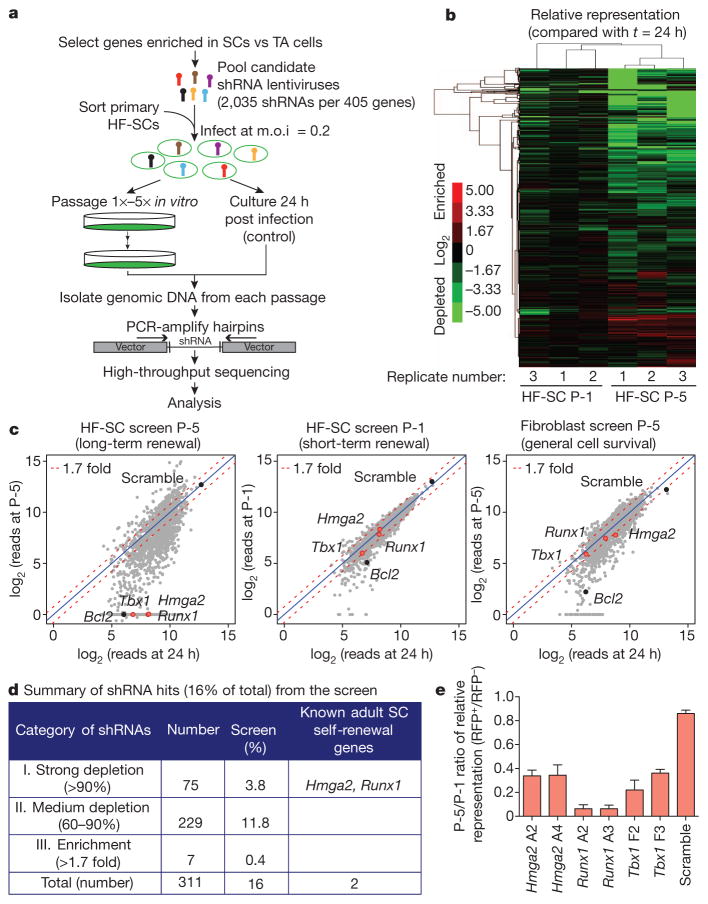
In the current study, we surmised that there might be two sources for finding intrinsic factors responsible for maintaining ‘stemness’ inside and outside the stem cell niche: self-renewal factors that have been identified in other stem cell studies; and nuclear proteins that we found to be enriched twofold in stem cells relative to their transit-amplifying progeny (Supplementary Fig. 1a, b). Focusing on about 400 such candidates, we devised an in vitro RNA interference (RNAi) screen for long-term versus short-term self-renewal (Fig. 1a). By choosing genes whose expression was enriched in stem cells relative to committed proliferative progeny, this pool of candidates should not contain housekeeping genes and general proliferation-associated genes. However, if short hairpin RNAs (shRNAs) target a gene that is essential for long-term but not short-term self-renewal, then cells expressing this gene should persist during early passages but then decrease in number or disappear with sequential passaging. Operating on this premise, we transduced purified primary HF-SCs in triplicate with a lentiviral pool encoding control (scramble) shRNAs and a pool of 2,035 candidate shRNAs (about five per gene) such that, on average, each stem cell expressed a single shRNA (Supplementary Fig. 1c). The transduced stem cells were cultured and, at 24 h and following each passage, shRNAs were amplified from the surviving cells and subjected to high-throughput sequencing.
Figure 1. In vitro RNAi screen for genes involved in stem cell long-term self-renewal.
a, In vitro RNAi screening strategy. b, Unsupervised hierarchical clustering of screening replicates. c, Scatter plots of normalized reads per shRNA between 24 h post infection and after one passage (P-1) or five passages (P-5) for HF-SCs (left and centre) and after five passages for fibroblasts (right). The data shown are from one replicate of each screening, highlighting the genes whose corresponding shRNAs were specifically depleted in the long-term passaging of stem cells (red dots) and control genes (black dots). The blue line is the diagonal line with a ratio of 1.0. The red dashed line shows the cut off for 1.7-fold change.
d, Screening statistics. e, Progressive selection against hairpins that target putative long-term self-renewal genes. Data are presented as mean ±s.d.; n =3. m.o.i., multiplicity of infection; RFP, red fluorescent protein; SC, stem cell; TA, transit-amplifying; vs, versus.
Data are shown for passage 1 (P-1) and P-5 (Fig. 1b, c, Supplementary Figs 2 and 3a, and Supplementary Tables 1 and 2). More than 96% of the initial shRNAs were detected at 24 h after transduction, and these shRNAs were used as a reference for changes in shRNA representation. Consistent with our strategic exclusion of housekeeping genes and general proliferation-associated genes, most cells that harboured shRNAs survived the first passage. By contrast, after five passages, many shRNAs were depleted or enriched, suggesting that the transduced cells had different long-term proliferative potentials. Using unsupervised hierarchical clustering, triplicates of individually transduced and passaged cells behaved strikingly similarly, suggesting that these changes reflected bona fide alterations in stem cell character.
Parallel screens with fibroblasts weeded out shRNAs corresponding to cell-survival genes such as Bcl2, which were selected against after five passages in both HF-SCs and fibroblasts (Fig. 1c, Supplementary Fig. 3b and Supplementary Table 3). Our refined short list of self-renewal candidates contained those whose cognates all showed similar trends and for which two or more shRNAs per gene displayed specific changes in P-5 stem cell cultures but not in P-1 stem cell cultures or in P-5 fibroblasts (Supplementary Fig. 2 and Supplementary Table 1). Category I shRNAs (Fig. 1d) were maintained in P-1 stem cell cultures but were underrepresented by more than 90% at P-5, meeting the criteria for an shRNA that suppresses a long-term self-renewal gene. Representing only 3.8% of the initial pool, category I included shRNAs targeting Hmga2, which is required for neural stem cell self-renewal10, and Runx1, which promotes HF-SC proliferation16.
Real-time PCR (rtPCR) of transcripts targeted by six of the most effective category I shRNAs confirmed that each shRNA blocked the expression of its intended target (Supplementary Fig. 4). Moreover, stem cells that were individually transduced with Hmga2, Runx1 or Tbx1 shRNA were progressively selected against over time (Fig. 1e). The transcription factor TBX1 was particularly intriguing because it has been implicated in tissue formation in other organs17,18. We selected it as our model for in vivo testing of the functional relevance of our RNAi screen.
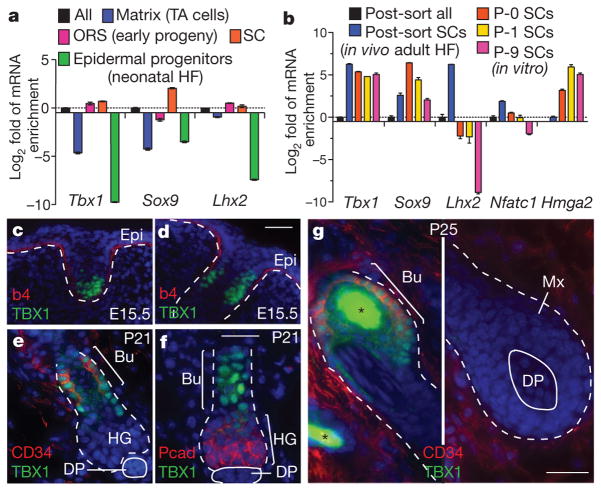
rtPCR and epigenetic chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) analyses19 of purified hair follicle populations revealed that Tbx1 was transcribed at higher levels in stem cells than in any of their progeny (Fig. 2a, b and Supplementary Fig. 5a). In vivo, the developmental expression of TBX1 protein most closely resembled that of two essential HF-SC transcription factors, SOX9 and LHX2 (Fig. 2c, d). The adult pattern of expression resembled that of CD34, which is a cell surface marker of HF-SCs (Fig. 2e, f). Nuclear TBX1 was not detected in self-renewing transit-amplifying cells or in terminally differentiating cells (Fig. 2g). Notably, in contrast to some other HF-SC transcription factors, TBX1 was also maintained in stem cells in long-term cultures.
Figure 2. The transcription factor TBX1 is highly enriched in stem cells in vivo and in vitro.
a, b, Tbx1 expression in HF-SCs, shown compared with established HF-SC transcription factor genes, as determined by rtPCR. The mRNAs were isolated from fluorescence-activated cell sorting (FACS)-purified stem cells or other populations, as indicated. Data are presented as mean ±s.d.; n =3. c–g, Nuclear TBX1 in the skin is restricted to developing and postnatal HF-SCs. The back skins of mice were processed for immunofluorescence at embryonic day 15.5 (E15.5), postnatal day 21 (P21) and P25 (P25). The white dashed line indicates the dermal–epithelial boundary, and the asterisk denotes hair shaft autofluorescence. Nuclei are shown in blue. Scale bars, 30 μm. b4, β4-integrin; Bu, bulge stem cells; DP, dermal papilla; Epi, epidermis; HF, hair follicle; HG, hair germ (early stem cell progeny); Mx, matrix (committed TA progeny); ORS, outer root sheath; Pcad, P-cadherin.
To evaluate TBX1 function in vivo, we conditionally targeted Tbx1 (Tbx1-cKO) in the skin epithelium of embryonic day 15.5 mice20,21. Tbx1-cKO mice were viable, and hair follicle morphogenesis appeared to be normal (Supplementary Fig. 5b, c). We tested for possible defects in stemness by analysing tissue regeneration during the normal hair cycle (Supplementary Fig. 6). For this purpose, same-sex littermates were shaved at the normal onset of the first hair cycle (postnatal day 21, P21) and the second hair cycle (P60). In both cases, Tbx1-cKO hair follicles remained quiescent longer than normal, but they eventually cycled. Maturation/differentiation was unaffected, as shown by the development of normal hair types and lengths.
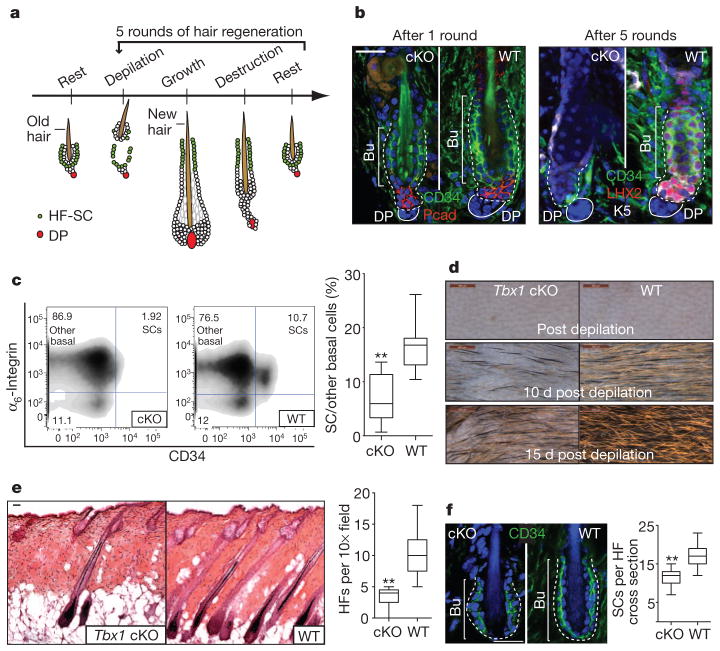
Self-renewal occurs briefly after anagen onset, replenishing the stem cells that are used during initiation15. To challenge stem cells to sustain long-term tissue regenerative potential, we repeatedly depilated the hair coat, a process that removes the old hair along with tightly adhering niche signalling cells that maintain stem cell quiescence14,15 (Fig. 3a). After depilation, more than 80% of the stem cells remained viable in their niche, where they became activated to enter a new hair cycle (Supplementary Fig. 7).
Figure 3. Tbx1-null stem cells fail in an in vivo assay for stem cell self-renewal and long-term tissue regeneration.
a, Schematic of the in vivo long-term hair regeneration assay. b, Decline in stem cell number with sequential rounds of hair regeneration in Tbx1-cKO hair follicles. c, FACS quantifications of stem cells after five rounds of hair regeneration.; n =11 for WT, n =12 for cKO. d, Close up of skin surface in the fifth cycle of hair regeneration. d, day. e, Representative haematoxylin and eosin images and quantifications of follicle densities in the fifth cycle of hair regeneration; n =4. f, Decline in stem cell number during normal ageing (1 year old) in Tbx1-cKO skin; n =5. c, e, f, Box-and-whisker plots: mid-line, median; box, 25th to 75th percentiles; and whiskers, minimum and maximum. **, P <0.001. b, d, e, f, The white dashed line indicates the dermal–epithelial boundary, and nuclei are shown in blue. Scale bars, 500 μm (d) and 30 μm (b, e, f).
Wild-type (WT) HF-SCs survived each round of depilation-induced hair regeneration, indicating the robust ability to sustain self-renewal and long-term tissue regeneration. By contrast, after five rounds, the Tbx1-cKO stem cell numbers had declined by more than 70% (P <0.001) (Fig. 3b, c). Their steady depletion was accompanied by a thinning of the hair coat and a reduction in hair follicle density (Fig. 3d, e and Supplementary Fig. 8). Histological analysis revealed that many Tbx1-cKO hair follicles were dormant and had lost their stem cell niche, retaining only sebaceous glands and dermal papillae. However, the few hair follicles that cycled appeared morphologically normal, reflecting the presence of active stem cell niches (Fig. 3e).
Similar results were obtained when hair cycles were monitored during natural ageing. Although the intervals between the hair cycles were longer than those in the depilation procedure, the Tbx1-cKO stem cell niche residents had declined by about 30% after 1 year (Fig. 3f). Thus, Tbx1-null stem cells seem to be specifically impaired in their long-term ability to replenish their niche during normal and depilation-induced tissue regeneration.
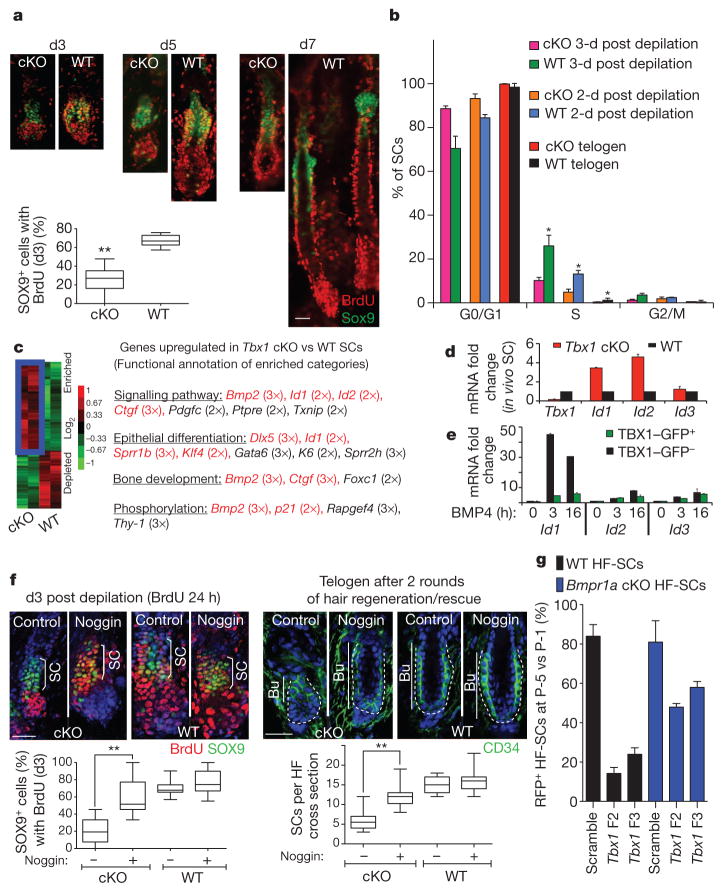
We used 5-bromodeoxyuridine (BrdU) incorporation to define the brief window of bulge stem cell proliferation that occurs following depilation. WT stem cell proliferation peaked at day 3 after depilation, and the cells returned to quiescence by day 7. Tbx1-cKO stem cells also proliferated but to a lesser extent during this time period (Fig. 4a). Within 2 to 3 days of depilation, only about 25% of Tbx1-null stem cells were BrdU-positive, whereas about 70% of WT stem cells were BrdU-positive (Fig. 4a). This proliferative decrease was verified by DNA-content-based cell-cycle analysis of purified stem cells, which showed that the decrease was accompanied by fewer stem cells being present in the niche (Fig. 4b). As discussed earlier, most hair follicles eventually produced hairs of WT length, reflecting an otherwise normal lineage program.
Figure 4. TBX1 controls stem cell proliferation in part by fine-tuning the response to BMP signalling.
a, Stem cell proliferative status post depilation, as assessed by BrdU incorporation (with BrdU added 24 h before analysis); n =3–5. b, Cell-cycle analysis of FACS-isolated HF-SCs; n =3–5; *, with respect to cKO. c, Gene expression analysis of HF-SCs. Genes upregulated in HF-SCs from Tbx1-cKO mice compared with WT HF-SCs are delineated in blue. Functional annotations for the enriched categories are shown. d, rtPCR of BMP targets in HF-SCs. e, rtPCR of Id1, Id2 and Id3 post BMP4 treatment with or without TBX1–GFP. f, Transient inhibition of BMP signalling by noggin in vivo rescues stem cell numbers in Tbx1-cKO mice; n =3 and ~50 hair follicles each. The white dashed line indicates the dermal–epithelial boundary, and nuclei are shown in blue. g, Long-term passage of Tbx1-shRNA-transduced HF-SCs; n =3. b, d, g, Data are presented as mean ±s.d. *, P <0.01. a, f, Box-and-whisker plots: mid-line, median; box, 25th to 75th percentiles; and whiskers, minimum and maximum. **, P <0.001. Scale bars, 30 μm.
We observed a similar trend when we monitored the normal hair cycle. In this case, fewer stem cells were expended than during depilation; thus, the demand for self-renewal was lower, as reflected by the natural bulge niche having a lower proliferative activity than the depilation-induced WT bulge niche. Consistent with a role for TBX1 in HF-SC self-renewal, the overall proliferative activity within the Tbx1-null niche was less than in the WT niche, and that in the natural niche was less than in the depilation-induced niche (Supplementary Fig. 9).
To understand how these differences arise, we transcriptionally profiled messenger RNAs that were isolated from purified HF-SCs 2 days after depilation (Fig. 4c, Supplementary Fig. 10 and Supplementary Table 4). Bioinformatic analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) functional gene annotation tool uncovered mostly cell-cycle regulators in the 123 genes that were downregulated by 1.8 fold or more in Tbx1-null stem cells compared with WT stem cells. The 188 genes that were upregulated by 1.8 fold or more were enriched for genes implicated in bone morpho-genetic protein (BMP) signalling (Fig. 4c, red), including Bmp2, which encodes a secreted ligand for BMP receptors, and Id genes, which are major targets of the transcriptional effectors of BMPs, namely SMAD4 in complex with phosphorylated SMAD1 (pSMAD1), pSMAD5 or pSMAD8 (denoted pSMAD1/5/8). Using rtPCR, Tbx1-cKO HF-SCs upregulated Id1 and Id2 (refs 22, 23) (Fig. 4d).
In cardiomyocytes, TBX1 seems to suppress BMP signalling by competitively interfering with SMAD4 for pSMAD1/5/8 binding24. Consistent with this idea, overexpression of a TBX1–green fluorescent protein (GFP) fusion protein in WT keratinocytes significantly suppressed BMP-induced Id1 transcription in vitro (Fig. 4e). Similar effects, albeit slower and weaker, were observed for Id2.
Transgenic overactivation of the BMP circuitry results in hair coat thinning with age25. If TBX1-deficient HF-SCs have a heightened sensitivity to BMP signalling, then BMP inhibitors might ameliorate the proliferative defect. We tested this hypothesis by plucking hair follicles from mice and then injecting them intradermally with beads soaked in the BMP antagonist noggin26,27. Within 3 days, Tbx1-cKO HF-SC proliferation had been restored to near WT levels (Fig. 4f, P <0.001, and Supplementary Fig. 11). As reflected by the bulge size, the expanded HF-SC pool was sustained throughout the hair cycle. However, additional treatments with noggin were necessary to maintain the stem cell pool through multiple rounds of depilation-induced hair regeneration. Interestingly, the self-renewal of TBX1-deficient stem cells was also elevated in vitro when BMP signalling was impaired by ablation of the BMP receptor BMPR1A (Fig. 4g). Together, these findings are consistent with a BMP-induced proliferative defect in Tbx1-null stem cells.
Given these inverse links between TBX1 and BMP signalling, it seemed paradoxical that Smad1 shRNAs surfaced in our self-renewal screen (Supplementary Table 1). Further analyses revealed that even though these shRNAs depleted Smad1 transcripts and SMAD1 protein, the SMAD1/5/8 target genes Id1, Id2 and Id3 were still expressed. Moreover, the transduced cells still responded to BMP signalling, as judged by reporter assays (Supplementary Fig. 12).
HF-SCs reside in a WNT-restricted, BMP-high micro-environment28,29 in which they must self-renew to replenish the stem cell pool. Therefore, HF-SCs must have an intrinsic mechanism to lower the BMP signalling threshold, and this mechanism fails to occur in the absence of TBX1. Entering the hair cycle also necessitates decreased BMP signalling; however, in this case, the proliferation is fuelled by early progeny (hair germ) with naturally low TBX1 levels that are present at the bulge base6,30. Because, paradoxically, hair cycle initiation was delayed in Tbx1-null mice, we surmise that the hair germ may be negatively influenced by BMP2 or other local effectors that are secreted by Tbx1-null HF-SCs.
In summary, we have discovered that TBX1 functions in the replenishment of HF-SCs during tissue regeneration. Our RNAi screens excluded roles for TBX1 in cell cycling, housekeeping and survival. The finding that once initiated, Tbx1-null hair cycles have a normal progression also ruled out roles for TBX1 in cell-fate determination or lineage progression. Instead, the TBX1 defect seems to be rooted in diminished stem cell self-renewal, coupled with enhanced sensitization to intrinsic BMP signalling. Together, these result in progressive HF-SC depletion and thinning of the hair coat. Although the effects of TBX1 are likely to be more complex than we have shown, its ability to intrinsically control these features poises it in the middle of a balancing act with the micro-environment to control stem cell behaviour in tissue homeostasis.
METHODS SUMMARY
RNAi screen
Candidate gene selection was based on previously published micro-array analyses5,6. HF-SCs isolated by fluorescence-activated cell sorting (FACS) were infected with an shRNA library (carried by TRC lentiviruses) targeting a set of ~400 candidate genes (2,035 shRNAs, with approximately 5 shRNAs per gene) to a final infection rate of approximately 20% (that is, about one virus per five HF-SCs). Twenty-four hours after infection, half of the infected HF-SCs were collected, and the other half were plated onto mitomycin-C-treated fibroblast feeder layers. Each week, nearly confluent cultures were trypsinized and replated (one passage). At each passage, a fraction of the cells were processed for genomic DNA isolation. Primers, including adaptors for Solexa sequencing, were used to amplify shRNA-encoding sequences from genomic DNA. Following the PCR amplification of shRNAs, sequencing was performed on an Illumina/Solexa Genome Analyzer II according to the manufacturer’s protocols. Analyses and plots of DNA sequencing data were performed in the R statistical environment. All shRNA identities, as well as primary screening data, are listed in Supplementary Tables 1–3.
Animal studies
Tbx1fl/fl mice were obtained from A. Baldini. To create conditional knockout mice, we mated hemizygous K14-Cre (CD1) mice with homozygous Tbx1fl/fl (C57BL/6) mice; F1 K14-Cre/Tbx1fl/+ (CD1/C57BL/6) progeny were subsequently bred with homozygous Tbx1fl/fl mice to generate K14-Cre;Tbx1fl/fl mice at a 25% Mendelian frequency. Depilation of mid-dorsal hair follicles was achieved on anaesthetized mice to provide a proliferative stimulus and to synchronize a population of anagen hair follicles. The mid-dorsum was coated with molten wax, which was peeled off after hardening. BrdU (Sigma-Aldrich) pulse experiments involved intraperitoneal injections (50 μg BrdU per g body weight) twice a day. After BrdU pulses for the indicated times, animals were killed. The skins were embedded in OCT compound, frozen in dry ice, sectioned using a cryostat (Richard-Allan Scientific) and stained for immunofluorescence using antibodies specific for BrdU.
METHODS
RNAi screen
Candidate gene selection was carried out based on previously published microarray analyses5,6. Fluorescence-activated cell sorting (FACS)-isolated HF-SCs were infected with an shRNA library (carried by TRC lentiviruses) targeting a set of ~400 candidate genes (2,035 shRNAs, with approximately 5 shRNAs per gene for most, if not all, candidates) to a final infection rate of approximately 20% (that is, about one virus per five HF-SCs). Twenty-four hours after infection, half of the infected HF-SCs were collected, and the other half were plated onto mitomycin-C-treated fibroblast feeder layers. Each week, nearly confluent cultures were trypsinized and replated (one passage). At each passage, a fraction of the cells were processed for genomic DNA isolation. Primers, including adaptors for Solexa sequencing, were used to amplify shRNAs from genomic DNA. Following the PCR amplification of hairpins, sequencing was performed on an Illumina/Solexa Genome Analyzer II according to the manufacturer’s protocols. Analyses and plots of DNA sequencing data were performed in the R statistical environment31. The fold change in shRNA representation after sequential passages in vitro was determined by comparing the shRNA representation in each sample to that in the control cell population collected 24 h after infection. The identities of all ‘hit’ shRNAs, as well as primary fold changes, are listed in Supplementary Tables 1–3.
Fluorescence based competition assay
shRNA-transduced (RFP+) HF-SCs (GFP+) were mixed with non-transduced HF-SCs and plated onto feeders (GFP−). At each passage, the cells were trypsinized and replated, and a fraction of the cells were used to measure the proportion of RFP+GFP+/RFP−GFP+ cells using flow cytometry on an LSRII FACS Analyzer (BD Biosciences). Analyses were carried out for five consecutive passages.
RNAi construct and sequences
The RNAi lentiviral constructs were from the RNAi Consortium (TRC) mouse lentiviral library. Scramble shRNA (Ctrl shRNA) that does not target any mouse mRNA was used as a control. Lentiviral vectors were packaged, concentrated and used for infection as previously described32. Sequences of individual shRNAs used in experiments are listed here, together with their symbol in oligoSeq. Tbx1 F2, CCGGGCTACCGGTATGCTTTCCATAC TCGAGTATGGAAAGCATACCGGTAGCTTTTTG; Tbx1 F3, CCGGCTGACC AATAACCTGCTGGATCTCGAGATCCAGCAGGTTATTGGTCAGTTTTTG; Hmga2 A2, CCGGGCCACAACAAGTCGTTCAGAACTCGAGTTCTGAACG ACTTGTTGTGGCTTTTTG; Hmga2 A4, CCGGAGACCTAGGAAATGGCCA CAACTCGAGTTGTGGCCATTTCCTAGGTCTTTTTTG; Runx1 A2 CCGGG CCCTCCTACCATCTATACTACTCGAGTAGTATAGATGGTAGGAGGGCT TTTTG; Runx1 A3, CCGGGTCTTTACAAATCCGCCACAACTCGAGTTGTG GCGGATTTGTAAAGACTTTTTG; Smad1 B3, CCGGGCCGAGTAACTGC GTCACCATCTCGAGATGGTGACGCAGTTACTCGGCTTTTT; Smad1 F1, CCGGCCCATTTGGTTCCAAGCAGAACTCGAGTTCTGCTTGGAACCAAA TGGGTTTTT; Smad1 F2, CCGGACCGTGTATGAACTCACCAAACTCGAGT TTGGTGAGTTCATACACGGTTTTTT; Smad1 F4, CCGGTGGTGCTCTATT GTGTACTATCTCGAGATAGTACACAATAGAGCACCATTTTT; and Smad1 F5, CCGGTCCTATTTCATCCGTGTCTTACTCGAGTAAGACACGGATGAA ATAGGATTTTT.
Mice, depilation and BrdU labelling
bx1fl/fl mice were obtained from A. Baldini. To create conditional knockout mice, we mated hemizygous K14-Cre (CD1) mice with homozygous Tbx1fl/fl(C57BL/6) mice; F1 K14-Cre/Tbx1fl/+ (CD1/C57BL/6) progeny were subsequently bred with homozygous Tbx1fl/fl mice to generate K14-Cre;Tbx1fl/fl mice at a 25% Mendelian frequency. Depilation of mid-dorsal hair follicles was achieved on anaesthetized mice to provide a proliferative stimulus and to synchronize a population of anagen hair follicles. The mid-dorsum was coated with molten wax, which was peeled off after hardening. BrdU (Sigma-Aldrich) pulse experiments involved intraperitoneal injections (50 μg BrdU per g body weight) twice a day. After BrdU pulses for the indicated times, animals were killed. The skins were embedded in OCT compound, frozen in dry ice, sectioned using a cryostat (Richard-Allan Scientific) and stained for immunofluorescence using antibodies specific for BrdU. All animals were maintained in an AAALAC-approved animal facility, and procedures were performed with IACUC-approved protocols.
Semi-quantitative RT–PCR
RNAs were isolated from cells using an RNeasy kit (QIAGEN), and DNase treatment was performed to remove genomic DNA. Equal RNA amounts were added to reverse-transcriptase reaction mix (Invitrogen) with oligo(dT)12 as a primer. Semi-quantitative PCR was conducted with a LightCycler system (Roche Diagnostics). Reactions were performed using the indicated primers and template mixed with the LightCycler DNA Master SYBR Green kit and were run for 45 cycles. The specificity of the reactions was determined by subsequent melting curve analysis. LightCycler analysis software was used to remove background fluorescence (noise band). The number of cycles needed to reach the crossing point for each sample was used to calculate the amount of each product using the 2(−Δ−ΔCt) method. The levels of PCR product were expressed as a function of Ppib. The primers were designed to produce a product spanning exon–intron boundaries. The sequences of the primers were as follows: Ppib sense, GTGAGCGCTTCCCAGATGAGA; Ppib antisense, TGCCGGAGTCGACAAT GATG; Sox9 sense, CGGCGGAGGAAGTCGGTGAAGAAC; Sox9 antisense, GGTGGGTGCGGTGCTGCTGATG; Tbx1 sense, GCTGTGGGACGAGTTCA ATC; Tbx1 antisense ACGTGGGGAACATTCGTCT; Lhx2 sense, CCAGCTTC GGACAATGAAGT; Lhx2 antisense, TTTCCTGCCGTAAAAGGTTG; Nfatc1 sense, AACGCCCTGACCACCGATAGCACT; Nfatc1 antisense, CCCGGCT GCCTTCCGTCTCATA; Runx1 sense, CTCCGTGCTACCCACTCACT; Runx1 antisense, ATGACGGTGACCAGAGTGC; Hmga2 sense, AAGGCAGCAAAA ACAAGAGC; Hmga2 antisense, CCGTTTTTCTCCAATGGTCT; Smad1 sense, AACACCAGGCGACATATTGG; Smad1 antisense, CACTGAGGCATTCCG CATA; Smad5 sense, GCAGTAACATGATTCCTCAGACC; Smad5 antisense, GCGACAGGCTGAACATCTCT; Smad8 sense, CGGATGAGCTTTGTGAA GG; Smad8 antisense, GGGTGCTCGTGACATCCT; Id1 sense, GAGTCTGAAG TCGGGACCAC; Id1 antisense, TTTTCCTCTTGCCTCCTGAA; Id2 sense, AATGGCCTTTTTGACACGAG; Id2 antisense, AAAGCAAGCAATCAACA TTCAA; Id3 sense, GAGGAGCTTTTGCCACGAC; and Id3 antisense, TGAAGAGGGCTGGGTTAAGA.
Histology and immunofluorescence
Tissues were embedded in OCT compound, and frozen sections were fixed in 4% paraformaldehyde and subjected to immunofluorescence microscopy or haematoxylin and eosin staining as previously described1. The antibodies (and their dilutions) used were as follows: anti-LHX2 (rabbit, 1:2,500, Fuchs lab), anti-SOX9 (rabbit, 1:1,000, Fuchs lab), anti-P-cadherin (goat, 1:100, R&D Systems), anti-α6-integrin (rat, 1:100, Pharmingen), anti-K5 (rabbit, 1:500, Fuchs lab), anti-CD34 (rat, 1:100, Pharmingen), anti-BrdU (rat, 1:500, Abcam), anti-active-caspase 3 (rabbit, 1:500, R&D Systems), anti-TBX1 (rabbit, 1:100, Zymed), and FITC-conjugated (1:100, Jackson) or Alexa594-conjugated (1:1,000, Molecular Probes) secondary antibodies. Nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI). Imaging was performed using either a Zeiss Axioskop equipped with Spot RT (Diagnostic Instruments) or a Zeiss LSM 510 laser-scanning microscope (Carl Zeiss MicroImaging) through a 40× water objective or a 25× objective. RGB images were assembled in Adobe Photoshop CS3, and panels were labelled in Adobe Illustrator CS3.
Isolation of HF-SCs and flow cytometry
Subcutaneous fat was removed from the skins with a scalpel, and the whole skin was placed dermis down on trypsin (Gibco) at 37 °C for 30 min. Single-cell suspensions were obtained by scraping the skin gently. The cells were then filtered with strainers (70 mm, followed by 40 mm). Cell suspensions were incubated with the appropriate antibodies for 30 min on ice. The following antibodies were used: anti-CD34–Alexa647 (1:100, eBioscience) and anti-α6-integrin–PE (1:100, BD Biosciences). DAPI was used to exclude dead cells. Cell isolations were performed on FACSAria sorters equipped with FACSDiva software (BD Biosciences). FACS analyses were performed using LSRII FACS Analyzers and then analysed with the FlowJo program.
RNA isolation and microarray analyses
RNAs from FACS-purified Tbx1-cKO mice and WT HF-SCs 2 days post depilation were provided to the Genomics Core Facility at Memorial Sloan-Kettering Cancer Center for quality control, quantification, reverse transcription, labelling, and hybridization to MOE430A 2.0 microarray chips (Affymetrix). Two entirely independent samples were used for data analyses. Arrays were scanned as per the manufacturer’s specifications for the Affymetrix MOE430v2 chip. Images were background-subtracted. Probe sets were identified as differentially expressed when the average fold change was ≥1.8 (P <0.1). Probe sets selected for visualization were log2 transformed, analysed with hierarchical clustering (Pearson correlation, average linkage) and visualized with heat maps to assist in interpretation.
Noggin intradermal injection
Recombinant mouse noggin (5 μg ml−1, R&D Systems) was injected intradermally, together with beads, into the back skin for 3 days post depilation. The skin was analysed on the third day. BrdU was injected twice in the last 24 h before harvesting the skin.
In vitro BMP4 treatment
For BMP4 treatment experiments, mouse keratinocytes grown in six-well plates and transfected with Tbx1-GFP (a Tbx1 expression vector obtained by cloning a mouse cDNA into the pCMV6-AC-GFP plasmid) were treated with 100 ng ml−1 mouse recombinant BMP4 for the indicated period. After treatment, cells were trypsinized, and GFP+ versus GFP− cells were isolated by FACS and separately processed for total RNA extraction and rtPCR analysis.
Immunoblotting
Mouse keratinocytes transduced with scramble or Smad1 shRNAs by lentiviral infection were selected in 1 μg ml−1 puromycin-containing media for 4 days. Cells were treated with or without BMP4 (100 ng ml−1) for 3 h and lysed directly in SDS sample buffer (50 mM Tris-HCl, pH 6.8, 100 mM DTT, 2% SDS, 0.1% bromophenol blue and 10% glycerol). Gel electrophoresis was performed using 4–12% NuPAGE Bis-Tris gradient gels (Invitrogen), and separated proteins were transferred to PVDF membranes (Millipore). Membranes were blocked for 1 h in Odyssey blocking buffer (LI-COR Biosciences), then incubated with primary antibodies in blocking buffer overnight at 4 °C. The primary antibodies used were as follows: mouse anti-SMAD1 (1:300, Abcam), rabbit anti-pSMAD1/5/8 (Ser463/465) (1:2,000, Millipore), mouse anti-SMAD2/3 (1:2,000, BD Biosciences), rabbit anti-pSMAD2 (Ser465/467) (1:2,000, Cell Signaling Technology), rabbit anti-SMAD5 (1:500, Abcam) and mouse anti-α-tubulin (1:2,000, Sigma). Secondary antibodies were conjugated to IRDye 680 or IRDye 800 (1:15,000, LI-COR Biosciences).
BMP-reporter experiments
Construction of a lentiviral BMP reporter (pLKO-H2B-CFP-BRE-ZsGreen) is described elsewhere30. Briefly, H2B-CFP works as a transduction marker, which is constitutively transcribed from the PGK promoter. As a reporter, ZsGreen is transcribed from a minimal CMV promoter conjugated to the BMP response element (BRE) in the presence of BMP. We transduced the BMP reporter into mouse keratinocytes, FACS-sorted the transduced cells based on H2B-CFP expression and transduced these cells with viral vectors carrying scramble or Smad1 shRNAs. After puromycin selection, we tested BMP-reporter activities in the presence or absence of BMP (100 ng ml−1) for 24 h. For visualization and quantification of direct fluorescent signals from the BMP reporter, keratinocytes were seeded onto coverslips and fixed with 4% paraformaldehyde for 10 min at room temperature.
Statistics
To determine the significance between two groups, indicated in the figures by asterisks, comparisons were made using Student’s t-test, performed by Prism5 software or Microsoft Excel. Box-and-whisker plots are used to describe the entire population without assumptions about the statistical distribution.
Supplementary Material
Acknowledgments
We are grateful to A. Baldini for the Tbx1fl/fl mice, the Comparative Biology Centre (AAALAC-accredited) for expert handling and care of the mice, The Rockefeller University FCRC for FACS sorting (supported by NYSTEM funds through NYSDOH, contract C023046), S. Dewell and The Rockefeller University Genomic Resource Centre for high-throughput sequencing, The Memorial Sloan Kettering Genomics Core Facility for RNA and microarray processing, Fuchs’ lab members Y. Hsu, B. Keyes, X. Wu, D. Devenport and L. Zhang for comments and suggestions, W. H. Lien for providing epigenetic ChIP-seq data for the Tbx1 gene in vivo and P. Janki for assisting in the production of the pooled virus for screening. This work was supported by grants from the National Institutes of Health (R01-AR050452; E.F.), the Empire State Stem Cell (NYSTEM N09G074; E.F.), a New York Stem Cell Foundation-Druckenmiller Fellowship (T.C.) and a NYSTEM Scholar Award (C026722; T.C.). E.F. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions T.C. and E.F. designed the study. T.C. carried out the experiments and analysed the data. E.H. analysed the sequencing data. S.B. contributed to optimizing and testing methods used in the screen. N.O. carried out the Smad1 shRNA-related experiments. N.S. participated in the experiments involving mouse handling. T.C. and E.F. co-wrote the paper. E.F. supervised the research.
Author Information Microarray data have been deposited in the Gene Expression Omnibus database under accession number GSE35575. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zon LI. Intrinsic and extrinsic control of haematopoietic stem-cell self-renewal. Nature. 2008;453:306–313. doi: 10.1038/nature07038. [DOI] [PubMed] [Google Scholar]
- 4.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 5.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Greco V, et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watt FM, Jensen KB. Epidermal stem cell diversity and quiescence. EMBO Mol Med. 2009;1:260–267. doi: 10.1002/emmm.200900033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- 9.Molofsky AV, et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewalin young but not old mice by reducingp16Ink4a andp19ArfExpression. Cell. 2008;135:227–239. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topley GI, Okuyama R, Gonzales JG, Conti C, Dotto GP. p21WAF1/Cip1 functions as a suppressor of malignant skin tumor formation and a determinant of keratinocyte stem-cell potential. Proc Natl Acad Sci USA. 1999;96:9089–9094. doi: 10.1073/pnas.96.16.9089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kippin TE, Martens DJ, van der Kooy D. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 2005;19:756–767. doi: 10.1101/gad.1272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito M, Kizawa K, Hamada K, Cotsarelis G. Hair follicle stem cells in the lower bulge form the secondary germ, a biochemically distinct but functionally equivalent progenitor cell population, at the termination of catagen. Differentiation. 2004;72:548–557. doi: 10.1111/j.1432-0436.2004.07209008.x. [DOI] [PubMed] [Google Scholar]
- 15.Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osorio KM, et al. Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development. 2008;135:1059–1068. doi: 10.1242/dev.012799. [DOI] [PubMed] [Google Scholar]
- 17.Aggarwal VS, et al. Mesodermal Tbx1 is required for patterning the proximal mandible in mice. Dev Biol. 2010;344:669–681. doi: 10.1016/j.ydbio.2010.05.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Fulcoli FG, Tang S, Baldini A. Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circ Res. 2009;105:842–851. doi: 10.1161/CIRCRESAHA.109.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lien WH, et al. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, et al. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131:3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- 21.Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korchynskyi O, ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J Biol Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 23.Hollnagel A, Oehlmann V, Heymer J, Ruther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274:19838–19845. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- 24.Fulcoli FG, Huynh T, Scambler PJ, Baldini A. Tbx1 regulates the BMP–Smad1 pathway in a transcription independent manner. PLoS ONE. 2009;4:e6049. doi: 10.1371/journal.pone.0006049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Blessing M, Nanney LB, King LE, Jones CM, Hogan BL. Transgenic mice as a model to study the role of TGF-β-related molecules in hair follicles. Genes Dev. 1993;7:204–215. doi: 10.1101/gad.7.2.204. [DOI] [PubMed] [Google Scholar]
- 26.Botchkarev VA, et al. Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nature Cell Biol. 1999;1:158–164. doi: 10.1038/11078. [DOI] [PubMed] [Google Scholar]
- 27.Kulessa H, Turk G, Hogan BL. Inhibition of Bmp signaling affects growth and differentiation in the anagen hair follicle. EMBO J. 2000;19:6664–6674. doi: 10.1093/emboj/19.24.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proc Natl Acad Sci USA. 2007;104:10063–10068. doi: 10.1073/pnas.0703004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andl T, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- 30.Oshimori N, Fuchs E. Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell. 2012;10:63–75. doi: 10.1016/j.stem.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Foundation for Statistical Computing. The R Project for Statistical Computing. 2011 〈 http://www.r-project.org〉.
- 32.Beronja S, Livshits G, Williams S, Fuchs E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. Nature Med. 2010;16:821–827. doi: 10.1038/nm.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.