Abstract
Introduction
Several candidate loci have been suggested as influencing mandibular prognathism (1p22.1, 1p22.2, 1p36, 3q26.2, 5p13-p12, 6q25, 11q22.2-q22.3, 12q23, 12q13.13, and 19p13.2). The goal of this study was to replicate these results in a well-characterized homogeneous sample set.
Methods
Thirty-three single nucleotide polymorphisms spanning all candidate regions were studied in 44 prognathic and 35 Class I subjects from the University of Pittsburgh School of Dental Medicine Dental Registry and DNA Repository. The 44 mandibular prognathism subjects had an average age of 18.4 years, 31 were females and 13 males, and 24 were White, 15 African American, two Hispanic, and three Asian. The 35 Class I subjects had an average age of 17.6 years, 27 were females and 9 males, and 27 were White, six African Americans, one Hispanic, and two Asian. Skeletal mandibular prognathism diagnosis included cephalometric values indicative of Class III such as ANB smaller than two degrees, negative Witts appraisal, and positive A–B plane. Additional mandibular prognathism criteria included negative OJ and visually prognathic (concave) profile as determined by the subject's clinical evaluation. Orthognathic subjects without jaw deformations were used as a comparison group. Mandibular prognathism and orthognathic subjects were matched based on race, sex and age. Genetic markers were tested by polymerase chain reaction using TaqMan chemistry. Chi-square and Fisher exact tests were used to determine overrepresentation of marker allele with alpha of 0.05.
Results
An association was unveiled between a marker in MYO1H (rs10850110) and the mandibular prognathism phenotype (p=0.03). MYO1H is a Class-I myosin that is in a different protein group than the myosin isoforms of muscle sarcomeres, which are the basis of skeletal muscle fiber typing. Class I myosins are necessary for cell motility, phagocytosis and vesicle transport.
Conclusions
More strict clinical definitions may increase homogeneity and aid the studies of genetic susceptibility to malocclusions. We provide evidence that MYO1H may contribute to mandibular prognathism.
Introduction
In orthodontics, one of the most challenging aspects in treating patients is predicting mandibular growth, especially on patients that show more pronounced characteristics of mandibular development. Through studies predominately conducted within family members and twin siblings, it is well documented that there is a strong link of mandibular prognathism and genetics.1,2 More specific, there is evidence that there is an autosomal-dominant inheritance, with incomplete penetrance associated with this phenotype.1,3 The expression of the phenotype is a product of genetics and environmental factors.4 The multifactorial nature of mandibular prognathism makes it very difficult to study and understand.
Mandibular prognathism has a prevalence of as low as 1% in Caucasians but as high as 15% in Asian populations.5,6
Dohmoto et al.7 found that in mice the size of the mandible was controlled by genes located at chromosomes 10 and 11 that correspond to human chromosomal regions 12q21 and 2p13 respectively.
In humans, genome wide linkage analysis provided evidence for linkage to mandibular prognathism at chromosomes 1p36, 6q25, and 19p13.2.8 The presence of P56IT variant in the growth hormone receptor gene (GHR) has been shown to affect mandibular morphology in Chinese and Japanese populations, especially in regards to mandibular height.9,10 In a study looking at a Hispanic cohort, Class III (due primarily to maxillary deficiency) was confirmed to be inherited in an autosomal-dominant pattern and linked to five loci: 1p22.1, 3q26.2, 11q22, 12q13.13, 12q23.11 Li et al.12 detected a specific locus, 14q24.3-31.2, associated with the mandibular prognathism phenotype on a Han Chinese population. In addition to the strong role of heredity there has been evidence suggesting the contribution of certain environmental factors to mandibular prognathism such as enlarged tonsils13, endocrine imbalances14, posture, trauma, and disease.15
It is well documented that the first studies of complex traits suggest a stronger genetic effect that is found by subsequent studies. Both bias and genuine population diversity might explain why early studies tend to overestimate the disease predisposition conferred by candidate gene polymorphisms.16 If there is a true effect of any of the previously described loci in mandibular prognathism, the expectation is that those results can be replicated. Therefore, we typed markers in eight loci and measured the association between genetic variation and mandibular prognathism using a population from Pittsburgh, USA.
Materials and Methods
Subjects
The subjects used in this study were active orthodontic patients from the Department of Orthodontics of the School of Dental Medicine at the University of Pittsburgh. Subjects were identified through the Dental Registry and DNA Repository project. In this project, since September of 2006, individuals who seek treatment at the University of Pittsburgh School of Dental Medicine have been invited to be part of the registry. They provide written informed consent authorizing the extraction of information from their dental records. Also, they provide a saliva sample from which DNA can be extracted. Unstimulated saliva samples were obtained from all participants (they were asked to spit) and stored in Oragene DNA Self-Collection kits (DNA Genotek Inc., Ottawa, ON, Canada) at room temperature until being processed. No centrifugation was performed on the saliva samples. DNA was extracted according to the manufacturer’s instructions. This project was approved by the University of Pittsburgh Institutional Review Board.
Clinical Definitions
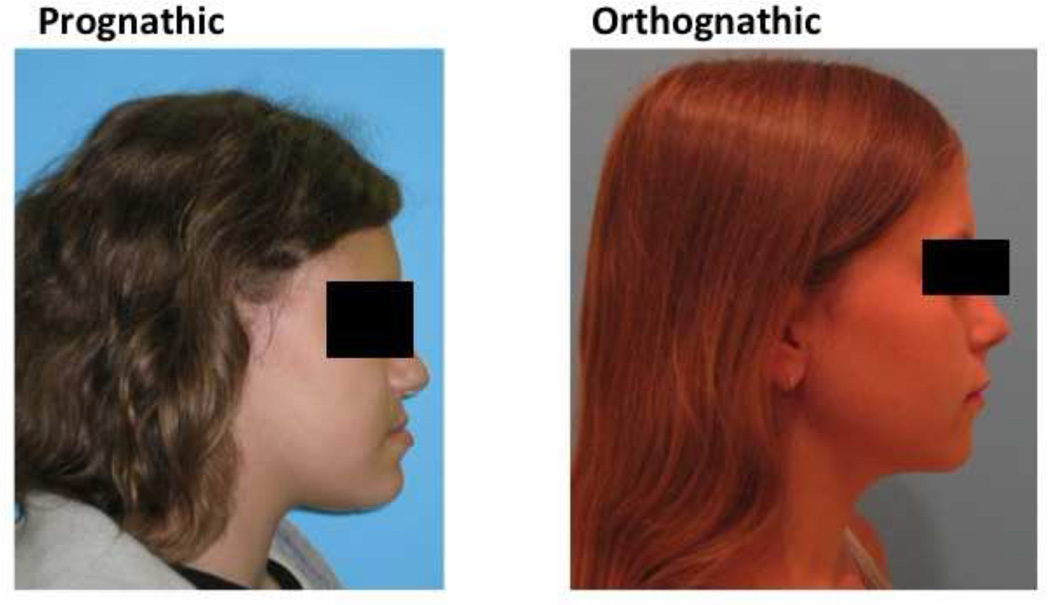
In September 2009, data from 1,630 individuals were extracted from the registry for this pilot project. The 95 individuals treated at the Orthodontics Department were considered eligible for this study. The assessment of the eligible subjects consisted of a careful review of each subject’s clinical and radiographic records. Clinical records consisted of digital orthodontic models (orthoCAD, Cadent, Inc., Carlstadt, NJ, USA), digital tracings of lateral cephalograms (Dolphin, Chatsworth, CA, USA) and digital photographs. The first step in the assessment process were the subject’s soft tissue profile photo (concave or straight profile) and certain cephalometric values in order to classify the patients as either orthognathic or prognathic. Specifically we looked at Steiners’ ANB, Wits Appraisal and Downs’ A–B plane. As described in the Steiner analysis, ANB angle values of less than 2 degrees indicate that the mandible is located ahead of the maxilla. For this study, subjects with smaller than 2 degrees ANB values were reviewed further to clarify if the discrepancy was attributed to a smaller than average size maxilla, by looking at the SNA values. In that case, the subjects were not representing true prognathism but rather a regular size mandible appearing protrusive due to a small maxilla and therefore were excluded from the true prognathic group. The Wits appraisal was another measurement we reviewed because this value has the advantage of indicating the anteroposterior jaw relationship in the facial complex regardless of intra-cranial references. A negative Wits indicates a Class III skeletal relationship and the more negative the value, the more severe the Class III. Downs’ A–B plane angle larger than −4.6 degrees indicates a skeletal Class III diagnosis, although this measurement may appear more severe in cases that present a pronounced bony pogonion. Additional Class III criteria included dental classification, such as Class III molar/canine relationships and negative OJ based on the digital models and clinical examination. Were excluded any subjects with facial clefting, abnormal anterior cranial base growth defects such as or similar to achondrodysplasia or midfacial growth deficiencies caused by or other pathologies such as tumors, cists or trauma which might scar midfacial periosteal surfaces and limit normal growth potentials. After analysis of clinical information, 44 mandibular prognathism and 35 orthognathic subjects were selected for this study. The 44 mandibular prognathism subjects had an average age of 18.4 years, 31 were females and 13 males, and 24 were White, 15 African American, two Hispanic, and three Asian. The 35 Class I subjects had an average age of 17.6 years, 27 were females and 9 males, and 27 were White, six African Americans, one Hispanic, and two Asian. Table 1 describes all measurements used in the study. Figure 1 exemplifies the individuals in each comparison group.
Table 1.
Summary of the subject measurements.
| Profile | Gender | Age | Ethnicity | Steiner's ANB (degrees) |
Wits Appraisal (mm) |
Down's A–B plane (degrees) |
|---|---|---|---|---|---|---|
| Prognathic | Female | 18 | Caucasian | −1.9 | −6.5 | 1.6 |
| Female | 20 | Caucasian | −4.9 | −6.8 | 6.6 | |
| Male | 17 | Caucasian | −8.1 | −12 | 12.7 | |
| Female | 23 | African-American | −1.2 | −8.4 | 1.3 | |
| Female | 20 | Caucasian | 0.7 | −7.6 | −0.1 | |
| Female | 23 | Caucasian | −10.4 | −33.4 | 14.1 | |
| Female | 14 | Hispanic | −2.7 | −6.2 | 2.7 | |
| Female | 22 | Caucasian | −6.6 | −15.7 | 4.5 | |
| Female | 19 | African-American | −3.4 | −6.4 | −2.8 | |
| Male | 19 | Caucasian | −1.1 | −7.3 | 1 | |
| Female | 30 | African-American | −1.2 | −12.2 | 3.7 | |
| Female | 15 | African-American | 2.4 | −3.4 | −2.4 | |
| Male | 15 | Caucasian | 0.3 | −1.3 | −1 | |
| Female | 16 | African-American | −0.9 | −5 | 2.4 | |
| Female | 12 | Asian | −2.3 | −5 | 2.9 | |
| Female | 12 | Hispanic | 0.6 | −8.1 | 0.7 | |
| Female | 21 | African-American | −1.9 | −4.5 | −1.7 | |
| Female | 13 | African-American | −4 | −12 | 4.9 | |
| Female | 21 | African-American | −4.8 | −4.3 | 4 | |
| Female | 15 | Caucasian | −7.5 | −13.3 | 9.1 | |
| Female | 28 | African-American | −1 | −5.9 | 2.3 | |
| Female | 14 | Caucasian | −0.3 | −4.7 | 2 | |
| Male | 11 | Caucasian | −4.4 | −7.8 | 7.3 | |
| Female | 18 | Caucasian | −4.7 | −10.6 | 4.1 | |
| Male | 18 | Caucasian | −1.9 | −7 | 0.6 | |
| Female | 25 | Caucasian | −1.6 | −5.5 | 2.1 | |
| Male | 26 | Caucasian | −8.8 | −12.1 | 4.3 | |
| Female | 18 | African-American | −6 | −10 | 7 | |
| Female | 11 | African-American | −2.3 | −8.8 | 2.7 | |
| Female | 18 | Caucasian | −2.9 | −4.4 | 5.5 | |
| Male | 24 | Asian | −3.4 | −4.1 | 4.7 | |
| Female | 13 | Caucasian | −0.9 | −5.4 | −0.3 | |
| Female | 15 | Caucasian | −0.7 | −4.6 | 0.4 | |
| Female | 15 | African-American | −1.2 | −7 | 2.6 | |
| Male | 22 | Asian | 0.3 | −6.6 | −1.7 | |
| Female | 15 | Caucasian | −3 | −10.6 | 3 | |
| Male | 28 | Caucasian | −1.9 | −8.3 | 2 | |
| Male | 12 | African-American | −3.8 | −8.2 | 4.6 | |
| Male | 19 | Caucasian | −2.6 | −7.7 | 1.2 | |
| Male | 17 | African-American | −2.2 | −3.6 | 1.8 | |
| Female | 13 | African-American | −4.1 | −12.5 | 5.8 | |
| Male | 25 | Caucasian | −0.3 | −4.1 | −0.9 | |
| Female | 15 | Caucasian | −4.7 | −10 | 5.6 | |
| Female | 24 | Caucasian | −1.1 | −3.5 | 2.1 | |
| Orthognathic | Female | 14 | Caucasian | 1.8 | −0.2 | −3.4 |
| Female | 14 | Caucasian | 1.7 | 1.3 | −4.5 | |
| Female | 13 | African-American | 2.4 | −0.9 | −2.7 | |
| Male | 14 | African-American | 2.6 | −1.7 | −2.7 | |
| Female | 17 | Caucasian | 2 | −0.6 | −4.1 | |
| Male | 36 | Caucasian | 1.4 | −0.7 | −3.7 | |
| Male | 52 | Caucasian | 0.8 | −1.3 | −5.4 | |
| Female | 14 | Caucasian | 2.8 | −0.5 | −5.3 | |
| Female | 13 | Caucasian | 2.5 | 1.4 | −5 | |
| Female | 18 | Caucasian | 2.6 | −1.5 | −3.2 | |
| Male | 19 | Caucasian | 2.9 | −0.3 | −6.8 | |
| Female | 14 | Caucasian | 3.8 | 1.4 | −8.7 | |
| Female | 15 | Caucasian | 1.8 | 1 | −5 | |
| Male | 11 | African-American | 2.3 | −2.9 | −3.4 | |
| Female | 24 | Caucasian | 2.4 | −2.7 | −3.7 | |
| Female | 15 | Caucasian | 0.1 | −2.7 | −0.3 | |
| Female | 15 | Caucasian | 2.5 | −3.2 | −2.5 | |
| Female | 15 | African-American | 2.8 | −1.3 | −3.9 | |
| Female | 12 | Caucasian | 0.8 | −0.1 | −1.9 | |
| Female | 17 | Caucasian | 0.7 | −0.9 | −1.7 | |
| Male | 16 | Caucasian | 1.9 | −0.6 | −3.6 | |
| Female | 13 | Caucasian | 1.6 | −0.2 | −4.8 | |
| Female | 14 | Caucasian | 3.3 | 0.1 | −7.4 | |
| Female | 12 | Caucasian | 1.7 | 0.5 | −3.6 | |
| Female | 24 | Caucasian | 3.3 | 0.5 | −7.7 | |
| Male | 16 | Caucasian | 2.1 | −1.1 | −5.4 | |
| Female | 17 | Asian | 2.2 | −0.4 | −4.6 | |
| Female | 15 | Caucasian | 1.2 | −0.6 | −3.1 | |
| Female | 15 | Caucasian | 1 | −1.2 | −3.5 | |
| Female | 11 | Hispanic | 3.7 | 0.1 | −6 | |
| Female | 13 | Caucasian | 1.7 | 1.8 | −4.5 | |
| Male | 28 | Asian | 3.7 | 0.9 | −5.1 | |
| Female | 11 | African-American | 1.9 | −1 | −1 | |
| Female | 11 | Caucasian | 3 | −0.1 | −4.6 | |
| Female | 16 | African-American | 3.8 | 0.1 | −4.8 | |
| Male | 22 | Caucasian | 2.7 | −4 | −2.6 |
Figure 1.
Example of individuals in each study group.
Selection of Single Nucleotide Polymorphisms and Genotyping Procedures
We selected 33 single nucleotide polymorphisms covering the eight candidate regions studied (Table 2) from the International HapMap Project database (http://www.hapmap.org). We used the function “Download tag SNP data” and selected 26 polymorphisms as representative of the polymorphisms in the region. We selected polymorphisms that maximally represent the linkage disequilibrium structure of a given region to avoid redundant information.17 Preference was given to polymorphisms with high heterozygosity levels and different minor allele frequencies to avoid intermarker linkage disequilibrium.
Table 2.
Markers studied.
| Marker | Locus | Gene | Original Work Suggesting as a Candidate Region |
|---|---|---|---|
| rs2503243 | 1p22.2 | Intergenic | Frazier-Bowers et al., 2009 |
| rs972054 | 1p22.2 | Intergenic | Frazier-Bowers et al., 2009 |
| rs1413533 | 1p22.2 | Intergenic | Frazier-Bowers et al., 2009 |
| rs4649030 | 1p36.11 | Intergenic | Yamaguchi et al., 2005 |
| rs2101560 | 3q26.32 | Intergenic | Frazier-Bowers et al., 2009 |
| rs1490055 | 3q26.32 | Intergenic | Frazier-Bowers et al., 2009 |
| rs2087312 | 3q26.32 | Intergenic | Frazier-Bowers et al., 2009 |
| rs987526 | 3q26.32 | Intergenic | Frazier-Bowers et al., 2009 |
| rs1601948 | 3q26.32 | Intergenic | Frazier-Bowers et al., 2009 |
| rs1387168 | 3q26.32 | Intergenic | Frazier-Bowers et al., 2009 |
| rs2940913 | 5p12 | Growth Hormone Receptor (GHR) | Yamaguchi et al., 2001; Zhou et al., 2005; Kang et al., 2009; Tomayasu et al., 2009 |
| rs2973015 | 5p12 | Growth Hormone Receptor (GHR) | Yamaguchi et al., 2001; Zhou et al., 2005; Kang et al., 2009; Tomayasu et al., 2009 |
| rs1509460 | 5p12 | Growth Hormone Receptor (GHR) | Yamaguchi et al., 2001; Zhou et al., 2005; Kang et al., 2009; Tomayasu et al., 2009 |
| rs2910875 | 5p12 | Growth Hormone Receptor (GHR) | Yamaguchi et al., 2001; Zhou et al., 2005; Kang et al., 2009; Tomayasu et al., 2009 |
| rs7718944 | 5p12 | Growth Hormone Receptor (GHR) | Yamaguchi et al., 2001; Zhou et al., 2005; Kang et al., 2009; Tomayasu et al., 2009 |
| rs7750085 | 6q26 | Parkinson Juvenile Disease Protein 2 (PARK2) | Yamaguchi et al., 2005 |
| rs3016534 | 6q26 | Parkinson Juvenile Disease Protein 2 (PARK2) | Yamaguchi et al., 2005 |
| rs12207168 | 6q26 | Parkinson Juvenile Disease Protein 2 (PARK2) | Yamaguchi et al., 2005 |
| rs9458378 | 6q26 | Parkinson Juvenile Disease Protein 2 (PARK2) | Yamaguchi et al., 2005 |
| rs1884153 | 6q26 | Parkinson Juvenile Disease Protein 2 (PARK2) | Yamaguchi et al., 2005 |
| rs571407 | 11q22.3 | Caspase 4 Isoform Gamma Precursor (CASP4) | Frazier-Bowers et al., 2009 |
| rs1902768 | 12q13.13 | Keratin 7 (KRT7) | Frazier-Bowers et al., 2009 |
| rs7300317 | 12q13.13 | Keratin 7 (KRT7) | Frazier-Bowers et al., 2009 |
| rs11113231 | 12q23.3 | Intergenic | Frazier-Bowers et al., 2009 |
| rs4964541 | 12q23.3 | Intergenic | Frazier-Bowers et al., 2009 |
| rs10850110 | 12q24.11* | Myosin 1H (MYO1H) | Frazier-Bowers et al., 2009 |
| rs10850364 | 12q24.21* | T-box 3 (TBX3) | Frazier-Bowers et al., 2009 |
| rs7351083 | 19p13.2 | Fibrilin 3 Precursor (FBN3) | Yamaguchi et al., 2005 |
| rs4804264 | 19p13.2 | Fibrilin 3 Precursor (FBN3) | Yamaguchi et al., 2005 |
| rs8103218 | 19p13.2 | Fibrilin 3 Precursor (FBN3) | Yamaguchi et al., 2005 |
| rs12327845 | 19p13.2 | Fibrilin 3 Precursor (FBN3) | Yamaguchi et al., 2005 |
| rs10411185 | 19p13.3 | Intergenic | Yamaguchi et al., 2005 |
Note:
These loci flank the region suggested by Frazier-Bowers et al., 2009 and were included to more comprehensively study this candidate region.
Selected SNPs were genotyped using Taqman chemistry18 on an automatic sequence-detection instrument (ABI Prism 7900HT, Applied Biosystems, Foster City, CA, USA). Assays and reagents were supplied by Applied Biosystems (Applied Biosystems, Foster City, CA, USA).
Statistical Analyses
Chi-square (χ2) or Fisher’s exact calculations were used to assess Hardy-Weinberg equilibrium and significance in all comparisons. The number of copies of each allele and each genotype per marker was compared between mandibular prognathism and Class I subjects (Table 2). Alpha of 0.05 was considered as statistically significant.
Results
Differences in gender and ethnic background between the two groups were not statistically significant. All makers were in Hardy-Weinberg equilibrium in both Class I and mandibular prognathism groups. Markers rs2503243, rs972054, rs1413533, rs1490055, rs2101560, rs1601948, rs1387168, rs2940913, rs7718944, rs3016534, rs9458378, and rs4964541were not informative and could not be analyzed. The G allele of marker rs10850110 (5’ of myosin 1H - MYO1H) was over-represented in mandibular prognathism subjects (p=0.03; Table 3). In Table 4, details of the samples contributing to the rs10850110 are displayed. The distribution of samples based on ethnic background suggests disproportionate number of samples from a specific subgroup is not influencing the results (among the 27 mandibular prognathism subjects 16 were White, eight were African American, and three belonged to other groups, whereas among Class I subjects, 19 were White, four were African Americans, and three belonged to other groups.
Table 3.
Summary of the association results. Values under “Mandibular Prognathism” and “Class I” columns indicate the number of individuals that possess each of the genotypes in each group and the number of alleles found in each group of subjects.
| Locus | Marker | Genotype | Allele | ||||
|---|---|---|---|---|---|---|---|
| Mandibular Prognathism |
Class I | Mandibular Prognathism |
Class I | ||||
| 1p22.2 | rs4649030 | AA | 3 | 3 | A | 13 | 18 |
| AG | 7 | 12 | G | 43 | 46 | ||
| GG | 18 | 17 | p=0.54 | ||||
| p=0.58 | |||||||
| 3q26.32 | rs2087312 | AA | 30 | 26 | A | 73 | 61 |
| AC | 13 | 9 | C | 13 | 19 | ||
| CC | 0 | 1 | p=0.16 | ||||
| p=0.41 | |||||||
| rs987526 | CC | 1 | 0 | C | 7 | 3 | |
| CT | 5 | 3 | T | 61 | 59 | ||
| CC | 28 | 28 | p=0.24 | ||||
| p=0.5 | |||||||
| 5p12 | rs2973015 | AA | 9 | 10 | A | 27 | 31 |
| AG | 9 | 11 | G | 27 | 21 | ||
| GG | 9 | 5 | p=0.32 | ||||
| p=0.5 | |||||||
| rs1509460 | GG | 5 | 3 | G | 35 | 29 | |
| GT | 25 | 23 | T | 39 | 43 | ||
| TT | 7 | 10 | p=0.4 | ||||
| p=0.58 | |||||||
| rs2910875 | AA | 4 | 5 | A | 18 | 27 | |
| AG | 10 | 11 | G | 20 | 31 | ||
| GG | 5 | 10 | p=0.94 | ||||
| p=0.69 | |||||||
| 6q26 | rs7750085 | AA | 8 | 13 | A | 35 | 39 |
| AT | 19 | 13 | T | 31 | 31 | ||
| TT | 6 | 9 | p=0.75 | ||||
| p=0.24 | |||||||
| rs12207168 | AA | 8 | 12 | A | 35 | 39 | |
| AG | 19 | 15 | G | 41 | 33 | ||
| GG | 11 | 9 | p=0.32 | ||||
| p=0.49 | |||||||
| rs1884153 | AA | 0 | 0 | A | 2 | 3 | |
| AG | 2 | 3 | G | 80 | 67 | ||
| GG | 39 | 32 | p=0.53 | ||||
| p=0.52 | |||||||
| 11q22.3 | rs571407 | CC | 1 | 0 | C | 7 | 3 |
| CT | 5 | 3 | T | 61 | 59 | ||
| TT | 28 | 28 | p=0.24 | ||||
| p=0.5 | |||||||
| 12q13.13 | rs1902768 | AA | 1 | 2 | A | 11 | 14 |
| AG | 9 | 10 | G | 59 | 50 | ||
| GG | 25 | 20 | p=0.36 | ||||
| p=0.67 | |||||||
| rs7300317 | AA | 8 | 7 | A | 35 | 33 | |
| AG | 19 | 19 | G | 35 | 37 | ||
| GG | 8 | 9 | p=0.92 | ||||
| p=0.94 | |||||||
| 12q23.3 | rs11113231 | AA | 6 | 5 | A | 38 | 30 |
| AG | 26 | 20 | G | 42 | 38 | ||
| GG | 8 | 9 | p=0.68 | ||||
| p=0.8 | |||||||
| 12q24.11 | rs10850110 | AA | 1 | 5 | A | 8 | 17 |
| AG | 6 | 7 | G | 46 | 35 | ||
| GG | 20 | 14 | p=0.03 | ||||
| p=0.15 | |||||||
| 12q24.21 | rs10850364 | AA | 3 | 7 | A | 19 | 30 |
| AG | 13 | 16 | G | 29 | 34 | ||
| GG | 8 | 9 | p=0.44 | ||||
| p=0.66 | |||||||
| 19p13.2 | rs7351083 | AA | 6 | 6 | A | 34 | 29 |
| AG | 22 | 17 | G | 42 | 41 | ||
| GG | 10 | 12 | p=0.69 | ||||
| p=0.7 | |||||||
| rs4804264 | CC | 9 | 6 | C | 33 | 27 | |
| CT | 15 | 15 | T | 49 | 45 | ||
| TT | 17 | 15 | p=0.73 | ||||
| p=0.82 | |||||||
| rs8103218 | CC | 6 | 7 | C | 25 | 30 | |
| CT | 13 | 16 | T | 25 | 28 | ||
| TT | 6 | 6 | p=0.86 | ||||
| p=0.96 | |||||||
| rs12327845 | CC | 25 | 15 | C | 63 | 45 | |
| CT | 13 | 15 | T | 23 | 23 | ||
| TT | 5 | 4 | p=0.34 | ||||
| p=0.42 | |||||||
| 19p13.3 | rs10411185 | AA | 11 | 5 | A | 43 | 29 |
| AG | 21 | 19 | G | 37 | 39 | ||
| GG | 8 | 10 | p=0.18 | ||||
| p=0.35 |
Note: Variations in the number of subjects or the number of alleles are due to PCR failure. These failures are related to the purity of the genomic DNA used and the uniqueness of PCR primer probes. These aspects are hard to be predicted but should not dramatically influence the data.
Table 4.
Demographic variables of the samples contributing to the rs10850110 marker analysis.
| Ethnicity | Gender | Number of Individuals with genotypes: Mandibular Prognathism |
Number of Individuals with genotypes: Class I |
||||
|---|---|---|---|---|---|---|---|
| AA | AG | GG | AA | AG | GG | ||
| White | Male | 1 | 2 | 4 | - | 1 | 2 |
| Female | - | 4 | 5 | 5 | 5 | 6 | |
| African-American | Male | - | - | 1 | - | - | 1 |
| Female | - | - | 7 | - | - | 3 | |
| Other | Male | - | - | - | - | - | 1 |
| Female | - | - | 3 | 1 | - | 1 | |
Discussion
This is the first report that independently attempted to replicate the recent results of two genome wide scans for mandibular prognathism and Class III (due primarily to maxillary deficiency).8,11 Our findings corroborate the previous suggestive linkage results with 12q23 (LOD score 2.9311). The most common allele of a marker flanking MYO1H (rs10850110) was over-represented in mandibular prognathism subjects. The frequency of the less common rs10850110 allele varies from 0.008 in Sub-Saharan Africans, to 0.089 in Japanese, 0.148 in Han Chinese, and 0.275 in Europeans (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=10850110). MYO1H is a class I myosin which is in a different protein grouping than the MHC (myosin heavy chain) isoforms found in the skeletal muscle sarcomere, which are the basis of fiber typing. Class I myosins are necessary for cell motility, phagocytosis and vesicle transport.19
Since the sample size of our study is modest and some genetic markers were uninformative, we cannot conclude the chromosomal regions studied here do not play a role in mandibular prognathism development. However, the definitions used to select subjects for this study likely increased homogeneity and the evidence of a role of 12q24 in mandibular prognathism is likely to be true. In an interval of 397,305 base pairs in 12q24.11, another four genes flank MYO1H. These genes are ACACB (acetyl-CoA carboxylase beta), FOXN4 (forkhead box N4), KCTD10 (potassium channel tetramerisation domain containing 10), and UBE3B (ubiquitin protein ligase E3B). These genes are involved in different aspects of metabolism, and although MYO1H is the best candidate for a role in mandibular prognathism, one cannot discard the possibility that these other genes have a role as well.
The cohort selected for this study reflects the ethnic breakdown of the city of Pittsburgh. According to the Pittsburgh Pennsylvania Census of 2000 (http://www.hellopittsburgh.com/Census.Cfm), the city was composed by 67.6% of European descent inhabitants and 27.1% of African descent inhabitants. The remaining 5.3% is composed by Hispanics, Asians, and other groups. There was no statistically significant difference between mandibular prognathism and orthognathic subjects in regards to the distribution of age, gender, and ethnic background. However, undetected population substructure could have revealed a spurious association, even though when this effect is present, we tend to see several positive associations. The small percentage of Asians in our sample likely unable us to detect the reported association between GHR (growth hormone receptor) and mandibular height that has been described primarily in Japanese, Chinese, and Koreans.9,10,20,21 Future investigations also should expand further the variants studied in the GHR locus. One ideal variant to be studied due to its allelic frequency is the d3/fl-GHR variant, which minor allele frequencies are 47.9% in African- Americans and 31.3% in European-Americans.20
While concerned about multiple testing, we did not apply the strict Bonferroni correction as it would increase type II errors and a major focus of this study was to replicate putative associations with the previously described contributing loci to mandibular prognathism. For example, under the Bonferroni correction, we would have lowered the alpha to 0.0025 (0.05/20) or 0.00125 (0.05/40) (number of tests in Table 3). Our previous work with other craniofacial defects has shown that true associations will be missed if nominal p-values are not considered as well. One example is the known association between IRF6 (interferon regulatory factor 6) and isolated cleft lip and palate that would have been missed in a project we performed in which we tested 1,489 genetic markers.22 Therefore we report here results with p-values below 0.05. However, our data must be carefully interpreted since it is expected that some of the p-values below 0.05 can be due to chance.
One of the biological pathways recently identified23 in contributing to skeletal height in humans is the nicotinic acetylcholine receptor signaling pathway with three associated genes included in the unconventional myosin grouping, MYO1F, MYO6 and MYO9B. MYO1F is a very similar gene to the MYO1H identified as associated with mandibular prognathism in our study and future studies should considered all the genes in the family as candidates for a role in mandibular prognathism development. In summary, we provide further evidence of a role of the 12q24 locus in mandibular prognathism development. The possible role on a type I myosin is intriguing since it would suggest that muscle function may have a more important role than previous thought in the development and deviations of the bone structures of the craniofacial complex. Since the sample size of this study is modest, replication of these findings in independent populations is necessary. We will continue our efforts to expand the population studied to allow the results of our study to be replicated as well as perform these analyses by gender, age groups, and ethnicity.
Acknowledgements
The authors thank the subjects for their participation in this study. Jacqueline Noel and Justin Bair provided support for subject recruitment. Data for this study were provided by the Dental Registry and DNA Repository (DRDR) of the School of Dental Medicine, University of Pittsburgh. The work was supported by an AAOF 2009 Grant (M.T-F.). The DRDR was in part supported by the NIH Grant 5TL1RR024155-02. This paper is the result of M.T-F.’s participation in the Clinical Research Scholars Program of the School of Dental Medicine, University of Pittsburgh.
References
- 1.Stiles KA, Luke JE. The inheritance of malocclusion due to mandibular prognathism. J Heredity. 1953;44:241–245. [Google Scholar]
- 2.Watanabe M, Suda N, Ohyama K. Mandibular prognathism in Japanese families ascertained through orthognathically treated patients. Am J Orthod Dentofacial Orthop. 2005;128:466–470. doi: 10.1016/j.ajodo.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 3.El-Gheriani AA, Maher BS, El-Gheriani AS, Sciote JJ, Abu-Shahba FA, Al-Azemi R, Marazita ML. Segregation analysis of mandibular prognathism in Libya. J Dent Res. 2003;82:523–527. doi: 10.1177/154405910308200707. [DOI] [PubMed] [Google Scholar]
- 4.Battagel J. The aetiological factors in Class III malocclusion. Eur J Ortho. 1993;15:347–370. doi: 10.1093/ejo/15.5.347. [DOI] [PubMed] [Google Scholar]
- 5.Allwright WC, Bundred WH. A survey of handicapping dentofacial anomalies among Chinese in Hong Kong. Int Dent J. 1964;14:505–519. [Google Scholar]
- 6.Emrich RE, Brodie AG, Blayney JR. Prevalence of Class I, Class II and Class III maloclussions (Angle) in an urban population; an epidemiological study. J Dent Res. 1965;44:947–953. doi: 10.1177/00220345650440053301. [DOI] [PubMed] [Google Scholar]
- 7.Dohmoto A, Shimizu K, Asada Y, Maeda T. Quantitative trait Loci on Chromosomes 10 and 11 influencing mandible size of SMXA RI mouse strains. J. Dent. Res. 2002;81:501–504. doi: 10.1177/154405910208100714. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi T, Park SB, Narita A, Maki K, Inoue I. Genome-wide linkage analysis of mandibular prognathism in Korean and Japanese patients. J. Dent. Res. 2005;84:255–259. doi: 10.1177/154405910508400309. [DOI] [PubMed] [Google Scholar]
- 9.Yamaguchi T, Maki K, Shibasaki Y. Growth hormone receptor gene variant and mandibular height in the normal Japanese population. Am J Orthod Dentofacial Orthop. 2001;119:650–653. doi: 10.1067/mod.2001.114536. [DOI] [PubMed] [Google Scholar]
- 10.Zhou J, Lu Y, Gao XH, Chen YC, Lu JJ, Bai YX, Shen Y, Wang BK. The growth hormone receptor gene is associated with mandibular height in a Chinese population. J Dent Res. 2005;84:1052–1056. doi: 10.1177/154405910508401116. [DOI] [PubMed] [Google Scholar]
- 11.Frazier-Bowers S, Rincon-Rodriquez R, Zhou J, Alexander K, Lange E. Evidence of linkage in a Hispanic cohort with a Class III dentofacial phenotype. J Dent Res. 2009;88:56–60. doi: 10.1177/0022034508327817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, Li X, Zhang F, Li J, Chen F. The identification of a novel locus for mandibular prognathism in the Han Chinese population. J Dent Res. 2010 doi: 10.1177/0022034510382546. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 13.Angle EH. Treatment of malocclusion of teeth. 7th ed. Philadelphia: S.S. White Manufacturing Company; 1907. [Google Scholar]
- 14.Downs WG. Studies in the causes of dental anomalies. Dent Res. 1928;28:267–379. [Google Scholar]
- 15.Gold JK. A new approach to the treatment of mandibular prognathism. Am J Orthod. 1949;35:893–912. doi: 10.1016/0002-9416(49)90085-8. [DOI] [PubMed] [Google Scholar]
- 16.Ioannidis JPA, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 17.Carlsson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ranade K, Chang MS, Ting CT, Pei D, Hsiao CF, Olivier M, Pesich R, Hebert J, Chen YD, Dzau VJ, Curb D, Olshen R, Risch N, Cox DR, Botstein D. High-throughput genotyping with single nucleotide polymorphisms. Genome Res. 2001;11:1262–1268. doi: 10.1101/gr.157801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowlerson A, Raoul G, Daniel Y, Close J, Maurage CA, Ferri J, Sciote JJ. Fiber-type differences in masseter muscle associated with different facial morphologies. Am J Orthod Dentofacial Orthop. 2005;127:37–46. doi: 10.1016/j.ajodo.2004.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang EH, Yamaguchi T, Tajima A, Nakajima T, Tomoyasu Y, Watanabe M, Yamaguchi M, Park SB, Maki K, Inoue I. Association of the growth hormone receptor gene polymorphisms with mandibular height in a Korean population. Arch Oral Biol. 2009;54:556–562. doi: 10.1016/j.archoralbio.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Tomayasu Y, Yamaguchi T, Tajima A, Nakajima T, Inoue I, Maki K. Furthe evidence for an association between mandibular height and the growth hormone receptor gene in a Japanese population. Am J Orthod Dentofacial Orthop. 2009;136:536–541. doi: 10.1016/j.ajodo.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 22.Vieira AR, McHenry TG, Daack-Hirsch S, Murray JC, Marazita ML. Candidate gene/loci studies in cleft lip/palate and dental anomalies finds novel susceptibility genes for clefts. Genet Med. 2008;10:668–674. doi: 10.1097/GIM.0b013e3181833793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segrè AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Mägi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Carola Zillikens M, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Hua Zhao J, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, König IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Müller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, Sanna S, Shi J, Sovio U, Thompson JR, Turchin MC, Vandenput L, Verlaan DJ, Vitart V, White CC, Ziegler A, Almgren P, Balmforth AJ, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan J, Elliott P, Elosua R, Eriksson JG, Freimer NB, Geus EJ, Glorioso N, Haiqing S, Hartikainen AL, Havulinna AS, Hicks AA, Hui J, Igl W, Illig T, Jula A, Kajantie E, Kilpeläinen TO, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu J, Lokki ML, Marusic A, Maschio A, Meitinger T, Mulas A, Paré G, Parker AN, Peden JF, Petersmann A, Pichler I, Pietiläinen KH, Pouta A, Ridderstråle M, Rotter JI, Sambrook JG, Sanders AR, Oliver Schmidt C, Sinisalo J, Smit JH, Stringham HM, Bragi Walters G, Widen E, Wild SH, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle WL, Nelis M, Peters MJ, Ripatti S, van Meurs JB, Aben KK, Ardlie KG, Beckmann JS, Beilby JP, Bergman RN, Bergmann S, Collins FS, Cusi D, den Heijer M, Eiriksdottir G, Gejman PV, Hall AS, Hamsten A, Huikuri HV, Iribarren C, Kähönen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer LJ, Lehtimäki T, Melander O, Mosley TH, Jr, Musk AW, Nieminen MS, O'Donnell CJ, Ohlsson C, Oostra B, Palmer LJ, Raitakari O, Ridker PM, Rioux JD, Rissanen A, Rivolta C, Schunkert H, Shuldiner AR, Siscovick DS, Stumvoll M, Tönjes A, Tuomilehto J, van Ommen GJ, Viikari J, Heath AC, Martin NG, Montgomery GW, Province MA, Kayser M, Arnold AM, Atwood LD, Boerwinkle E, Chanock SJ, Deloukas P, Gieger C, Grönberg H, Hall P, Hattersley AT, Hengstenberg C, Hoffman W, Mark Lathrop G, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright AF, Assimes TL, Barroso I, Hofman A, Mohlke KL, Boomsma DI, Caulfield MJ, Adrienne Cupples L, Erdmann J, Fox CS, Gudnason V, Gyllensten U, Harris TB, Hayes RB, Jarvelin MR, Mooser V, Munroe PB, Ouwehand WH, Penninx BW, Pramstaller PP, Quertermous T, Rudan I, Samani NJ, Spector TD, Völzke H, Watkins H, Wilson JF, Groop LC, Haritunians T, Hu FB, Kaplan RC, Metspalu A, North KE, Schlessinger D, Wareham NJ, Hunter DJ, O'Connell JR, Strachan DP, Wichmann HE, Borecki IB, van Duijn CM, Schadt EE, Thorsteinsdottir U, Peltonen L, Uitterlinden AG, Visscher PM, Chatterjee N, Loos RJ, Boehnke M, McCarthy MI, Ingelsson E, Lindgren CM, Abecasis GR, Stefansson K, Frayling TM, Hirschhorn JN. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]