Summary
DNA methylation has long been considered a very stable DNA modification in mammals that could only be removed by replication in the absence of re-methylation, i.e. by mere dilution of this epigenetic mark (so-called passive DNA demethylation). However, in recent years, a significant number of studies have revealed the existence of active processes of DNA demethylation in mammals, with important roles in development and transcriptional regulation, allowing the molecular mechanisms of active DNA demethylation to be unraveled. Here we review the recent literature highlighting the prominent role played in active DNA demethylation by base excision repair and especially by Thymine DNA Glycosylase.
Keywords: DNA methylation, DNA demethylation, histone modification, base excision repair, Thymine DNA Glycosylase, embryonic lethality, 5-hydroxymethylcytosine, 5-hydroxymethyluracil, 5-formylcytosine, 5-carboxylcytosine
DNA methylation and demethylation
The transformation of cytosine to 5-methylcytosine (5mC) is used by mammals as an epigenetic modification to regulate transcription [1-3]. Maintenance of proper methylation patterns is crucial for development, and alterations can result in embryonic lethality [4, 5]. Incorrect methylation patterns have been implicated in cancer, where overall hypomethylation of the genome and hypermethylation and silencing of tumor suppressor genes have been observed [6, 7]. DNA methylation is a well-studied process, and as a result, much is known about the molecular mechanisms: de novo DNA methyltransferases DMNT3a and DMNT3b are responsible for establishing methylation patterns, and maintenance DNA methyltransferase DMNT1 that targets newly replicated, transiently hemimethylated DNA, maintains methylation patterns.
In stark contrast, little is known about the DNA demethylation mechanisms or the enzymes responsible. Previously, cytosine methylation was thought to be a very stable modification, and therefore demethylation could only occur in a passive fashion, i.e. by progressive dilution when replication of methylated DNA is not followed by remethylation by DNMT1. Although passive demethylation does take place, over the years several instances of active demethylation in mammals have been described, all involving processes that occur in the absence of DNA replication and presumably via direct enzymatic actions. Such well-documented examples include demethylation of the paternal genome following fertilization [8, 9], removal of the imprinting marks in primordial germ cells [8-11], and demethylation during embryonic development and somatic differentiation in order to initiate tissue-specific gene expression [12, 13]. A related, more recently described phenomenon is the DNA demethylation occurring in the dentate gyrus of the hippocampus of adult mice as a consequence of neuronal activity, implying a role in memory and learning [14, 15].
Interestingly, several non-mammalian models have demonstrated the involvement of DNA repair mechanisms in the process of active demethylation. The nucleotide excision repair enzymes XPG and XPB are required for demethylation in Xenopus, where the demethylation process is initiated by Gadd45a, a genome stability and stress sensor protein [16, 17]. Similarly, Gadd45a promotes demethylation in zebrafish embryos, a process that occurs in two steps: activation-induced deaminase (AID) or apolipoprotein B RNA-editing catalytic component 2b and 2a (Apobec2b/2a) deaminate 5-methylcytosine to thymine which is in turn removed by base excision repair, specifically by the zebrafish thymine glycosylase MBD4 [18, 19]. Finally, in the flowering plant Arabidopsis, direct removal of 5-methylcytosine has been ascertained, and base excision repair 5-methylcytosine glycosylases, such as DEMETER (DME) and REPRESSOR OF SILENCING 1 (ROS1), play a key role in regulation of imprinting, and prevention of transcriptional gene silencing of endogenous genes, transgenes and transposons, respectively [20-24]
Thymine DNA Glycosylase and base excision repair
Thymine DNA Glycosylase (TDG), like the class of enzymes from which it derives the name, catalyzes the first step in the base excision repair pathway, i.e. removal of the mismatched/damaged base, thus leaving an abasic, apurinic/apyriminic (AP) site, which is then cleaved by AP endonuclease, for subsequent incorporation of the correct nucleotide by DNA polymerases. Specifically, TDG removes thymine or uracil from G:T or G:U mismatches [25-28]. Since TDG and the sequence-unrelated but biochemically related glycosylase MBD4/MED1 have a preference for mismatches located in the context of CpG sites, the presumption is that these two enzymes protect CpG sites from the potentially mutagenic consequences of spontaneous deamination of cytosine and 5-methylcytosine to uracil and thymine, respectively [18, 29-32]. In fact, removal of the latter two bases prior to the next round of DNA replication prevents misincorporation of adenine opposite thymine or uracil, with the consequent fixation of a transition mutation (CpG to CpA or CpG to TpG).
In addition to this presumed antimutagenic role, TDG has also been implicated in transcriptional regulation, as it was found to interact with a host of transcription factors, including retinoic acid receptor (RAR), retinoid X receptor (RXR) [33], estrogen receptor (ERα) [34], c-jun [35] and thyroid transcription factor 1 (TTF1) [36]. Furthermore, TDG interacts with and is acetylated by the histone acetyl-transferases (HAT) p300 and the CREB-binding protein (CBP) [37], and binds to the transcriptional activator p160 [38]. In vitro transactivation assays of reporter constructs following TDG overexpression, suggested a role of TDG in modulating the activity of nuclear hormone receptors (ERα, retinoic acid and retinoic X receptors), as well as of transcriptional activators (CBP and p160 CHECK) [33, 34, 36, 37].
Biochemical studies had initially suggested an involvement of both chicken and human TDG in DNA demethylation associated with transcriptional activation, and this effect was ascribed to a modest glycosylase activity of TDG on 5-methylcytosine [39-41]. This mechanism would be similar to the direct removal of 5-methylcytosine in Arabidopsis by the 5-methylcytosine glycosylases DME and ROS1 [20-23], mentioned above. The modest glycosylase activity of TDG on 5-methylcytosine has not been confirmed and likely was due to impurities in the substrate DNA.
More recent evidence suggested a role of TDG in active promoter demethylation. In fact, dynamic cycles of DNA methylation and demethylation at some promoters have been described in breast cancer lines during ERα activation, and TDG loading on these promoters was synchronous with the demethylation phase [42, 43]. TDG is known to interact with the de novo DNA methyltransferases DNMT3a [44] and DNMT3b [45], and it was proposed that TDG could mediate demethylation by acting on G:T mismatches created by the controlled deaminase activity of DNMT3a and DNMT3b [43].
Embryonic lethality associated with TDG inactivation in the murine germ line
A better understanding of the role of TDG in DNA demethylation, epigenetic regulation and mammalian development could only be obtained when mice with targeted inactivation of TDG in the germ line were derived and characterized by three research groups, including the groups of Primo Schär, Yoshihiko Uehara and ours [46-48].
During the process of creating a knockout mouse for TDG, we observed no live births of Tdg-/- mice. Timed matings between Tdg+/- mice revealed that Tdg null embryos were developmentally arrested at embryonic day (E) 11.5 with a complex phenotype. Macroscopic abnormalities included liver and pericardial hemorrhage, hypoplastic branchial arches, delayed limb development, prominent telencephalic vesicles and diffuse hemorrhagic lesions, while microscopic investigation showed specific patterning defects of the heart, stenosis of the dorsal aorta and abnormal vascular branching of the internal carotids and coronaries [47]. The null allele we generated bears a deletion of TDG exons 3-7. Embryonic lethality of Tdg-null embryos was also described by Cortazar et al. who deleted exons 6 and 7 [46], and by Saito and coworkers who used a targeting vector that replaced parts of exons 8 and 9, corresponding to a portion of the domain required for glycosylase activity, as well as in vitro interactions with transcription factors RARα and RXRα [47].
We noted that some specific phenotypic features of the Tdg-null embryos, including the cardiovascular defects, resemble developmental defects previously noted in embryos null for the histone acetyltransferases p300 and CBP, and for factors of the retinoic acid signaling pathway, such as RAR, RXR, and Raldh2 [49-52]. This observation was substantiated by molecular analyses demonstrating a role of TDG in RAR/RXR- and p300-dependent transcription (vide infra). Thus, the lethality phenotype is most likely due, at least in part, to the absence of this transcription-related function of TDG required for proper embryonic development.
On the other hand, Saito and colleagues commented on the similarity of the embryonic lethality with that of GATA3 deficient mice. Embryonic lethality of the GATA3 mutant mice is reported to be caused by lack of dopamine and noradrenaline [53] catecholamines confirmed to be required for normal development [54-56]. In order to explore this similarity further, Saito and colleagues detected reduced levels of dopamine and, especially, noradrenaline in Tdg-null embryos. They then measured mRNA levels of the catecholamine biosynthetic enzymes, tyrosine hydroxylase (TH, that converts L-tyrosine to L-DOPA), L-aromatic amino acid decarboxylase (AADC, that decarboxylates L-DOPA to dopamine) and dopamine beta-hydroxylase (DBH, that converts dopamine to noradrenaline). They reported a significant depletion of DBH mRNA in Tdg-null embryos [48]. It has been shown that GATA3-mutant embryos with noradrenaline deficiency can be rescued by feeding dams with precursors to noradrenaline, such as D,L threo-3,4-dihydroxyphenylserine (DOPS) that is directly converted to noradrenaline by AADC [53]. Saito and colleagues found that DOPS fed to pregnant Tdg heterozygous females was able to partially rescue Tdg-null embryos, prolonging their survival up to 14.5dpc [48]. The partial rescue by DOPS suggests that the lethality of the TDG mutant embryos is due only in part to reduced noradrenaline levels.
Role of TDG in transcription and chromatin regulation
In order to conduct functional investigations in a cell culture system, we and others established mouse embryonic fibroblast (MEF) lines with different Tdg genotypes. We detected significant changes in the expression profile of Tdg-null and control MEFs, with retinoic acid signaling being the main pathway compromised. Mechanistically, we found that transactivation of p300 and RAR/RXR reporters was reduced in Tdg-null MEFs, confirming the proposed role of TDG as a transcriptional coactivator for nuclear hormone receptors previously demonstrated only in TDG overexpression systems [33, 34, 36, 37]. We also found that TDG is required for the formation of complexes of RAR/RXR with p300 both off and on the DNA, the latter phenomenon consistent with a direct role in the transcriptional regulation of the Crabp2 and Rbp1 genes. Since, in chromatin immunoprecipitation (ChIP) experiments, RAR/RXR were bound equally well to these two promoters in both Tdg-null and wild type MEFs, TDG appears to be required at a later step that triggers the recruitment of p300, presumably via the direct interactions of TDG with both the nuclear hormone receptors and the histone acetyltransferases [33, 34, 36, 37], and the consequent histone H3 acetylation. This suggests an important general function of TDG in retinoic acid-dependent transcription [47].
Cortazar and coworkers also used ChIP to conduct a detailed characterization of the association of TDG with promoters of differentially expressed genes Hoxa10, Hoxd13, Sfrp2, Twist2 and Rarb, which were found to be downregulated in MEFs deficient for TDG. They found that in comparison to random intergenic sequences and silent promoters of Oct4 and Tuba3, the promoters for these genes were enriched in TDG precipitates, indicating that TDG may be targeted to certain promoters to prevent silencing. The data also revealed promoter-specific patterns of the loss of activating (H3K4me2) and the increase of repressive histone marks H3K27me3 and H3K9me3. Further evidence of these specific patterns was illustrated when Sfrp2 and Twist2 genes had restored activity by the expression of a TDG cDNA, but Hoxa10 and Hoxd13 did not. Therefore, in the case of the latter two, the loss of H3K4 methylation paired with H3K9 and H3K27 methylation and aberrant CpG methylation (vide infra) maintains chromatin in a repressive state even in the presence of re-expressed Tdg [46].
Again using ChIP, Cortazar and colleagues demonstrated that the promoters of differentially expressed genes in embryonic stem (ES) cells undergoing differentiation into neuronal progenitor cells were enriched with TDG. They found that TDG associates with promoters of Oct4 and Nanog in ES cells, but not in neuronal progenitor cells and MEFs, presumably losing the interaction during heterochromatinization. They hypothesize that an inability to associate with heterochromatized promoters could explain why Hoxa10 and Hoxd13 activity could not be rescued by re-expression of TDG in Tdg-null MEFs. Upon further experimentation, the authors discovered that loss of TDG interrupts the association of H3K4-specific methyltransferase MLL1 with the promoters of Hoxa10, Hoxd13, Sfrp2 and Twist2 [46]. In addition, the binding to these promoters by CBP/p300 was decreased in TDG-deficient MEFs, consistent with the data mentioned above of the lack of p300 recruitment onto the Crabp2 and Rbp1 promoters in the absence of TDG. CBP/p300 has also been shown to protect gene promoters from polycomb-mediated H3K27 trimethylation. Thus, TDG contributes to the maintenance of active and bivalent chromatin during differentiation [46]. All of these data indicate that TDG acts functionally as an important transcriptional co-activator, forming complexes with activating histone modifiers MLL and CBP/p300 in order to maintain chromatin in an active state during differentiation.
In keeping with a primarily “transcriptional” rather than “anti-mutagenic” defect in TDG-mutant embryos, the three research groups failed to detect the expected increase in CpG site mutation frequency in Tdg-null cells and embryos. Indeed, a decrease in G:T repair efficiency was noted in extracts of Tdg-null embryos, MEFs and ES cells. Although lack of G:T mismatch repair activity should lead to an increase in mutation frequency, the latter was unchanged in mutant embryos at 10.5dpc or MEFs [46-48]. It is presently unclear whether the methods used in these studies are sensitive enough to conclude in a definitive manner that TDG does not have any anti-mutagenic function at CpG sites. One possibility is that MBD4 provides a redundant genome-stability function, efficiently processing G:T mismatches in the absence of TDG. Yet another possibility is that initial enzymatic steps in demethylation (e.g. deamination, see below) do not happen in the absence of TDG, and therefore that TDG may have a role in the initiation of the demethylation process [47].
Role of TDG in protecting CpG islands from hypermethylation and in mediating DNA demethylation
Given the possible role of TDG in demethylation [41-43], we and others examined the DNA methylation patterns of promoters of select genes differentially expressed for wild-type and Tdg null MEFs. By using sodium bisulfite sequencing, we found that the CpG island less than 2 kb upstream of the transcriptional start site of Efs, Crabp2, Hoxa5 and H19 (all down regulated in Tdg null MEFs) was hypermethylated in Tdg-null cells [47]. By using even more detailed sodium bisulfite pyrosequencing, Cortazar and colleagues detected increased methylation levels of the CpG island of the Hoxa10, Hoxd13, Sfrp2, Twist2 and Rarb genes [46]. These results demonstrate that sequences normally kept unmethylated become hypermethylated in the absence of TDG, indicating aberrant, unscheduled de novo methylation. Further experimentation using the imprinted gene Igf2 in wild-type and Tdg mutant primordial germ cells confirmed that the previous observations were not due to in vitro culture stress of the MEFs, and that TDG does in fact serve to prevent hypermethylation during development [47].
Our group also sought to determine if TDG is responsible for DNA demethylation; to this end, we utilized a heterologous in vitro-methylated Oct4 pluripotency gene for transcriptional reactivation in embryonic carcinoma P19 cells, as well as P19 cells expressing a shRNA targeting TDG. An Oct4 promoter∷EGFP reporter assay showed a lack of EGFP expression reactivation in the Tdg knock down cells, and bisulfite sequencing confirmed that demethylation of the Oct4 promoter is reduced, demonstrating the direct involvement of TDG in demethylation. Due to demethylation in the parental P19 cells occurring within 12 hours and lack of origin of replication in the reporter plasmid, we concluded that TDG is playing a role in active demethylation [47].
Furthermore, sodium bisulfite sequencing revealed that demethylation of the enhancer of two liver-specific genes, Alb encoding albumin and Tat encoding tyrosine amino transferase, is impaired in Tdg-null embryos [47].
It should be emphasized that presently it is not clear whether the hypemethylation at CpG island-containing promoters, detected in the absence of TDG, is caused by a deficiency of TDG in promoting an active, antagonizing demethylation, similar to its proposed role in active demethylation at enhancers.
As these results suggested that TDG plays an enzymatic role in demethylation, we next determined that the catalytic functionality of TDG is responsible for its role in demethylation. We predicted that inactivating its glycosylase active site would cause the embryonic lethality observed in the Tdg null mice. This was accomplished using a knock-in mouse strain which expressed a point mutation (N151A)[47], abolishing the glycosylase function of TDG [57]. There were no live births of TdgN151A/N151A mice, and further analysis showed that embryonic lethality occurred one day earlier than the Tdg null embryos, at E10.5 with general developmental delay and other abnormalities. Additionally, the Tat enhancer remained methylated. These results corroborate that of the Oct4 promoter∷EGFP reporter assay, and show that the catalytic function of TDG is crucial for development and DNA demethylation [47]. From these observations, it is clear that TDG plays a dual epigenetic role in controlling both DNA methylation and chromatin modifications (Fig. 1).
Figure 1.
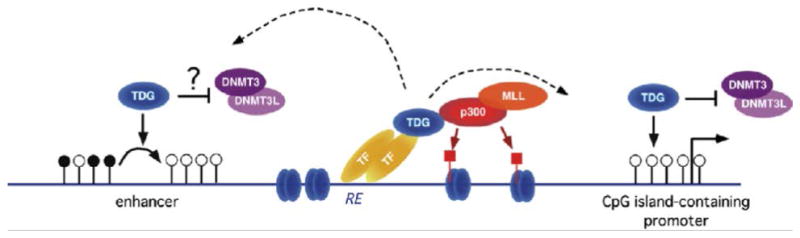
Schematic of the dual epigenetic role of TDG in regulation of DNA methylation patterns and chromatin modification. Depicted are an enhancer (left) and a CpG island-containing promoter (right). In this model, binding to transcription factors (TF) at responsive elements (RE) tethers TDG to DNA; in turn, TDG promotes recruitment of MLL and p300, with consequent production of p300-acetylated nucleosomes (marked by red-squared lollipops). The recruited TDG mediates DNA demethylation of CpG sites, indicated by the white lollipops, at enhancers (left). The recruited TDG also helps maintaining CpG islands at promoters in their unmethylated state (right). Both functions may be aided by TDG inhibition of de novo DNA methyltransferases (DNMTs) (modified from [47], with permission from Elsevier).
Mechanisms of TDG-mediated DNA demethylation
The mechanisms by which TDG mediates DNA demethylation are not fully characterized and it is possible that multiple pathways are involved (Fig. 2). We wanted to determine if TDG mediates DNA demethylation in a manner similar to that in zebrafish embryos, where demethylation is initiated by deamination of 5mC to thymine by AID, Apobec2a or Apobec2b and then the excision of thymine by the MBD4 glycosylase, a process mediated by GADD45 [19]. We found by Co-IP experiments using the same P19 cell lines described above, that TDG forms a complex with AID and GADD45a, and also that AID interacts with GADD45a, independent of TDG. We also observed that there was a decrease in AID expression in the TDG-downregulated cells, so it is possible that this in vivo interaction has functional consequences for AID and perhaps TDG regulates levels of AID or its stability [47].
Figure 2.
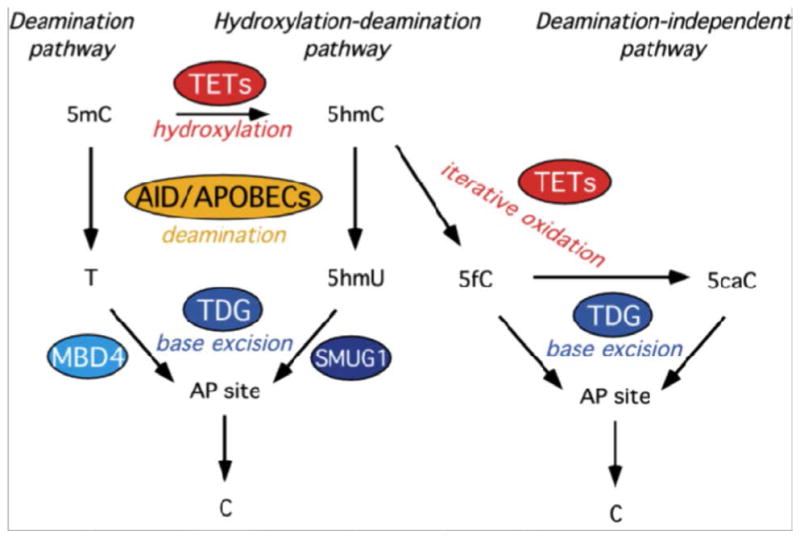
Schematic of the central role of TDG in DNA demethylation pathways: the deamination (left), hydroxylation-deamination (center) and deamination-independent (right) pathways are shown; 5mC: 5-methylcytosine; 5hmC: 5-hydroxymethylcytosine; T: thymine; 5hmU: 5-hydroxymethyluracil; 5fC: 5-formylcytosine; 5caC: 5-carboxylcytosine; AP site: apurinic/apyrimidinic site; C: cytosine (modified from [47], with permission from Elsevier).
A related demethylation pathway in which TDG and AID/APOBECs might be involved concerns the newly identified DNA base, 5-hydroxymethylcytosine (5hmC), an oxidation product of 5-methylcytosine, generated by TET oxidases (TET1-3) [58, 59]. TET1 was identified as the gene involved in the t(10;11) (ten eleven translocation) in acute myelogenous leukemia [60, 61]; TET2 is frequently mutated in myelodysplastic syndrome [62]. TET proteins are involved in the active DNA demethylation occurring in the adult brain, a process that is also dependent on the AID/APOBEC deaminases [14]. Thus, it is possible that 5hmC is deaminated to 5-hydroxymethyluracil (5hmU), generating a G:5hmU mismatch [14]. TDG has a strong glycosylase activity on 5hmU [47, 63, 64]. In fact, while two other base excision repair glycosylases, MBD4 and SMUG1 are efficient in removal of the mismatched T and 5hmU, respectively, TDG appears to be the only glycosylase with strong activity on both deaminated bases [47].
More recently, work in other laboratories identified a third, deaminase-independent demethylation pathway, in which 5hmC is sequentially oxidized to 5-formylcytosine (fC) and 5-carboxylcytosine (caC) by TET oxidases [65-67]. While it is possible that a putative decarboxylase exists that directly converts caC into cytosine, it was shown that TDG exhibits a specific glycosylase activity on fC and caC [65, 68]. In fact, crystallographic evidence indicates that caC is secured in the TDG active site by polar interactions involved in the recognition of the 5-carboxyl moiety. Due to these exclusive structural properties, TDG is the first and only glycosylase able to selectively bind and excise 5caC and 5fC from duplex DNA [69]. Figure 2 describes the involvement of TDG in several DNA demethylation pathways.
While current studies indicate that TDG is not involved in direct excision of 5-methylcytosine, it should be noted that there is evidence that the protein kinase A- and protein kinase C-phosphorylated MBD4 does acquire glycosylase activity on 5-methylcytosine, mediating demethylation and transcriptional de-repression of the CYP27B1 gene [70]The role of MBD4 in DNA demethylation (discussed in [18]) brings about the possibility of functional redundancy of TDG and MBD4 in this process. However, it is likely that TDG and MBD4 have distinct roles in DNA demethylation, due to their marked difference in nuclear distribution: in fact TDG and MBD4 associate with euchromatin and heterochromatin, respectively [37]
TDG alterations in cancer
The important role of TDG in epigenetic stability suggests a potential involvement in cancer and indeed initial evidence on the role of TDG in tumorigenesis is accumulating. TDG shows frequently reduced mRNA expression in multiple myeloma [71] and pancreatic adenocarcinoma [72]. Loss of TDG expression in rectal cancer may synergize with deficiency of the mismatch repair protein PMS2, creating a supermutator phenotype at CpG sites [73]. Finally, TDG appears to be involved in the TGFβ-dependent active demethylation and expression of the tumor suppressor gene p15-INK4b [74].
Future Perspective
The field of active DNA demethylation underwent a significant acceleration in the past few years with the identification of the critical role played by base excision repair and the new modified cytosine bases. Thus, it is easy to predict that in the next 5-10 years, this area of research will continue to flourish. Future multidisciplinary studies will lead to a better characterization of the interaction of TDG with other transcription and DNA repair factors. Additional studies will determine the molecular mechanisms explaining the essential role of TDG in development and further define its importance in transcription by identifying the genes that more critically depend on TDG for activation, either as a transcriptional co-activator or as a factor involved in DNA demethylation. A mechanistic understanding of enzymatic demethylation could lead to strategies for targeted demethylation of genes. The relative importance of TDG in the various pathways of DNA demethylation outlined above will be defined, and this knowledge, paired with the use of animal systems to recreate physiopathological mechanisms and model human disease, will reveal the disease relevance of defective DNA demethylation.
Executive Summary
DNA methylation and demethylation
DNA methylation is used by vertebrates to repress transcription and is mediated by DNA methyltransferases
Accumulating evidence indicates that active mechanisms of DNA demethylation exist and are mediated by DNA repair systems
Thymine DNA Glycosylase and base excision repair
TDG is a base excision repair enzyme presumably involved in protecting CpG sites from transition mutations caused by deamination of cytosine and 5mC to uracil and thymine
TDG interacts with several transcription factors and co-activators
Past literature suggested a role of TDG in DNA demethylation
Embryonic lethality associated with TDG inactivation in the murine germ line
TDG is required for mammalian development
Phenotype of Tdg-null embryos suggests an impairment of RAR-RXR- and CBP-p300-dependent transcription and of catecholamine production
Role of TDG in transcription and chromatin regulation
TDG is required for efficient RAR-RXR- and p300-dependent transcription and for p300 recruitment
TDG is required for MLL recruitment
TDG contributes to the maintenance of active and bivalent chromatin
There is no reported increase in CpG site mutation frequency in Tdg-null cells and embryos
Role of TDG in protecting CpG islands from hypermethylation and in mediating DNA demethylation
TDG in involved in protection of CpG islands from de novo aberrant DNA methylation
TDG is involved in active DNA demethylation of enhancers and promoters
The catalytic activity of TDG is required for development and DNA demethylation
Mechanisms of TDG-mediated DNA demethylation
TDG forms a complex with the deaminase AID and the stress response protein GADD45a
TDG could mediate DNA demethylation by removing thymine and 5hmU originated by enzymatic deamination of 5mC and 5hmC, respectively
TDG could mediate DNA demethylation by removing 5fC and 5caC originated by sequential oxidation of 5hmC by Tet proteins
TDG alterations in cancer
Loss of TDG expression may be important for the genetic and epigenetic instability occurring in cancer
Bibliography
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 2.Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 3.Siegfried Z, Cedar H. DNA methylation: a molecular lock. Curr Biol. 1997;7:R305–R307. doi: 10.1016/s0960-9822(06)00144-8. [DOI] [PubMed] [Google Scholar]
- 4.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 5.Okano M, Bell DW, Haber DA, et al. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 8.Mayer W, Niveleau A, Walter J, et al. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 9.Oswald J, Engemann S, Lane N, et al. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 10.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 11.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 12.Kress C, Thomassin H, Grange T. Local DNA demethylation in vertebrates: how could it be performed and targeted? FEBS Lett. 2001;494:135–140. doi: 10.1016/s0014-5793(01)02328-6. [DOI] [PubMed] [Google Scholar]
- 13.Niehrs C. Active DNA demethylation and DNA repair. Differentiation. 2009;77:1–11. doi: 10.1016/j.diff.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Guo JU, Su Y, Zhong C, et al. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma DK, Jang MH, Guo JU, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barreto G, Schafer A, Marhold J, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 17.Niehrs C, Schafer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012 doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Buontempo S, Sannai M, Bellacosa A. MBD4/MED1 protein in DNA repair and demethylation, cancer, and other diseases. In: Giordano A, Macaluso M, editors. Cancer Epigenetics - Biomolecular Therapeutics in Human Cancer. Wiley VCH; 2012. in press. [Google Scholar]
- 19.Rai K, Huggins IJ, James SR, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi Y, Gehring M, Johnson L, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110:33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 21.Gehring M, Huh JH, Hsieh TF, et al. DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell. 2006;124:495–506. doi: 10.1016/j.cell.2005.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morales-Ruiz T, Ortega-Galisteo AP, Ponferrada-Marin MI, et al. DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc Natl Acad Sci U S A. 2006;103:6853–6858. doi: 10.1073/pnas.0601109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng X, Pontes O, Zhu J, et al. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu J, Kapoor A, Sridhar VV, et al. The DNA glycosylase/lyase ROS1 functions in pruning DNA methylation patterns in Arabidopsis. Curr Biol. 2007;17:54–59. doi: 10.1016/j.cub.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 25.Cortazar D, Kunz C, Saito Y, et al. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Neddermann P, Gallinari P, Lettieri T, et al. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J Biol Chem. 1996;271:12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 27.Neddermann P, Jiricny J. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J Biol Chem. 1993;268:21218–21224. [PubMed] [Google Scholar]
- 28.Neddermann P, Jiricny J. Efficient removal of uracil from G·U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc Natl Acad Sci U S A. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellacosa A. Role of MED1 (MBD4) Gene in DNA repair and human cancer. J Cell Physiol. 2001;187:137–144. doi: 10.1002/jcp.1064. [DOI] [PubMed] [Google Scholar]
- 30.Bellacosa A, Cicchillitti L, Schepis F, et al. MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc Natl Acad Sci U S A. 1999;96:3969–3974. doi: 10.1073/pnas.96.7.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendrich B, Hardeland U, Ng HH, et al. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 32.Petronzelli F, Riccio A, Markham GD, et al. Biphasic kinetics of the human DNA repair protein MED1 (MBD4), a mismatch-specific DNA N-glycosylase. J Biol Chem. 2000;275:32422–32429. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- 33.Um S, Harbers M, Benecke A, et al. Retinoic acid receptors interact physically and functionally with the T:G mismatch-specific thymine-DNA glycosylase. J Biol Chem. 1998;273:20728–20736. doi: 10.1074/jbc.273.33.20728. [DOI] [PubMed] [Google Scholar]
- 34.Chen D, Lucey MJ, Phoenix F, et al. T:G mismatch-specific thymine-DNA glycosylase potentiates transcription of estrogen-regulated genes through direct interaction with estrogen receptor alpha. J Biol Chem. 2003;278:38586–38592. doi: 10.1074/jbc.M304286200. [DOI] [PubMed] [Google Scholar]
- 35.Chevray PM, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Missero C, Pirro MT, Simeone S, et al. The DNA glycosylase T:G mismatch-specific thymine DNA glycosylase represses thyroid transcription factor-1-activated transcription. J Biol Chem. 2001;276:33569–33575. doi: 10.1074/jbc.M104963200. [DOI] [PubMed] [Google Scholar]
- 37.Tini M, Benecke A, Um SJ, et al. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 38.Lucey MJ, Chen D, Lopez-Garcia J, et al. T:G mismatch-specific thymine-DNA glycosylase (TDG) as a coregulator of transcription interacts with SRC1 family members through a novel tyrosine repeat motif. Nucleic Acids Res. 2005;33:6393–6404. doi: 10.1093/nar/gki940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jost JP, Siegmann M, Sun L, et al. Mechanisms of DNA demethylation in chicken embryos.Purification and properties of a 5-methylcytosine-DNA glycosylase. J Biol Chem. 1995;270:9734–9739. doi: 10.1074/jbc.270.17.9734. [DOI] [PubMed] [Google Scholar]
- 40.Zhu B, Benjamin D, Zheng Y, et al. Overexpression of 5-methylcytosine DNA glycosylase in human embryonic kidney cells EcR293 demethylates the promoter of a hormone-regulated reporter gene. Proc Natl Acad Sci U S A. 2001;98:5031–5036. doi: 10.1073/pnas.091097298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu B, Zheng Y, Hess D, et al. 5-methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc Natl Acad Sci USA. 2000;97:5135–5139. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kangaspeska S, Stride B, Metivier R, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 43.Metivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 44.Li YQ, Zhou PZ, Zheng XD, et al. Association of Dnmt3a and thymine DNA glycosylase links DNA methylation with base-excision repair. Nucleic Acids Res. 2007;35:390–400. doi: 10.1093/nar/gkl1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boland MJ, Christman JK. Characterization of Dnmt3b:thymine-DNA glycosylase interaction and stimulation of thymine glycosylase-mediated repair by DNA methyltransferase(s) and RNA. J Mol Biol. 2008;379:492–504. doi: 10.1016/j.jmb.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cortazar D, Kunz C, Selfridge J, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 47.Cortellino S, Xu J, Sannai M, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saito Y, Ono T, Takeda N, et al. Embryonic lethality in mice lacking mismatch-specific thymine DNA glycosylase is partially prevented by DOPS, a precursor of noradrenaline. Tohoku J Exp Med. 2012;226:75–83. doi: 10.1620/tjem.226.75. [DOI] [PubMed] [Google Scholar]
- 49.Mark M, Ghyselinck NB, Chambon P. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol. 2006;46:451–480. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka Y, Naruse I, Hongo T, et al. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech Dev. 2000;95:133–145. doi: 10.1016/s0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 51.Vermot J, Niederreither K, Garnier JM, et al. Decreased embryonic retinoic acid synthesis results in a DiGeorge syndrome phenotype in newborn mice. Proc Natl Acad Sci USA. 2003;100:1763–1768. doi: 10.1073/pnas.0437920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao TP, Oh SP, Fuchs M, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 53.Lim KC, Lakshmanan G, Crawford SE, et al. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat Genet. 2000;25:209–212. doi: 10.1038/76080. [DOI] [PubMed] [Google Scholar]
- 54.Thomas SA, Matsumoto AM, Palmiter RD. Noradrenaline is essential for mouse fetal development. Nature. 1995;374:643–646. doi: 10.1038/374643a0. [DOI] [PubMed] [Google Scholar]
- 55.Zhou QY, Quaife CJ, Palmiter RD. Targeted disruption of the tyrosine hydroxylase gene reveals that catecholamines are required for mouse fetal development. Nature. 1995;374:640–643. doi: 10.1038/374640a0. [DOI] [PubMed] [Google Scholar]
- 56.Kobayashi K, Morita S, Sawada H, et al. Targeted disruption of the tyrosine hydroxylase locus results in severe catecholamine depletion and perinatal lethality in mice. The Journal of biological chemistry. 1995;270:27235–27243. doi: 10.1074/jbc.270.45.27235. [DOI] [PubMed] [Google Scholar]
- 57.Hardeland U, Bentele M, Jiricny J, et al. Separating substrate recognition from base hydrolysis in human thymine DNA glycosylase by mutational analysis. The Journal of biological chemistry. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 58.Ito S, D'alessio AC, Taranova OV, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ono R, Taki T, Taketani T, et al. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23) Cancer Res. 2002;62:4075–4080. [PubMed] [Google Scholar]
- 61.Lorsbach RB, Moore J, Mathew S, et al. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 62.Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. Journal of clinical oncology:official journal of the American Society of Clinical Oncology. 2011;29:504–515. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bennett MT, Rodgers MT, Hebert AS, et al. Specificity of human thymine DNA glycosylase depends on N-glycosidic bond stability. J Am Chem Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hardeland U, Bentele M, Jiricny J, et al. The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res. 2003;31:2261–2271. doi: 10.1093/nar/gkg344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfaffeneder T, Hackner B, Truss M, et al. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angewandte Chemie. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- 68.Maiti A, Drohat AC. Thymine DNA Glycosylase Can Rapidly Excise 5-Formylcytosine and 5-Carboxylcytosine: POTENTIAL IMPLICATIONS FOR ACTIVE DEMETHYLATION OF CpG SITES. The Journal of biological chemistry. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang L, Lu X, Lu J, et al. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat Chem Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim MS, Kondo T, Takada I, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 71.Peng B, Hurt EM, Hodge DR, et al. DNA hypermethylation and partial gene silencing of human thymine- DNA glycosylase in multiple myeloma cell lines. Epigenetics. 2006;1:138–145. doi: 10.4161/epi.1.3.2938. [DOI] [PubMed] [Google Scholar]
- 72.Yatsuoka T, Furukawa T, Abe T, et al. Genomic analysis of the thymine-DNA glycosylase (TDG) gene on 12q22-q24.1 in human pancreatic ductal adenocarcinoma. Int J Pancreatol. 1999;25:97–102. doi: 10.1385/IJGC:25:2:97. [DOI] [PubMed] [Google Scholar]
- 73.Vasovcak P, Krepelova A, Menigatti M et al. Unique mutational profile associated with a loss of TDG expression in the rectal cancer of a patient with a constitutional PMS2 deficiency. DNA Repair. 2012 doi: 10.1016/j.dnarep.2012.04.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thillainadesan G, Chitilian JM, Isovic M et al. TGF-beta-Ddependent actives demethylation and expression of the p15(ink4b) tumor suppressor are impaired by the ZNF217/CoREST complex. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.03.027. in press. [DOI] [PubMed] [Google Scholar]


