Abstract
SNPs mapped to 8q24.21 have been shown to be associated with glioma development. By means of tag SNP genotyping/imputation, pooled next-generation sequencing (NGS) using long-range PCR, and subsequent validation SNP genotyping we identified seven low-frequency SNPs that were consistently and highly associated with glioma risk (p=10−25 to 10−14). The most associated SNP, rs55705857, remained highly significant after individual adjustment for the other top six and two previously published SNPs. After stratifying by histologic and tumor genetic subtype, the most significant associations were with oligodendroglial tumors and IDH1 or IDH2 mutated gliomas, (ORrs55705857 = 5.1, p=1.1x10−31 and ORrs55705857 = 4.8, p=6.6 x10−22, respectively). Strong associations were observed for IDH1 or IDH2 mutated astrocytomas (grades II–IV) (OR rs55705857=5.16–6.66; p=4.7x10−12 to 2.2x10−8), but not IDH1 or IDH2 wild-type astrocytomas (smallest p=0.26). The conserved sequence block that includes rs55705857 is consistently modeled as a microRNA.
Keywords: Oligodendroglioma, Glioblastoma, IDH1 and IDH2 mutation, single nucleotide polymorphism
We and others have previously shown that SNPs in 8q24.21 near CCDC26 are risk loci for oligodendroglial tumors and IDH1 or IDH2 mutated gliomas (1–4). To identify higher risk/low frequency loci within 8q24.21 we used a two-stage study design consisting of tag SNP array genotyping/imputation in parallel with long-range PCR/pooled NGS (Stage 1), followed by validation custom genotyping (Stage 2) (see Online Methods). Stage 1 utilized both imputation and pooled NGS because we were concerned that reliance on a single method might miss potentially important SNPs. We used two independent groups of cases (total n=1657; Mayo n=852 and UCSF n=805) and controls (total n=1301; Mayo n=789 and UCSF n=512). Supplementary Table 1 summarizes the subjects used in each stage. Seven of 157 candidate SNPs genotyped in Stage 2 (rs72714236, rs72714295, rs72714302, rs72716319, rs72716328, rs147958197, rs55705857) were highly significantly associated with glioma risk (see Methods and Supplementary Table 2). Importantly, rs55705857 was detected by the pooled NGS method; the remaining six SNPs were detected by imputation alone or by both imputation and pooled NGS (Supplementary Table 2). The minor allele frequencies (MAF) for these seven SNPs among all glioma cases ranged from 0.11–0.14, while in controls the MAFs ranged from 0.04 to 0.06 (p=5x10−25 to 3x10−14, Supplementary Table 3). Importantly, rs55705857 was detected by the pooled NGS method; the remaining six SNPs were detected by imputation alone or by both imputation and pooled NGS (Supplementary Table 2). This observation suggests that, until the human genome is mapped in greater detail, a combination of sequencing and imputation will continue to be necessary to identify uncommon risk loci.
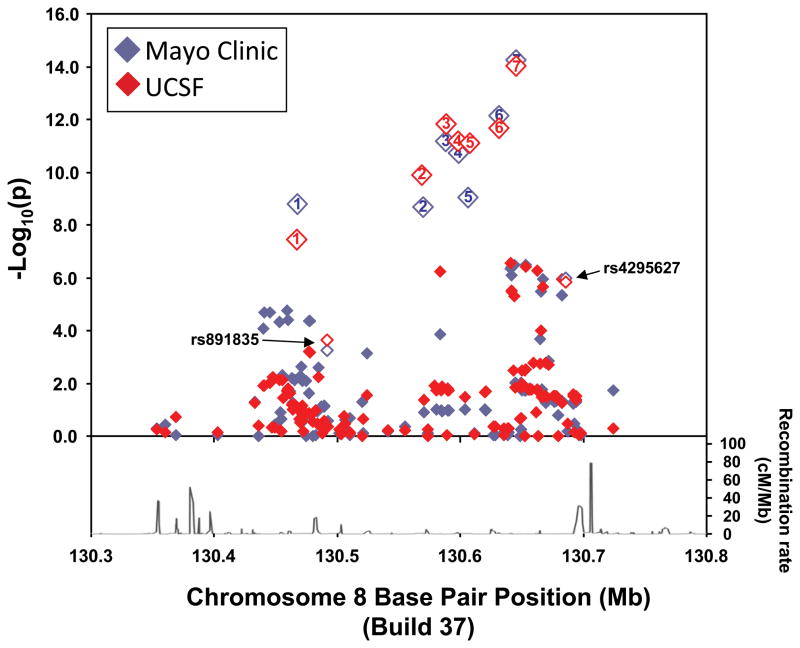
Stratification by histologic subtype showed differences in the strength of the SNP associations (Supplementary Table 3). Figure 1 illustrates the Stage 2 associations of oligodendroglioma risk for the 157 SNPs within 8q24.21 (including the 7 most highly associated loci). The results were remarkably consistent between study sites (see also Supplementary Tables 2 and 3). The strongest association, as measured by the size of the OR, was for the subgroup of oligodendroglioma cases versus controls with the G allele of rs55705857 (SNP7 in Figure 1) with an OR of 6.3 (95% CI = 4.6 – 8.8; p=2.2x10−28). All the other glioma subtypes examined (oligodendrogliomas, mixed oligoastrocytomas, grade II–III astrocytomas and glioblastoma (grade IV astrocytoma)) were also significantly associated with the 7 SNPs (Supplementary Table 3) with the strongest associations being with rs55705857 (Figure 2).
Figure 1.
Case-control SNP associations with oligodendroglioma risk for 157 SNPs within the 8q24.21 (CCDC26) region by study site, Mayo Clinic (red) and University of California San Francisco (UCSF) (blue). The −log10 p-value of the association of each SNP versus chromosome 8q24 position is plotted. Only Stage 2 validation genotyping data are illustrated. The recombination rate for the region using HapMap is plotted in the lower part of the figure.
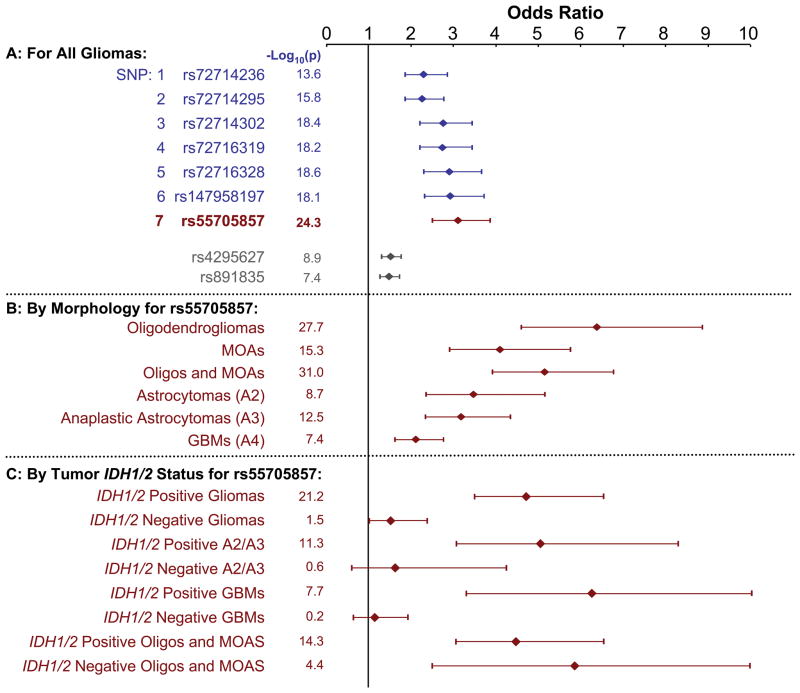
Figure 2.
Glioma case-control odds ratios (ORs), 95% confidence intervals (CIs), and (−log10p) for SNPs within the 8q24.21 (CCDC26) region. A: Associations of all gliomas with 7 most strongly and significantly associated SNPs along with two SNPs from prior studies (1, 2) (rs4295627 and rs891835). B: Associations of rs55705857 G allele with various glioma morphologic subtypes. C: Association of rs55705857 G allele with glioma morphologic subtypes further stratified by presence or absence of tumor IDH1 or IDH2 mutation. Only Stage 2 validation genotyping data are illustrated. Abbreviations: MOA=mixed oligoastrocytoma, Oligo=oligodendroglioma, GBM=glioblastoma
IDH1 or IDH2 mutations occur in approximately 50–80% of grade II–III gliomas and secondary glioblastomas, but in less than 10% of primary glioblastomas (5–8). IDH mutation has been associated with younger age of onset and better survival among glioblastoma patients, and with other somatic genetic and epigenetic alterations (9). Interestingly, although rs55705857 (and the other six SNPs) were associated with risk of both IDH mutated and IDH wild-type oligodendroglial tumors (Figure 2 and Supplementary Table 4), these SNPs were only associated with IDH mutated astrocytic gliomas (WHO grades II and III astrocytoma and glioblastoma) but not with IDH wild type astrocytic gliomas. Specifically, ORrs55705857= 6.7 (95% CI = 3.4 – 12.9; p=2.2x10−8) and ORrs55705857=5.2 (95% CI = 3.2 – 8.2; p=4.7x10−12) for IDH1 or IDH2 mutated glioblastoma and grade II or III astrocytoma, respectively, in Stage 2. However, the ORs were close to 1.0 and not significant for astrocytic glioma patients with IDH1 and IDH2 wild-type tumors. While the samples sizes were relatively small for some glioma subtypes, the results were consistent between the two independent study sites (Supplementary Table 4). Furthermore, the MAF for rs55705857 in the IDH1 or IDH2 mutant astrocytic cases was nearly identical to the MAF for oligodendroglioma cases (MAF~0.20).
1p/19q co-deletion is observed in 50–70% of oligodendrogliomas and mixed oligoastrocytomas and is associated with superior therapeutic response and survival (10, 11). Of the 264 Mayo Clinic oligodendrogliomas and mixed oligoastrocytomas, 172 had 1p/19q deletion data. For these gliomas we observed an ORrs55705857 =6.5 (95% CI = 4.2 – 10; p=9.5x10−18) for the development of 1p/19q codeleted oligodendrogliomas and mixed oligoastrocytomas and an ORrs55705857=4.0 (95% CI = 2.4 – 6.8; p=2.2x10−7) for 1p/19q non-codeleted oligodendrogliomas and mixed oligoastrocytomas in Stage 2. Together these results show that rs55705857 is associated with risk of oligodendroglial tumors regardless of tumor 1p/19q and IDH mutation status but only with risk of astrocytic gliomas harboring IDH mutations. Approximately 40% of patients with these glioma subtypes carry one or more of the G risk alleles for rs55705857 compared to only about 8% of the controls.
The seven low frequency SNPs identified herein and the two SNPs reported by Shete et al. (1) are all highly correlated. When individually adjusting for the other six low frequency SNPs and the two reported SNPs, only rs55705857 remained significant (Supplementary Table 5), with ORs ranging from 4.8–7.0 for gliomas of oligodendroglial lineage. Thus, the primary association signal is due to rs55705857.
Subsequent imputation on the validation panel did not identify additional SNPs with statistically stronger associations than the original 7 SNPs we identified. Thus, the reported association does not extend beyond the boundaries of the region genotyped (data not shown).
Associations of similar magnitude and significance to those observed here for rs55705857 with glioma subtypes have rarely been reported in cancer genome wide association and subsequent fine mapping studies (12–15). Indeed, the NHGRI catalogue (See URL link below) lists only two cancer studies with associations greater than 4.0 (12, 13); one of those was a variant for melanoma identified within high risk pedigrees and another was for a variant associated with lung cancer survival.
The risk region maps within a gene-poor region of 8q24 (Supplementary Figure 1). Some of the loci reside within the introns of CCDC26, a predicted long noncoding RNA (lncRNA) gene. The most significant risk locus, rs55705857, resides in a conserved cluster (PhastCons track – UCSC browser) from 130,645,483 to 130,645,975 bases. Fifteen bases (including rs55705857) are 100% conserved from platypus to human (see Supplementary Figure 1). The functional relevance of the conserved region is not known and the functional annotation of the region is limited. However, structural modeling indicates that the conserved region may also encode a novel lncRNA or a miRNA (data not shown). The conserved elements include the predicted unpaired loops of a miRNA and rs55705857 lies within the seed sequence of the putative miRNA; an observation that may have functional relevance (16). Alternatively, there may be long-and short-range interactions between this highly conserved region with other critical regions of the genome, or there may be other causative variants that our methods have not detected.
Our data show that variants within the 8q24.21 region are associated with specific morphologic and molecular glioma subtypes. Similar subtype specific associations have been recently observed in other cancers. Variants within 5p12, 8q24 (~1.5Mb proximal to the variants reported here), 9p21, 10q21 and 11q13 are associated with estrogen receptor positive but not estrogen receptor negative breast cancers (reviewed in 17). Variants within 19p13 are associated with triple-negative breast cancer (17). Variants within 5p15 are associated with different histologic types of lung cancer (18). Serous ovarian cancer is associated with variants in 8q24 (also ~1.5Mb proximal to the variants reported here) (19). Other variants are associated with other ovarian cancer subtypes (reviewed in 20). The ORs for all these associations are less than 1.5. In aggregate, the implication of these data is that morphologic and molecular subtyping is critical to cancer genetic epidemiology.
Variants within the 8q24 gene desert – including the CCDC26 region – are associated with risk of multiple cancers (19,21–23). The ORs for these associations do not approach the strength we observed for rs55705857 and glioma risk. While most of these other cancer variants are ~1.5Mb proximal to the region reported here (19,21–23), one possible hypothesis is that synthetic association/long-range linkage disequilibrium may be the basis for some of these associations (24).
Acquired IDH1 or IDH2 mutations in glioma are associated with a specific DNA methylation pattern (6,25), a specific histone modification pattern (26,27), and a stem cell phenotype (25,26). Our results strongly suggest that the glioma risk locus in the 8q24/CCDC26 region identified in this present study might interact with IDH mutations and/or the downstream effects of such mutations to facilitate the development and progression of gliomas. An alternative explanation might be that variants in this region may foster formation of IDH1 or IDH2 mutations. That oligodendroglial tumors without IDH1 or IDH2 mutations are also strongly associated with this risk locus suggests that alterations within these tumors may arise or be maintained by mechanisms similar to (but distinct from) IDH mutation.
Online Methods
Supplementary Figure 2 summarizes the overall study design. The study had two stages: Stage 1 consisted of imputation of a prior 8q24 tag SNP genotyping dataset (2) (Stage 1A) and pooled next-generation sequencing (NGS) using long range PCR (Stage 1B). Stage 2 consisted of custom validation genotyping of alterations detected in Stage 1.
Study populations/Ethical considerations
Mayo Clinic and UCSF case and control characteristics are summarized in Supplementary Table 1. These studies were approved by the Mayo Clinic Office for Human Research Protection and the UCSF Committee on Human Research. Informed consent was obtained from all participants.
Mayo Clinic case-control study
Details of subject recruitment for the Mayo Clinic case-control series have been described previously (2,28). A total of 860 cases and 795 controls from the Mayo Clinic were used in this study; Stage 1A used 582 glioma cases and 532 controls, Stage 1B used 220 oligodendroglioma cases and 274 controls, and Stage 2 used 852 glioma cases and 789 controls (693 of these cases and 578 controls were also used in Stage 1).
UCSF case-control study
UCSF cases and controls were taken from the San Francisco Bay Area Adult Glioma Study (AGS). Details of subject recruitment for AGS have been described previously (28–30). A total of 953 cases and 1079 controls from the UCSF Adult Glioma Study were used in this study; Stage 1A used 191 oligodendroglioma cases and 192 controls, Stage 1B used 177 oligodendroglioma cases and 547 controls, and Stage 2 used 805 glioma cases and 512 controls (182 of these cases, but none of the controls were also used in Stage 1).
Pathology Review and IDH1 and IDH2 Mutation and 1p/19q Deletion
Pathology review was performed by two pathologists (TT and CG) as previously described (28). IDH1 and IDH2 mutation analysis and 1p/19q deletion testing were performed using previously described methods (6,31).
Long Range PCR/Pooled Next Generation Sequencing (NGS) (Stage 1B)
Two peaks of association within 8q24.21 between 130.435 – 130.526 Mbp and 130.624 –130.699 Mbp were previously observed (2). Four pools of DNAs were prepared - two oligodendroglial case pools (n=177 for UCSF and 220 for Mayo), and two control pools (n=547 for UCSF and 274 for Mayo) - and each subjected to long-range PCR covering the two peaks, followed by NGS (to a target depth of 2000X) by deCODE Genetics (22). Supplementary Figure 3 summarizes the final sequence coverage for each pool.
For quality-control, MAFs for common variants with prior genotyping data (2) and MAFs estimated by NGS were compared (Supplementary Figure 4). Concordant MAFs (i.e. MAFs that differed by less than 0.10 in frequency) were found for 74 of 77 of these common SNPs (96%).
Validation Custom Genotyping (Stage 2)
Candidate selected SNPs (See Statistical Methods) from imputation of the tag SNP panel and the long-range PCR/pooled NGS were validated using custom genotyping. GoldenGate assays were designed by Illumina (San Diego, CA) and performed using Illumina’s VeraCode platform. Genotyping was performed by Mayo Clinic Genotyping and the UCSF Genomics Core Facilities. Samples were submitted in 96-well plates. Each plate contained several intra- and inter-plate replicates.
Statistical Methods
Stage 1A: Prior 8q24.21 region Tag SNP panel - Imputation and Statistical Analysis
Quality control and statistical analysis methods for the 96 SNPs genotyped on the prior 8q24 tag SNP panel were previously reported for Mayo Clinic and UCSF (2). The 96 SNPs in this previously published dataset were non-redundant, had MAFs greater than 5%, and covered a 322kb region encompassing the association peak defined by Shete et al. (1). For this study, imputations were performed separately for the Mayo and UCSF data using MACH (32) with 1000 Genomes as the reference population. The region imputed spanned both peaks of the prior publication (2), the “valley” between the peaks, and 250 Kb centromeric and telomeric to the peaks. The imputation also included areas not covered by the final long-range PCR/pooled NGS results. Over 4000 SNPs with a quality score >0.25 (r2) were separately imputed using both the UCSF and Mayo Clinic cases with gliomas of oligodendroglial lineage.
Stage 1B: Pooled NGS of 8q24 region - Statistical Analysis
Two statistical methods were used to analyze the deCODE NGS data. First, deCODE provided association results based on a likelihood-ratio test comparing the MAF of a variant in the case and control pools (22). Second, the deCODE data were locally reanalyzed. Allele frequencies were calculated based on read counts; separately for reads generated from each case and control pool. For quality control, variants were removed that had fewer than 1000 read counts (poorly covered regions generate inaccurate allele frequency estimates). Variants whose allele frequency estimates differed more than 10% between the two case pools and between the two control pools were also removed. Based on the derived allele frequencies, the number of chromosomes in the original pools carrying various alleles was estimated. Association tests were conducted using Fisher’s exact test for the estimated number of chromosomes carrying the minor alleles in the corresponding case and control pools.
SNP Selection for Validation Genotyping
Results from both the observed and imputed prior 8q24 tag SNP panel (2) and the pooled NGS data were used to identify SNPs for validation genotyping. We chose SNPs that were significantly associated with glioma across 4 analyses: (i) imputation of Mayo data within the prior 8q24 tag SNP panel (2), (ii) imputation of UCSF data within the prior 8q24 tag SNP panel (2), (iii) deCODE analysis of NGS data, and (iv) UCSF/Mayo reanalysis of NGS data. Significance was defined as p<0.05. Candidates for validation included those meeting any of the following 5 criteria:
SNPs significant in all 4 analyses.
SNPs significant in 3 of 4 analyses.
SNPs significant in 2 of 4 analyses, where 1 was from imputation and 1 was from the NGS data.
NGS SNPs with case or control frequencies <10% that were significant in both NGS analyses only; this criteria aimed to identify rare variants that were not genotyped by the prior 8q24 tag SNP panel (2).
SNPs significant in both of the imputation analyses only; this criteria aimed to identify SNPs that failed NGS.
There were a total of 129 SNPs in the 8q24.21 region that met 1 of these 5 criteria (see Supplementary Figure 5). Of these SNPs, 104 were on the prior tag array (2), were in high LD with the previously reported SNPs or had high MAFs (>0.10). We selected the remaining 25 SNPs, 46 8q24 associated literature and tag SNPs, as well as 155 additional SNPs that were highly significant in a single imputation or NGS analysis. A total of 226 SNPs were selected for further custom SNP array design and analysis. Of these, 182 SNPs passed the design phase (44 SNPs had low design scores or were adjacent to other SNPs).
Stage 2: 8q24.21 region Validation Genotyping Panel - Quality Control and Statistical Analysis
Samples with call rates <0.9 and <0.975 in the Mayo (n=12) and UCSF (n=3) series, respectively, were excluded from analysis. Subsequently, SNPs with call rates <0.9 and <0.975 in the Mayo (n=31) and UCSF (n=31) series, respectively, were excluded from analysis (16 in common between sites). Because a custom chip containing infrequent variants was used, no MAF exclusions were made. SNPs with HWE p-values <0.001 in control subjects were excluded (4 Mayo and 4 UCSF; 0 in common). Identity-by-descent was evaluated (33) and 2 individuals appeared related between Mayo and UCSF; these 2 individuals were removed from the UCSF series for all pooled analyses. Informative custom Illumina genotyping was successful at Mayo Clinic and/or UCSF for 157 SNPs (Supplementary Table 2).
An additive logistic regression model for 0, 1, or 2 copies of the minor allele was used to investigate the association of glioma risk for each SNP. The Mayo and UCSF series were first analyzed separately and subsequently a pooled analysis was performed. Analyses were performed for all cases versus controls and subsequently cases were stratified by tumor histology, IDH mutation status, and 1p/19q deletion status. All analyses were adjusted for both age and gender; pooled analyses were also adjusted for institution.
For the 7 low frequency SNPs identified, dominant and genotypic models were also used to test association and no important differences from the additive model were found (data not shown). There were too few risk homozygotes to test a recessive model. To determine if the most significant SNP rs55705857 explained the association of the other 6 SNPs and the two Shete et al. (1) SNPs, logistic models were fit individually conditioning on rs55705857 and adjusted for age, gender and study site. Last, there were subjects who were genotyped in both Stage 1 and Stage 2. For these subjects, the rs55705857 association was validated across both Stage 1 and Stage 2; the association was further replicated using independent subjects in Stage 2 (data not shown).
Imputation of the validation genotyping panel was performed separately for Mayo and UCSF data using Beagle (34) with 1000 Genomes as reference population.
miRNA Modeling
miRNA modeling was performed using Sfold (35).
Supplementary Material
Acknowledgments
Work at the Mayo Clinic was supported by the National Institutes of Health (grant numbers P50CA108961 and P30 CA15083), National Institute of Neurological Disorders and Stroke (grant number RC1NS068222Z), the Bernie and Edith Waterman Foundation and the Ting Tsung and Wei Fong Chao Family Foundation. Work at University of California, San Francisco was supported by the National Institutes of Health (grant numbers R01CA52689, P50CA097257, R25CA126831 and CA112355), as well as the National Brain Tumor Foundation, the UCSF Lewis Chair in Brain Tumor Research and by donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen. The authors wish to acknowledge study participants, the clinicians and research staff at the participating medical centers, deCODE Genetics, the late Dr. Bernd Scheithauer, the Mayo Clinic Comprehensive Cancer Center Biospecimens and Processing and Genotyping Shared Resources and the UCSF Diller Cancer Center Genomics Core.
Abbreviations
- OR
odds ratios
- SNP
single nucleotide polymorphism
- MAF
minor allele frequency
Footnotes
URLs
NHGRI catalogue: http://www.genome.gov/gwastudies/
Competing Financial Interests
The authors declare no competing financial interests.
Author Contributions
R.B.J. and D.H.L. led the study at Mayo Clinic and M.R.W. and J.K.W. led the study at UCSF. R.B.J., M.R.W., D.H.L., J.K.W., J.E.E.-P., Y.X., H.S., T.M.K., T.R., H.M.H. and P.B. contributed to manuscript preparation. Study coordination was the responsibility of S.F. and T.M.K. at Mayo Clinic and T.R. and L.S.M. at UCSF. Y.X., J.E.E.-P., S.H., K.M.W. and B.L.F. co-directed and conducted biostatistics and bioinformatic analyses with additional support from P.A.D., M.K., I.S. and A.R.P. Laboratory work was performed by T.M.K., A.L.R., C.H., A.A.C. and S.R.F. under the direction of R.B.J. at Mayo Clinic and by H.M.H., S.Z., J.H.P. and G.H. under the direction of J.K.W., J.L.W. and M.R.W. at UCSF. Pathology support was provided by C.G. and T.T. Subject enrollment and clinical record review was performed or facilitated by M.D.P., S.M.C., M.S.B.,J.C.B., B.P.O., and D.H.L.
References
- 1.Shete S, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenkins RB, et al. Distinct germ line polymorphisms underlie glioma morphologic heterogeneity. Cancer Genet. 2011;204:13–18. doi: 10.1016/j.cancergencyto.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanson M, et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum Mol Genet. 2011;20:2897–2904. doi: 10.1093/hmg/ddr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rice T, et al. Associations of glioma risk loci by IDH1 mutation status. NeuroOncology. 2011;13(Suppl 3):iii27. [Google Scholar]
- 5.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen BC, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst. 2011;103:143–153. doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houillier C, et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology. 2010;75:1560–1566. doi: 10.1212/WNL.0b013e3181f96282. [DOI] [PubMed] [Google Scholar]
- 8.van den Bent MJ, et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors, a report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin Cancer Res. 2010;16:1597–1604. doi: 10.1158/1078-0432.CCR-09-2902. [DOI] [PubMed] [Google Scholar]
- 9.Hartmann C, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age, a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 10.Cairncross G, et al. Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma, Intergroup Radiation Therapy Oncology Group Trial 9402. J Clin Oncol. 2006;24:2707–2714. doi: 10.1200/JCO.2005.04.3414. [DOI] [PubMed] [Google Scholar]
- 11.Idbaih A, et al. Genomic aberrations associated with outcome in anaplastic oligodendroglial tumors treated within the EORTC phase III trial 26951. J Neurooncol. 2011;103:221–230. doi: 10.1007/s11060-010-0380-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teerlink C, et al. A unique genome-wide association analysis in extended Utah high-risk pedigrees identifies a novel melanoma risk variant on chromosome arm 10q. Hum Genet. 2012;131:77–85. doi: 10.1007/s00439-011-1048-z. [DOI] [PubMed] [Google Scholar]
- 13.Sato Y, et al. Genome-wide association study on overall survival of advanced non-small cell lung cancer patients treated with carboplatin and paclitaxel. J Thorac Oncol. 2011;6:132–138. doi: 10.1097/JTO.0b013e318200f415. [DOI] [PubMed] [Google Scholar]
- 14.Low SK, et al. Genome-wide association study of pancreatic cancer in Japanese population. PLoS One. 2010;5:e11824. doi: 10.1371/journal.pone.0011824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Treviño LR, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mu W, Zhang W. Bioinformatic resources of microRNA sequences, gene targets, and genetic variation. Frontiers Genet. 2012;3:1–8. doi: 10.3389/fgene.2012.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens KN, et al. 19p13.1 is a triple negative-specific breast cancer susceptibility locus. Cancer Res. 2012 doi: 10.1158/0008-5472.CAN-11-3364. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Truong T, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. J Natl Cancer Inst. 2010;102:959–971. doi: 10.1093/jnci/djq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goode EL, et al. A genome-wide association study identifies susceptibility loci fro ovarian cancer at 2q31 and 8q24. Nature Genet. 2010;42:874–881. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gayther SA, Pharoah PD. The inherited genetics of ovarian and endometrial cancer. Curr Opin Genet Dev. 2010;20:231–238. doi: 10.1016/j.gde.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al Olama AA, et al. Multiple loci on 8q24 assocaited with prostater cacner susceptibility. Nature Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 22.Gudmundsson J, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nature Genet. 2009;41:1122–1128. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghoussaini M, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K, et al. Interpretation of Association Signals and Identification of Causal Variants from Genome-wide Association Studies. Am J Hum Genet. 2010;86:730–742. doi: 10.1016/j.ajhg.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noushmehr H, et al. Cancer Genome Atlas Research Network. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wrensch M, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41:905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Felini MJ, et al. Reproductive factors and hormone use and risk of adult gliomas. Cancer Causes Control. 2009;20:87–96. doi: 10.1007/s10552-008-9220-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wrensch M, et al. Nonsynonymous coding single-nucleotide polymorphisms spanning the genome in relation to glioblastoma survival and age at diagnosis. Clin Cancer Res. 2007;13:197–205. doi: 10.1158/1078-0432.CCR-06-1199. [DOI] [PubMed] [Google Scholar]
- 31.Smith JS, et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J of Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–23. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.