Abstract
S. aureus is a pathogenic bacterium that requires iron to carry out vital metabolic functions and cause disease. The most abundant reservoir of iron inside the human host is heme, which is the cofactor of hemoglobin. To acquire iron from hemoglobin, S. aureus utilizes an elaborate system known as the iron-regulated surface determinant (Isd) system1. Components of the Isd system first bind host hemoglobin, then extract and import heme, and finally liberate iron from heme in the bacterial cytoplasm2,3. This pathway has been dissected through numerous in vitro studies4-9. Further, the contribution of the Isd system to infection has been repeatedly demonstrated in mouse models8,10-14. Establishing the contribution of the Isd system to hemoglobin-derived iron acquisition and growth has proven to be more challenging. Growth assays using hemoglobin as a sole iron source are complicated by the instability of commercially available hemoglobin, contaminating free iron in the growth medium, and toxicity associated with iron chelators. Here we present a method that overcomes these limitations. High quality hemoglobin is prepared from fresh blood and is stored in liquid nitrogen. Purified hemoglobin is supplemented into iron-deplete medium mimicking the iron-poor environment encountered by pathogens inside the vertebrate host. By starving S. aureus of free iron and supplementing with a minimally manipulated form of hemoglobin we induce growth in a manner that is entirely dependent on the ability to bind hemoglobin, extract heme, pass heme through the bacterial cell envelope and degrade heme in the cytoplasm. This assay will be useful for researchers seeking to elucidate the mechanisms of hemoglobin-/heme-derived iron acquisition in S. aureus and possibly other bacterial pathogens.
Keywords: Infection, Issue 72, Immunology, Microbiology, Infectious Diseases, Cellular Biology, Pathology, Micronutrients, Bacterial Infections, Gram-Positive Bacterial Infections, Bacteriology, Staphylococcus aureus, iron acquisition, hemoglobin, bacterial growth, bacteria
Protocol
1. Purification of Hemoglobin from Fresh Blood
Acquire fresh human blood supplemented with an anticoagulant. Keep blood on ice or at 4 °C throughout the purification.
Centrifuge blood for 20 min at 1,500 x g. The red blood cells (RBCs) will be at the bottom of the tube. Carefully aspirate the supernatant and gently resuspend the pellet in ice-cold 0.9% (w/v) NaCl solution. Repeat the centrifugation and wash 3 times.
Resuspend the pellet in 1 volume of ice-cold 10 mM Tris-HCl (pH 8.0). This will induce lysis of the RBCs due to osmotic pressure. Add toluene to ~ 20% final volume.
Incubate at 4 °C on a rotisserie overnight.
Centrifuge the lysate at 20,000 x g for 1 hr. Collect the middle hemolysate fraction leaving the toluene (floating on top) and pellet untouched. Use a long-neck pipette to collect the middle fraction.
Pass through 0.44 μm syringe filter. If the solution contains particulate matter and cannot be passed through the filter, repeat step 1.5.
Purify hemoglobin (Hb) with a high-performance liquid chromatography (HPLC) anion exchange column (Varian, PL-SAX 1,000 Å 8 μm, 150 mm × 4.6 mm). The mobile phase A is 10 mM Tris-HCl (pH 8.0) and mobile phase B is 10 mM Tris-HCl (pH 8.0) + 0.5 M NaCl. A 0%-100% gradient of solvent B is run over 2 min at 2.0 ml/min flow rate. The elution is monitored based on absorption (λ: 410 nm and 280 nm). Collect only the fraction characterized by a bright red color and a prominent absorption peak (Figure 1).
Dialyze the elution against phosphate-buffered saline (PBS) overnight and then for a few hours again. Sterilize by passing through a 0.22 μm syringe filter.
To measure Hb concentration, prepare standard Hb solutions of known concentrations in PBS. Determine the concentration of Hb in the sample by mixing the standard solutions (see the table with reagents used) or the sample solution with 2x Drabkin's reagent (prepared from powder) at a 1:1 ratio. For example, mix 100 μl Hb solution with 100 μl 2x Drabkin's reagent in 96-well plates. Incubate for 15 min and measure absorbance at 540 nm. Plot a standard curve and determine the Hb concentration in the sample. Five to fifteen mg/ml yields are typical.
Run 15-20 μg purified hemoglobin on a 15% SDS-PAGE in duplicate. Stain one of the gels; transfer the proteins from another gel onto a nitrocellulose membrane and immunoblot for hemoglobin (Figure 2).
Freeze and store 1 ml aliquots of hemoglobin in liquid nitrogen.
2. Preparation of Iron-deplete Growth Media
Prepare Roswell Park Memorial Institute (RPMI) broth by dissolving the RPMI powder in water, add sodium bicarbonate as recommended by the manufacturer and 1% cassamino acids (CA) (w/v). Sterilize by passing through a 0.2 μm filter and store refrigerated.
Prepare metal-depleted RPMI (NRPMI) by adding 7% (w/v) Chelex 100 and mixing overnight on a stir plate. Remove Chelex 100 by passing through a 0.2 μm filter and store refrigerated. Supplement media with essential non-iron metals: 25 μM ZnCl2, 25 μM MnCl2, 100 mM CaCl2 and 1 mM MgCl2 prepared in advance as a sterile 1,000x solution. Use disposable plastic containers for this step to avoid iron contamination from re-usable supplies.
3. Staphylococcus aureus Growth using Hemoglobin as a Sole Iron Source
Streak S. aureus for isolation on tryptic soy agar (TSA) from a frozen stock. Incubate at 37 °C for 20-24 hr.
Resuspend ethylenediamine-N,N'-bis(2-hydroxyphenylacetic acid) (EDDHA) in anhydrous ethanol to 100 mM. EDDHA does not go into solution but is sterilized by ethanol.
Add EDDHA to RPMI to a final concentration of 0.5 mM. Allow EDDHA to dissolve for at least 30 min before proceeding to the next step. Due to batch-to-batch variation, final EDDHA concentration may need to be lowered to 0.25 mM to allow bacterial growth.
Inoculate single colonies of S. aureus into 5 ml of RPMI containing EDDHA in 15 ml screw-cap conical tubes. Incubate at 37 °C with shaking at 180 rotations per minute (rpm) for 16-20 hr.
Centrifuge the overnight cultures for 5 min at 7,500 x g and resuspend the pellet in NRPMI containing 0.5 mM EDDHA. Normalize OD600 to ~3.
Prepare NRPMI containing 2.5 μg per ml Hb and 0.1-1.0 mM EDDHA. Due to variation between the batches, the EDDHA concentration required to chelate free iron in NRPMI may vary.
Subculture 10 μl of bacterial suspension from step 3.5 into 1 ml NRPMI + EDDHA + Hb in a 15 ml screw-cap conical tube.
Incubate the cultures at 37 °C for up to 48 hr with shaking at 180 rpm or on a rolling drum.
Every 6-12 hr take OD600 readings by removing 50 μl of the culture and mixing it with 150 μl of PBS in 96-well plates.
Representative Results
We have purified human hemoglobin from hemolysate with HPLC (Protocol step 1.7). Figure 1 shows recorded absorbance of eluate at 280 and 410 nm wavelengths. Fraction 5 was collected and other fractions were discarded. Yields of five to fifteen milligrams of hemoglobin per milliliter of eluate are typically acquired. Purified hemoglobin was analyzed by SDS-PAGE in duplicate and the gels were either stained for proteins or transferred onto nitrocellulose and immunoblotted (Protocol step 1.10, Figure 2).
We have assessed the ability of purified human hemoglobin to support the growth of wild type S. aureus and S. aureus that lacks the IsdB component of the Isd system (ΔisdB). IsdB is a hemoglobin receptor that is required for hemoglobin-derived iron acquisition12. Wild type and ΔisdB were grown in iron-depleted medium supplemented with human hemoglobin (NRPMI + EDDHA + Hb) as described in section 3 of the protocol. When grown in NRPMI + EDDHA + Hb, wild type but not ΔisdB is able to proliferate as indicated by an increase in optical density of the cultures over time (Figure 3A). In contrast, the ability to utilize supplemented free iron (+FeCl3) is identical in wild type and ΔisdB (Figure 3B). Neither wild type nor ΔisdB proliferate in the absence of an iron source (Figure 3B).
We have compared the growth of S. aureus in NRPMI + EDDHA supplemented with hemoglobin purified from the fresh blood or lyophilized hemoglobin. Figure 4 illustrates that while hemoglobin purified from blood requires IsdB for growth, lyophilized hemoglobin enables proliferation of ΔisdB.
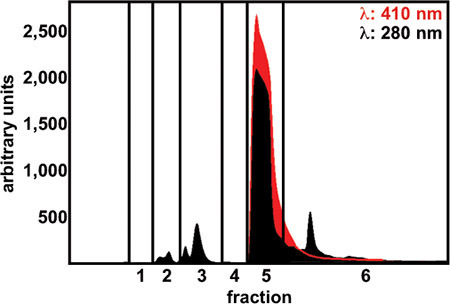 Figure 1. Absorption of the elution fractions during hemoglobin purification. Absorption at 410 nm (specific to heme-binding proteins) and 280 nm (characteristic for all proteins) was monitored throughout the elution. Fraction number five contained hemoglobin and was collected.
Figure 1. Absorption of the elution fractions during hemoglobin purification. Absorption at 410 nm (specific to heme-binding proteins) and 280 nm (characteristic for all proteins) was monitored throughout the elution. Fraction number five contained hemoglobin and was collected.
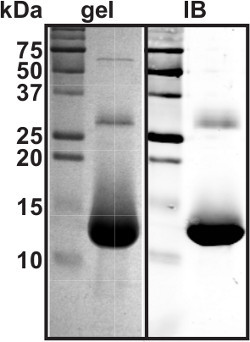 Figure 2. Purified human hemoglobin Twenty micrograms of purified hemoglobin was separated using denaturing 15% SDS-PAGE. A. Gel stained for proteins with Bio-Rad protein assay dye reagent. B. Nitrocellulose membrane immunoblotted for hemoglobin. The intense lower band is a hemoglobin monomer, while the more faint upper bands are hemoglobin dimers and tertramers, which did not get completely denatured.
Figure 2. Purified human hemoglobin Twenty micrograms of purified hemoglobin was separated using denaturing 15% SDS-PAGE. A. Gel stained for proteins with Bio-Rad protein assay dye reagent. B. Nitrocellulose membrane immunoblotted for hemoglobin. The intense lower band is a hemoglobin monomer, while the more faint upper bands are hemoglobin dimers and tertramers, which did not get completely denatured.
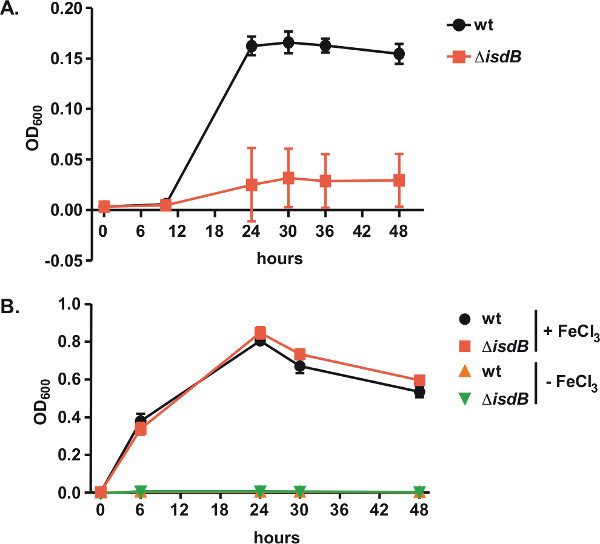 Figure 3. Use of human hemoglobin as an iron source by S. aureusA. Growth of wild type (wt) or isogenic hemoglobin receptor isdB mutant (ΔisdB) in NRPMI supplemented with iron chelator EDDHA and human hemoglobin is statistically different as determined by Student's two-tailed t test, p < 0.05. B. Growth in NRPMI supplemented with 10 μM iron chloride (+FeCl3) or EDDHA (-FeCl3) is not different between wt and ΔisdB. Graphs depict data from a representative experiment, which included three biological replicates. Error bars denote standard deviation of optical density readings at indicated time points between the replicates.
Figure 3. Use of human hemoglobin as an iron source by S. aureusA. Growth of wild type (wt) or isogenic hemoglobin receptor isdB mutant (ΔisdB) in NRPMI supplemented with iron chelator EDDHA and human hemoglobin is statistically different as determined by Student's two-tailed t test, p < 0.05. B. Growth in NRPMI supplemented with 10 μM iron chloride (+FeCl3) or EDDHA (-FeCl3) is not different between wt and ΔisdB. Graphs depict data from a representative experiment, which included three biological replicates. Error bars denote standard deviation of optical density readings at indicated time points between the replicates.
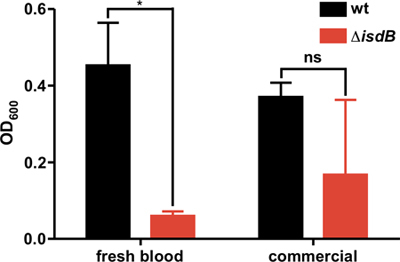 Figure 4. Growth of wild type (wt) or isogenic hemoglobin receptor mutant (ΔisdB) in media supplemented with hemoglobin purified from fresh blood or lyophilized hemoglobin. The graph depicts data from a representative experiment, which included three biological replicates. The error bars represent standard deviation of optical density readings at indicated time points between the replicates. Asterisks denote values that are statistically different as determined by Student's two-tailed t test, p < 0.05.
Figure 4. Growth of wild type (wt) or isogenic hemoglobin receptor mutant (ΔisdB) in media supplemented with hemoglobin purified from fresh blood or lyophilized hemoglobin. The graph depicts data from a representative experiment, which included three biological replicates. The error bars represent standard deviation of optical density readings at indicated time points between the replicates. Asterisks denote values that are statistically different as determined by Student's two-tailed t test, p < 0.05.
Discussion
Iron is an essential nutrient required by organisms from all kingdoms of life15. In vertebrates, iron is sequestered to avoid toxicity caused by this element. This sequestration also conceals iron from invading microbes in a process known as nutritional immunity16. In response, pathogens have evolved strategies that circumvent nutritional immunity. One such mechanism relies on hemoglobin, which is the most abundant source of iron within the host17. Hemoglobin is contained within red blood cells. Hemoglobin released from damaged red blood cells is bound by host haptoglobin, which signals rapid removal of haptoglobin-hemoglobin complexes by macrophages18. In order to gain access to hemoglobin, S. aureus expresses hemolysins that lyse red blood cells19. Haptoglobin-induced hemoglobin removal is countered by high affinity binding of hemoglobin by the cell surface receptors expressed by S. aureus. S. aureus extracts heme from the peptide component of hemoglobin and imports heme into the cytoplasm by passing it from cell wall proteins to membrane proteins. In the cytoplasm, heme is degraded by oxygenases to release free iron or is utilized intact in the electron transport chain20.
Numerous studies have demonstrated the contribution of the Isd system to infection8,10-14. Further, biochemical studies have assigned the functions of its individual components in hemoglobin-derived iron acquisition4-9. Here we describe an assay that monitors the growth of S. aureus with hemoglobin as the only source of available iron. In this regard, we need to point out certain conditions that are pivotal for the success of the experiment.
The quality and type of reagents utilized in hemoglobin-derived iron acquisition growth assays can dramatically influence the results obtained in these assays. In contrast to hemoglobin obtained from fresh blood, hemoglobin that is stored in lyophilized form allows bacterial growth independent of the components of the Isd system (Figure 4)21. This is likely due to the changes in the structure of hemoglobin that are induced by lyophylization. Specifically, lyophilized hemoglobin contains tetramers, dimers, and monomers of hemoglobin, as well as heme in its free form22. Purification of hemoglobin from fresh blood and its preservation in liquid nitrogen ensure integrity of the protein22. Extensive research on hemoglobin has facilitated the development of protocols for the purification of high-quality hemoglobin from fresh blood or in recombinant form, which can be used in our assay23-25.
The choice of iron chelator may affect the growth assay. For example, 2,2-dipyridyl, which is frequently used as an iron chelator, is toxic to bacterial cells26. These two effects make it hard to discern if the inhibition of bacterial growth by 2,2-dipyridyl is due to iron chelation or toxicity. Additionally, 2,2-dipyridyl is likely to be membrane permeable, and thus penetrates the bacterial cell and chelates iron intracellularly27. We have found that growth inhibition by EDDHA is reversed by supplementation of an iron source, making EDDHA a more suitable iron chelator for bacterial growth assays. However, the concentration of EDDHA that needs to be added to the growth medium may vary. This is due to variation of both EDDHA potency and growth medium iron content from batch to batch. The iron chelating activity can be normalized between batches of EDDHA by chemically removing residual iron28. However, due to variation between batches of RPMI, we found that the final EDDHA concentration may still need to be adjusted to allow growth in the presence of an iron source. We found that the highest concentration of EDDHA that is permissive for growth of wild type S. aureus in the presence of hemoglobin is optimal for this experiment.
Modifications can be made to the current protocol to more closely resemble the environment encountered by bacteria during infection. For example, within the host, heme that is released from hemoglobin is quickly bound by hemopexin. Hemopexin cannot be used as an iron source by S. aureus, therefore its addition would prevent acquisition of heme released from hemoglobin during incubation with S. aureus12.
We feel that this assay may be used for research into metal acquisition by a variety of bacterial pathogens. Furthermore, this method may be used to measure iron acquisition from non-hemoglobin sources of iron that are present within the host.
Disclosures
We have nothing to disclose.
Acknowledgments
This research was supported by U.S. Public Health Service grants AI69233 and AI073843 from the National Institute of Allergy and Infectious Diseases. E.P.S. is a Burroughs Wellcome Fellow in the Pathogenesis of Infectious Diseases. K.P.H. was funded by the Cellular and Molecular Microbiology Training grant Program 5 T32 A107611-10.
References
- Mazmanian S, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- Pishchany G, Skaar EP. Taste for blood: hemoglobin as a nutrient source for pathogens. PLOS Pathogens. 2012;8:e1002535. doi: 10.1371/journal.ppat.1002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley KP, Skaar EP. A battle for iron: host sequestration and Staphylococcus aureus acquisition. Microbes and infection. Institut Pasteur. 2012;14:217–227. doi: 10.1016/j.micinf.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna Kumar K, et al. Structural basis for hemoglobin capture by Staphylococcus aureus cell-surface protein. IsdH. The Journal of biological chemistry. 2011;286:38439–38447. doi: 10.1074/jbc.M111.287300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg JC, Mao CX, Murphy ME. Iron-coordinating tyrosine is a key determinant of NEAT domain heme transfer. Journal of Molecular Biology. 2011;413:684–698. doi: 10.1016/j.jmb.2011.08.047. [DOI] [PubMed] [Google Scholar]
- Villareal VA, et al. Transient weak protein-protein complexes transfer heme across the cell wall of Staphylococcus aureus. Journal of the American Chemical Society. 2011;133:14176–14179. doi: 10.1021/ja203805b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muryoi N, et al. Demonstration of the iron-regulated surface determinant (Isd) heme transfer pathway in Staphylococcus aureus. J. Biol. Chem. 2008;283:28125–28136. doi: 10.1074/jbc.M802171200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reniere ML, Skaar EP. Staphylococcus aureus haem oxygenases are differentially regulated by iron and haem. Mol. Microbiol. 2008;69:1304–1315. doi: 10.1111/j.1365-2958.2008.06363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, et al. Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J. Biol. Chem. 2008;283:6668–6676. doi: 10.1074/jbc.M708372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishchany G, et al. Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host Microbe. 2010;8:544–550. doi: 10.1016/j.chom.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishchany G, Dickey SE, Skaar EP. Subcellular localization of the Staphylococcus aureus heme iron transport components IsdA and IsdB. Infect. Immun. 2009;77:2624–2634. doi: 10.1128/IAI.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J. Bacteriol. 2006;188:8421–8429. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, et al. IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine. 2010;28:6382–6392. doi: 10.1016/j.vaccine.2010.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AG, et al. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. Faseb J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorg. Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- Weinberg ED. Iron availability and infection. Biochimica et Biophysica Acta (BBA) - General Subjects. 2009;1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Drabkin D. Metabolism of the Hemin Chromoproteins. Physiological Reviews. 1951;31:345–431. doi: 10.1152/physrev.1951.31.4.345. [DOI] [PubMed] [Google Scholar]
- Graversen JH, Madsen M, Moestrup SK. CD163: a signal receptor scavenging haptoglobin-hemoglobin complexes from plasma. The international journal of biochemistry & cell biology. 2002;34:309–314. doi: 10.1016/s1357-2725(01)00144-3. [DOI] [PubMed] [Google Scholar]
- Torres VJ, et al. Staphylococcus aureus Fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect. Immun. 2010;78:1618–1628. doi: 10.1128/IAI.01423-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer ND, Skaar EP. Molecular Mechanisms of Staphylococcus aureus Iron Acquisition. Annu. Rev. Microbiol. 2011. [DOI] [PMC free article] [PubMed]
- Hurd AF, et al. The iron-regulated surface proteins IsdA, IsdB, and IsdH are not required for heme iron utilization in Staphylococcus aureus. Fems. Microbiology Letters. 2012;329:93–100. doi: 10.1111/j.1574-6968.2012.02502.x. [DOI] [PubMed] [Google Scholar]
- Boys BL, Kuprowski MC, Konermann L. Symmetric behavior of hemoglobin alpha- and beta- subunits during acid-induced denaturation observed by electrospray mass spectrometry. Biochemistry. 2007;46:10675–10684. doi: 10.1021/bi701076q. [DOI] [PubMed] [Google Scholar]
- Williams RC, Jr, Tsay KY. A convenient chromatographic method for the preparation of human hemoglobin. Analytical Biochemistry. 1973;54:137–145. doi: 10.1016/0003-2697(73)90256-x. [DOI] [PubMed] [Google Scholar]
- Shen TJ, et al. Production of unmodified human adult hemoglobin in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1993;90:8108–8112. doi: 10.1073/pnas.90.17.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjula BN, Acharya SA. Purification and molecular analysis of hemoglobin by high-performance liquid chromatography. Methods Mol. Med. 2003;82:31–47. doi: 10.1385/1-59259-373-9:031. [DOI] [PubMed] [Google Scholar]
- Neilands JB. Microbial envelope proteins related to iron. Annual review of microbiology. 1982;36:285–309. doi: 10.1146/annurev.mi.36.100182.001441. [DOI] [PubMed] [Google Scholar]
- Chart H, Buck M, Stevenson P, Griffiths E. Iron regulated outer membrane proteins of Escherichia coli: variations in expression due to the chelator used to restrict the availability of iron. Journal of General Microbiology. 1986;132:1373–1378. doi: 10.1099/00221287-132-5-1373. [DOI] [PubMed] [Google Scholar]
- Rogers HJ. Iron-Binding Catechols and Virulence in Escherichia coli. Infection and Immunity. 1973;7:445–456. doi: 10.1128/iai.7.3.445-456.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]


