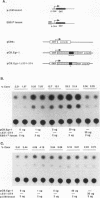
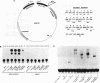
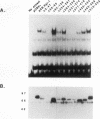
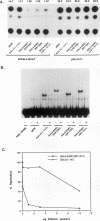
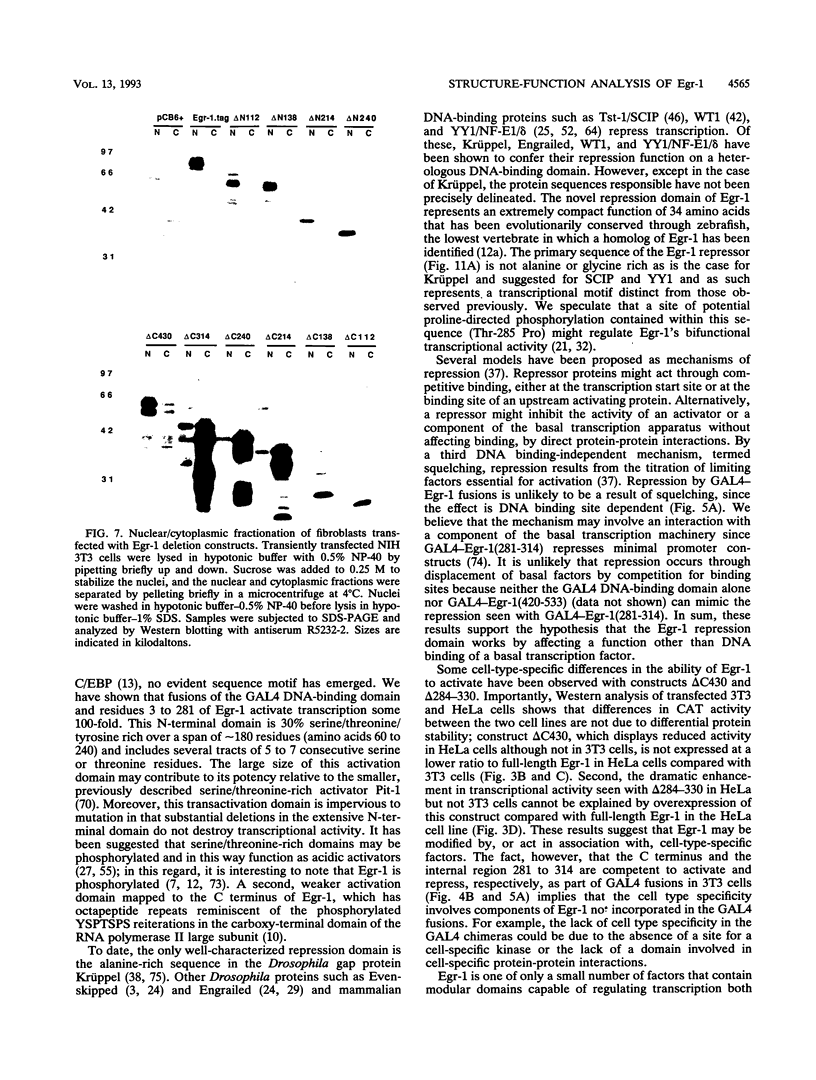
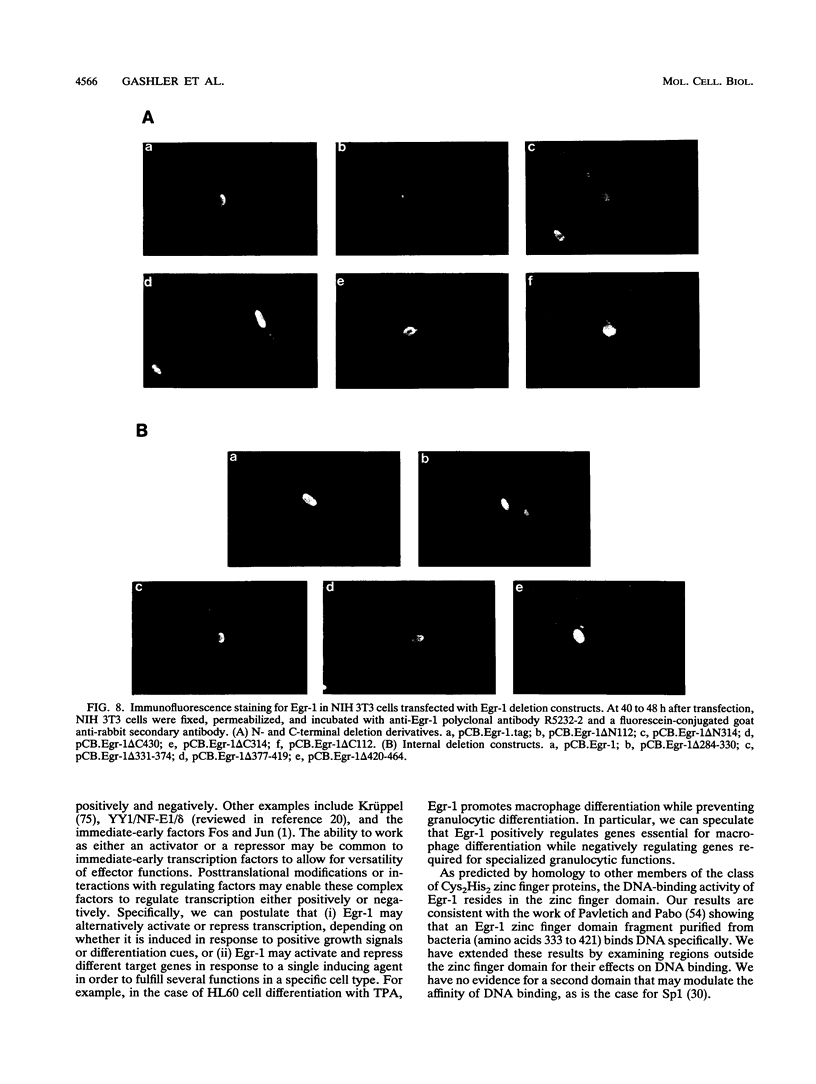
Abstract
Egr-1 is an immediate-early response gene induced transiently and ubiquitously by mitogenic stimuli and also regulated in response to signals that initiate differentiation. The Egr-1 gene product, a nuclear phosphoprotein with three zinc fingers of the Cys2His2 class, binds to the sequence CGCCCCCGC and transactivates a synthetic promoter construct 10-fold in transient-transfection assays. We have analyzed the structure and function of the Egr-1 protein in detail, delineating independent and modular activation, repression, DNA-binding, and nuclear localization activities. Deletion analysis, as well as fusions to the DNA-binding domain of GAL4, indicated that the activation potential of Egr-1 is distributed over an extensive serine/threonine-rich N-terminal domain. In addition, a novel negative regulatory function has been precisely mapped 5' of the zinc fingers: amino acids 281 to 314 are sufficient to confer the ability to repress transcription on a heterologous DNA-binding domain. Specific DNA-binding activity was shown to reside in the three zinc fingers of Egr-1, as predicted by homology to other known DNA-binding proteins. Finally, nuclear localization of Egr-1 is specified by signals in the DNA-binding domain and basic flanking sequences, as determined by subcellular fractionation and indirect immunofluorescence. Basic residues 315 to 330 confer partial nuclear localization on the bacterial protein beta-galactosidase. A bipartite signal consisting of this basic region in conjunction with either the second or third zinc finger, but not the first, suffices to target beta-galactosidase exclusively to the nucleus. Our work shows that Egr-1 is a functionally complex protein and suggests that it may play different roles in the diverse settings in which it is induced.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abate C., Luk D., Curran T. Transcriptional regulation by Fos and Jun in vitro: interaction among multiple activator and regulatory domains. Mol Cell Biol. 1991 Jul;11(7):3624–3632. doi: 10.1128/mcb.11.7.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A. E., Pringle J. R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984 Mar;98(3):934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989 Aug 11;58(3):433–440. doi: 10.1016/0092-8674(89)90424-8. [DOI] [PubMed] [Google Scholar]
- Bonventre J. V., Sukhatme V. P., Bamberger M., Ouellette A. J., Brown D. Localization of the protein product of the immediate early growth response gene, Egr-1, in the kidney after ischemia and reperfusion. Cell Regul. 1991 Mar;2(3):251–260. doi: 10.1091/mbc.2.3.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. M., Koski R. A., Gashler A., McKiernan M., Morris C. F., Gaffney R., Hay R. V., Sukhatme V. P. Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Mol Cell Biol. 1990 May;10(5):1931–1939. doi: 10.1128/mcb.10.5.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy B. A., Lau L. F., Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with "zinc finger" sequences. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy B., Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8737–8741. doi: 10.1073/pnas.86.22.8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole A. J., Saffen D. W., Baraban J. M., Worley P. F. Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature. 1989 Aug 10;340(6233):474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- Corden J. L. Tails of RNA polymerase II. Trends Biochem Sci. 1990 Oct;15(10):383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Day M. L., Fahrner T. J., Aykent S., Milbrandt J. The zinc finger protein NGFI-A exists in both nuclear and cytoplasmic forms in nerve growth factor-stimulated PC12 cells. J Biol Chem. 1990 Sep 5;265(25):15253–15260. [PubMed] [Google Scholar]
- Garcia-Bustos J., Heitman J., Hall M. N. Nuclear protein localization. Biochim Biophys Acta. 1991 Mar 7;1071(1):83–101. doi: 10.1016/0304-4157(91)90013-m. [DOI] [PubMed] [Google Scholar]
- Gerster T., Balmaceda C. G., Roeder R. G. The cell type-specific octamer transcription factor OTF-2 has two domains required for the activation of transcription. EMBO J. 1990 May;9(5):1635–1643. doi: 10.1002/j.1460-2075.1990.tb08283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman M. Z., Wilson R. N., Weinberg R. A. Multiple protein-binding sites in the 5'-flanking region regulate c-fos expression. Mol Cell Biol. 1986 Dec;6(12):4305–4316. doi: 10.1128/mcb.6.12.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. E., Ziff E. B. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984 Oct 4;311(5985):433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Guiochon-Mantel A., Lescop P., Christin-Maitre S., Loosfelt H., Perrot-Applanat M., Milgrom E. Nucleocytoplasmic shuttling of the progesterone receptor. EMBO J. 1991 Dec;10(12):3851–3859. doi: 10.1002/j.1460-2075.1991.tb04954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S. The Yin and the Yang of mammalian transcription. Curr Biol. 1992 Mar;2(3):152–154. doi: 10.1016/0960-9822(92)90268-f. [DOI] [PubMed] [Google Scholar]
- Hall F. L., Vulliet P. R. Proline-directed protein phosphorylation and cell cycle regulation. Curr Opin Cell Biol. 1991 Apr;3(2):176–184. doi: 10.1016/0955-0674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Craik C., Hiraoka Y. Homeodomain of yeast repressor alpha 2 contains a nuclear localization signal. Proc Natl Acad Sci U S A. 1990 Sep;87(18):6954–6958. doi: 10.1073/pnas.87.18.6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. N., Hereford L., Herskowitz I. Targeting of E. coli beta-galactosidase to the nucleus in yeast. Cell. 1984 Apr;36(4):1057–1065. doi: 10.1016/0092-8674(84)90055-2. [DOI] [PubMed] [Google Scholar]
- Han K., Levine M. S., Manley J. L. Synergistic activation and repression of transcription by Drosophila homeobox proteins. Cell. 1989 Feb 24;56(4):573–583. doi: 10.1016/0092-8674(89)90580-1. [DOI] [PubMed] [Google Scholar]
- Hariharan N., Kelley D. E., Perry R. P. Delta, a transcription factor that binds to downstream elements in several polymerase II promoters, is a functionally versatile zinc finger protein. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9799–9803. doi: 10.1073/pnas.88.21.9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hunter T., Karin M. The regulation of transcription by phosphorylation. Cell. 1992 Aug 7;70(3):375–387. doi: 10.1016/0092-8674(92)90162-6. [DOI] [PubMed] [Google Scholar]
- Inostroza J. A., Mermelstein F. H., Ha I., Lane W. S., Reinberg D. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell. 1992 Aug 7;70(3):477–489. doi: 10.1016/0092-8674(92)90172-9. [DOI] [PubMed] [Google Scholar]
- Jaynes J. B., O'Farrell P. H. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991 Jun;10(6):1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Courey A. J., Ladika J., Tjian R. Distinct regions of Sp1 modulate DNA binding and transcriptional activation. Science. 1988 Dec 16;242(4885):1566–1570. doi: 10.1126/science.3059495. [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. A short amino acid sequence able to specify nuclear location. Cell. 1984 Dec;39(3 Pt 2):499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kemp B. E., Pearson R. B. Protein kinase recognition sequence motifs. Trends Biochem Sci. 1990 Sep;15(9):342–346. doi: 10.1016/0968-0004(90)90073-k. [DOI] [PubMed] [Google Scholar]
- Kleinschmidt J. A., Seiter A. Identification of domains involved in nuclear uptake and histone binding of protein N1 of Xenopus laevis. EMBO J. 1988 Jun;7(6):1605–1614. doi: 10.1002/j.1460-2075.1988.tb02986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W., Cooper J. A., Hunter T., Verma I. M. Platelet-derived growth factor induces rapid but transient expression of the c-fos gene and protein. Nature. 1984 Dec 20;312(5996):711–716. doi: 10.1038/312711a0. [DOI] [PubMed] [Google Scholar]
- Lemaire P., Revelant O., Bravo R., Charnay P. Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4691–4695. doi: 10.1073/pnas.85.13.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire P., Vesque C., Schmitt J., Stunnenberg H., Frank R., Charnay P. The serum-inducible mouse gene Krox-24 encodes a sequence-specific transcriptional activator. Mol Cell Biol. 1990 Jul;10(7):3456–3467. doi: 10.1128/mcb.10.7.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Manley J. L. Transcriptional repression of eukaryotic promoters. Cell. 1989 Nov 3;59(3):405–408. doi: 10.1016/0092-8674(89)90024-x. [DOI] [PubMed] [Google Scholar]
- Licht J. D., Grossel M. J., Figge J., Hansen U. M. Drosophila Krüppel protein is a transcriptional repressor. Nature. 1990 Jul 5;346(6279):76–79. doi: 10.1038/346076a0. [DOI] [PubMed] [Google Scholar]
- Lillie J. W., Green M. R. Transcription activation by the adenovirus E1a protein. Nature. 1989 Mar 2;338(6210):39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- Lim R. W., Varnum B. C., Herschman H. R. Cloning of tetradecanoyl phorbol ester-induced 'primary response' sequences and their expression in density-arrested Swiss 3T3 cells and a TPA non-proliferative variant. Oncogene. 1987;1(3):263–270. [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Madden S. L., Cook D. M., Morris J. F., Gashler A., Sukhatme V. P., Rauscher F. J., 3rd Transcriptional repression mediated by the WT1 Wilms tumor gene product. Science. 1991 Sep 27;253(5027):1550–1553. doi: 10.1126/science.1654597. [DOI] [PubMed] [Google Scholar]
- Martin D. I., Orkin S. H. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev. 1990 Nov;4(11):1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987 Nov 6;238(4828):797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- Monuki E. S., Kuhn R., Weinmaster G., Trapp B. D., Lemke G. Expression and activity of the POU transcription factor SCIP. Science. 1990 Sep 14;249(4974):1300–1303. doi: 10.1126/science.1975954. [DOI] [PubMed] [Google Scholar]
- Morin N., Delsert C., Klessig D. F. Nuclear localization of the adenovirus DNA-binding protein: requirement for two signals and complementation during viral infection. Mol Cell Biol. 1989 Oct;9(10):4372–4380. doi: 10.1128/mcb.9.10.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath S. T., Nayak D. P. Function of two discrete regions is required for nuclear localization of polymerase basic protein 1 of A/WSN/33 influenza virus (H1 N1). Mol Cell Biol. 1990 Aug;10(8):4139–4145. doi: 10.1128/mcb.10.8.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette A. J., Malt R. A., Sukhatme V. P., Bonventre J. V. Expression of two "immediate early" genes, Egr-1 and c-fos, in response to renal ischemia and during compensatory renal hypertrophy in mice. J Clin Invest. 1990 Mar;85(3):766–771. doi: 10.1172/JCI114502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K., Atchison M. L. Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3' enhancer and the immunoglobulin heavy-chain mu E1 site. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan S., Gashler A., Siegel M. G., Chang L. C., Joseph L. J., Shows T. B., Le Beau M. M., Sukhatme V. P. EGR3, a novel member of the Egr family of genes encoding immediate-early transcription factors. Oncogene. 1991 Jun;6(6):917–928. [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Rihs H. P., Peters R. Nuclear transport kinetics depend on phosphorylation-site-containing sequences flanking the karyophilic signal of the Simian virus 40 T-antigen. EMBO J. 1989 May;8(5):1479–1484. doi: 10.1002/j.1460-2075.1989.tb03531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal N. Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol. 1987;152:704–720. doi: 10.1016/0076-6879(87)52075-4. [DOI] [PubMed] [Google Scholar]
- Roux P., Blanchard J. M., Fernandez A., Lamb N., Jeanteur P., Piechaczyk M. Nuclear localization of c-Fos, but not v-Fos proteins, is controlled by extracellular signals. Cell. 1990 Oct 19;63(2):341–351. doi: 10.1016/0092-8674(90)90167-d. [DOI] [PubMed] [Google Scholar]
- Ryseck R. P., Hirai S. I., Yaniv M., Bravo R. Transcriptional activation of c-jun during the G0/G1 transition in mouse fibroblasts. Nature. 1988 Aug 11;334(6182):535–537. doi: 10.1038/334535a0. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988 Oct 6;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Sanes J. R., Rubenstein J. L., Nicolas J. F. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986 Dec 1;5(12):3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastry B. S. Xenopus transcription factor IIIA (XTFIIIA): after a decade of research. Prog Biophys Mol Biol. 1991;56(2):135–144. doi: 10.1016/0079-6107(91)90017-m. [DOI] [PubMed] [Google Scholar]
- Shi Y., Seto E., Chang L. S., Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991 Oct 18;67(2):377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- Silver P. A. How proteins enter the nucleus. Cell. 1991 Feb 8;64(3):489–497. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- Silver P. A., Keegan L. P., Ptashne M. Amino terminus of the yeast GAL4 gene product is sufficient for nuclear localization. Proc Natl Acad Sci U S A. 1984 Oct;81(19):5951–5955. doi: 10.1073/pnas.81.19.5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaete R. R., Mocarski E. S. Regulation of cytomegalovirus gene expression: alpha and beta promoters are trans activated by viral functions in permissive human fibroblasts. J Virol. 1985 Oct;56(1):135–143. doi: 10.1128/jvi.56.1.135-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhatme V. P., Cao X. M., Chang L. C., Tsai-Morris C. H., Stamenkovich D., Ferreira P. C., Cohen D. R., Edwards S. A., Shows T. B., Curran T. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988 Apr 8;53(1):37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- Sukhatme V. P. Early transcriptional events in cell growth: the Egr family. J Am Soc Nephrol. 1990 Dec;1(6):859–866. doi: 10.1681/ASN.V16859. [DOI] [PubMed] [Google Scholar]
- Theill L. E., Castrillo J. L., Wu D., Karin M. Dissection of functional domains of the pituitary-specific transcription factor GHF-1. Nature. 1989 Dec 21;342(6252):945–948. doi: 10.1038/342945a0. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tratner I., Verma I. M. Identification of a nuclear targeting sequence in the Fos protein. Oncogene. 1991 Nov;6(11):2049–2053. [PubMed] [Google Scholar]
- Waters C. M., Hancock D. C., Evan G. I. Identification and characterisation of the egr-1 gene product as an inducible, short-lived, nuclear phosphoprotein. Oncogene. 1990 May;5(5):669–674. [PubMed] [Google Scholar]
- Zuo P., Stanojević D., Colgan J., Han K., Levine M., Manley J. L. Activation and repression of transcription by the gap proteins hunchback and Krüppel in cultured Drosophila cells. Genes Dev. 1991 Feb;5(2):254–264. doi: 10.1101/gad.5.2.254. [DOI] [PubMed] [Google Scholar]