Abstract
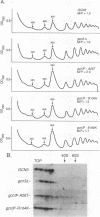
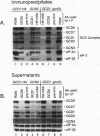
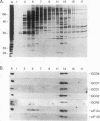
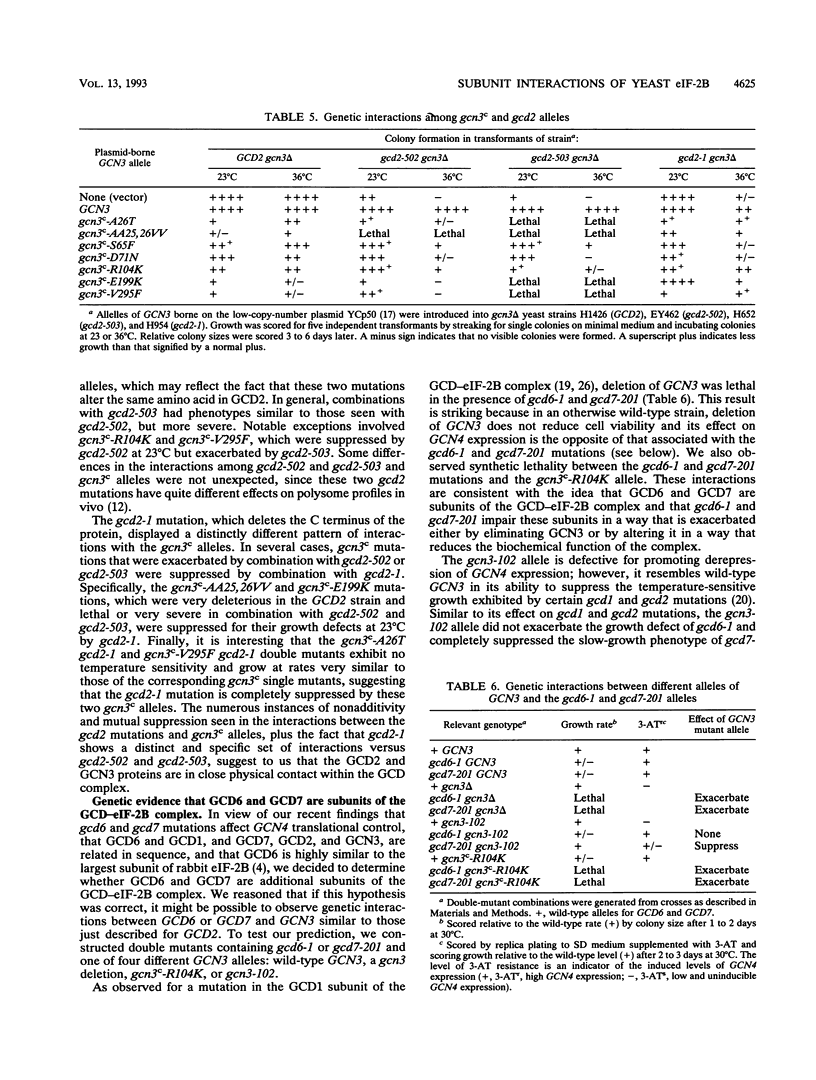
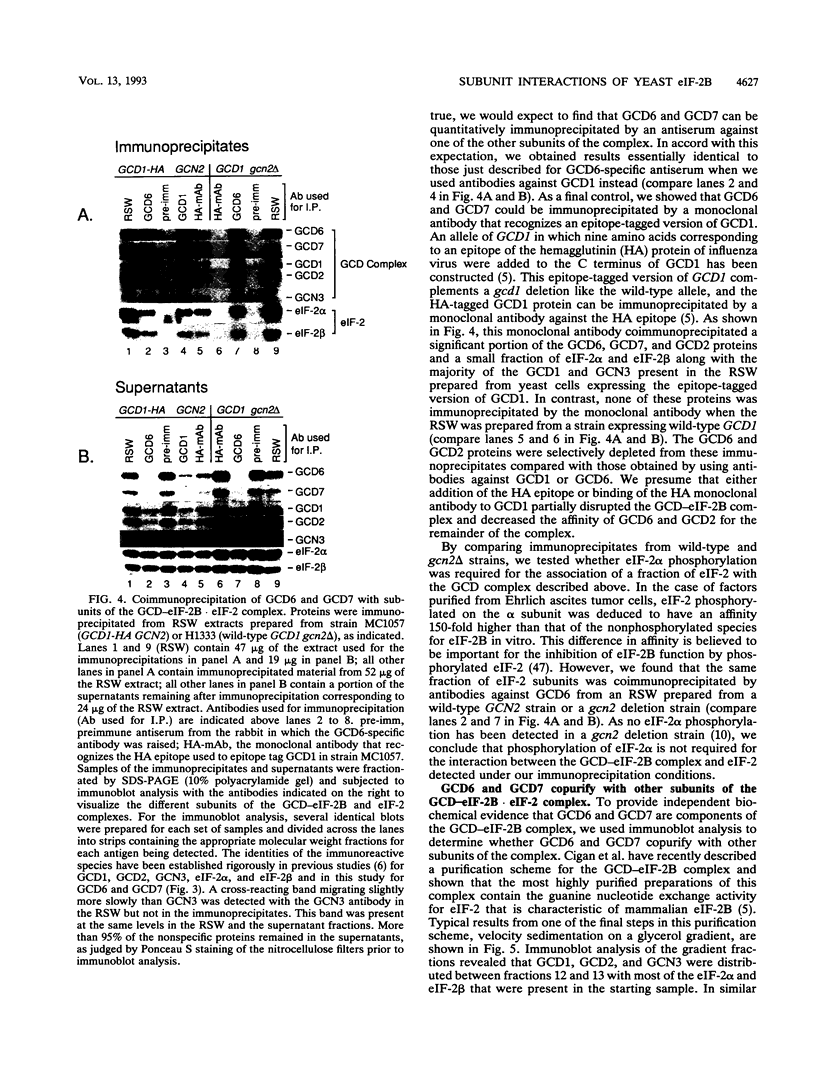
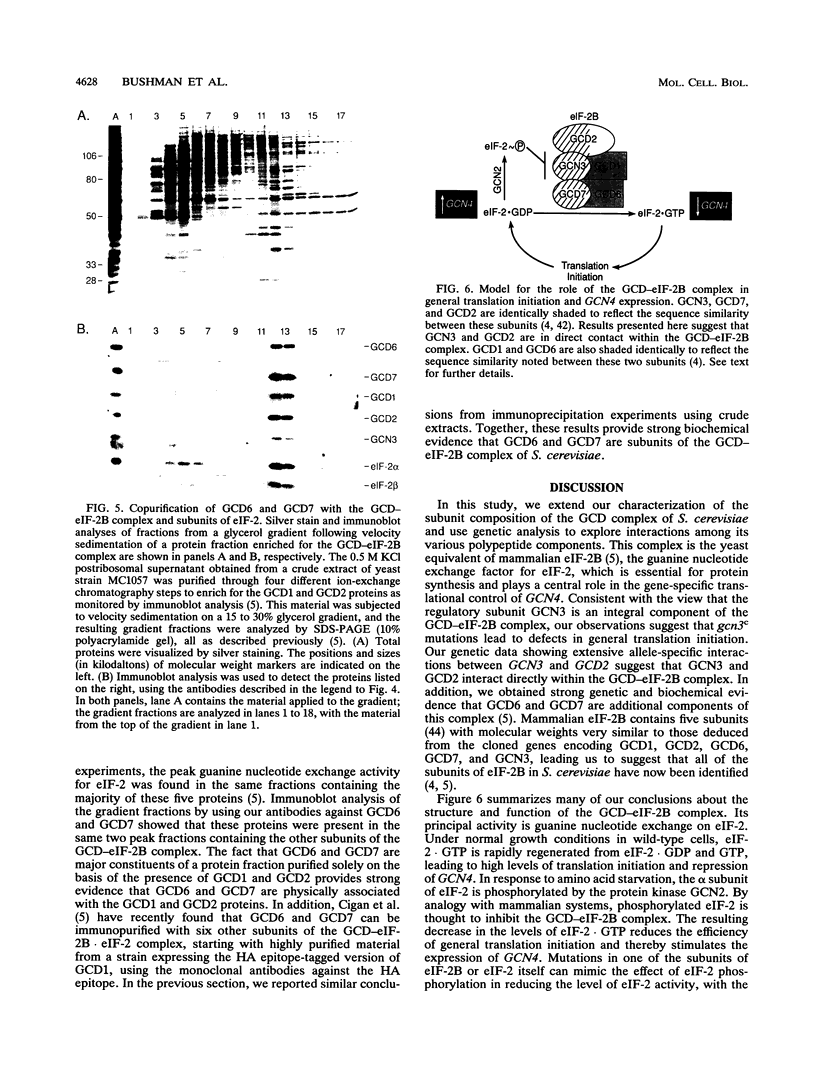
Phosphorylation of eukaryotic translation initiation factor 2 (eIF-2) in amino acid-starved cells of the yeast Saccharomyces cerevisiae reduces general protein synthesis but specifically stimulates translation of GCN4 mRNA. This regulatory mechanism is dependent on the nonessential GCN3 protein and multiple essential proteins encoded by GCD genes. Previous genetic and biochemical experiments led to the conclusion that GCD1, GCD2, and GCN3 are components of the GCD complex, recently shown to be the yeast equivalent of the mammalian guanine nucleotide exchange factor for eIF-2, known as eIF-2B. In this report, we identify new constituents of the GCD-eIF-2B complex and probe interactions between its different subunits. Biochemical evidence is presented that GCN3 is an integral component of the GCD-eIF-2B complex that, while dispensable, can be mutationally altered to have a substantial inhibitory effect on general translation initiation. The amino acid sequence changes for three gcd2 mutations have been determined, and we describe several examples of mutual suppression involving the gcd2 mutations and particular alleles of GCN3. These allele-specific interactions have led us to propose that GCN3 and GCD2 directly interact in the GCD-eIF-2B complex. Genetic evidence that GCD6 and GCD7 encode additional subunits of the GCD-eIF-2B complex was provided by the fact that reduced-function mutations in these genes are lethal in strains deleted for GCN3, the same interaction described previously for mutations in GCD1 and GCD2. Biochemical experiments showing that GCD6 and GCD7 copurify and coimmunoprecipitate with GCD1, GCD2, GCN3, and subunits of eIF-2 have confirmed that GCD6 and GCD7 are subunits of the GCD-eIF-2B complex. The fact that all five subunits of yeast eIF-2B were first identified as translational regulators of GCN4 strongly suggests that regulation of guanine nucleotide exchange on eIF-2 is a key control point for translation in yeast cells just as in mammalian cells.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol. 1991 Jan;11(1):486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A. E., Botstein D. Dominant suppressors of yeast actin mutations that are reciprocally suppressed. Genetics. 1989 Apr;121(4):675–683. doi: 10.1093/genetics/121.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Bushman J. L., Asuru A. I., Matts R. L., Hinnebusch A. G. Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Mar;13(3):1920–1932. doi: 10.1128/mcb.13.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Foiani M., Hannig E. M., Hinnebusch A. G. Complex formation by positive and negative translational regulators of GCN4. Mol Cell Biol. 1991 Jun;11(6):3217–3228. doi: 10.1128/mcb.11.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Feng L., Donahue T. F. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti A., Baglioni C. Phosphorylation of initiation factor eIF-2 alpha, binding of mRNA to 48 S complexes, and its reutilization in initiation of protein synthesis. J Biol Chem. 1983 Dec 10;258(23):14556–14562. [PubMed] [Google Scholar]
- Dever T. E., Chen J. J., Barber G. N., Cigan A. M., Feng L., Donahue T. F., London I. M., Katze M. G., Hinnebusch A. G. Mammalian eukaryotic initiation factor 2 alpha kinases functionally substitute for GCN2 protein kinase in the GCN4 translational control mechanism of yeast. Proc Natl Acad Sci U S A. 1993 May 15;90(10):4616–4620. doi: 10.1073/pnas.90.10.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992 Feb 7;68(3):585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [PubMed] [Google Scholar]
- Foiani M., Cigan A. M., Paddon C. J., Harashima S., Hinnebusch A. G. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M., Redman R., Kaplansky D. A. Evidence that the primary effect of phosphorylation of eukaryotic initiation factor 2(alpha) in rabbit reticulocyte lysate is inhibition of the release of eukaryotic initiation factor-2.GDP from 60 S ribosomal subunits. J Biol Chem. 1985 Aug 5;260(16):9491–9500. [PubMed] [Google Scholar]
- Gross M., Wing M., Rundquist C., Rubino M. S. Evidence that phosphorylation of eIF-2(alpha) prevents the eIF-2B-mediated dissociation of eIF-2 X GDP from the 60 S subunit of complete initiation complexes. J Biol Chem. 1987 May 15;262(14):6899–6907. [PubMed] [Google Scholar]
- Haffter P., McMullin T. W., Fox T. D. A genetic link between an mRNA-specific translational activator and the translation system in yeast mitochondria. Genetics. 1990 Jul;125(3):495–503. doi: 10.1093/genetics/125.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P., McMullin T. W., Fox T. D. Functional interactions among two yeast mitochondrial ribosomal proteins and an mRNA-specific translational activator. Genetics. 1991 Feb;127(2):319–326. doi: 10.1093/genetics/127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Cigan A. M., Freeman B. A., Kinzy T. G. GCD11, a negative regulator of GCN4 expression, encodes the gamma subunit of eIF-2 in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jan;13(1):506–520. doi: 10.1128/mcb.13.1.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Hinnebusch A. G. Molecular analysis of GCN3, a translational activator of GCN4: evidence for posttranslational control of GCN3 regulatory function. Mol Cell Biol. 1988 Nov;8(11):4808–4820. doi: 10.1128/mcb.8.11.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Williams N. P., Wek R. C., Hinnebusch A. G. The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics. 1990 Nov;126(3):549–562. doi: 10.1093/genetics/126.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima S., Hannig E. M., Hinnebusch A. G. Interactions between positive and negative regulators of GCN4 controlling gene expression and entry into the yeast cell cycle. Genetics. 1987 Nov;117(3):409–419. doi: 10.1093/genetics/117.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima S., Hinnebusch A. G. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Nov;6(11):3990–3998. doi: 10.1128/mcb.6.11.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy K. M., Lee A., Chen E., Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991 Jun;5(6):958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- Hershey J. W. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Repeated DNA sequences upstream from HIS1 also occur at several other co-regulated genes in Saccharomyces cerevisiae. J Biol Chem. 1983 Apr 25;258(8):5238–5247. [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A., Safer B. Purification of the eukaryotic initiation factor 2-eukaryotic initiation factor 2B complex and characterization of its guanine nucleotide exchange activity during protein synthesis initiation. J Biol Chem. 1983 Mar 10;258(5):3402–3408. [PubMed] [Google Scholar]
- Koromilas A. E., Roy S., Barber G. N., Katze M. G., Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992 Sep 18;257(5077):1685–1689. doi: 10.1126/science.1382315. [DOI] [PubMed] [Google Scholar]
- Lanker S., Bushman J. L., Hinnebusch A. G., Trachsel H., Mueller P. P. Autoregulation of the yeast lysyl-tRNA synthetase gene GCD5/KRS1 by translational and transcriptional control mechanisms. Cell. 1992 Aug 21;70(4):647–657. doi: 10.1016/0092-8674(92)90433-d. [DOI] [PubMed] [Google Scholar]
- Lucchini G., Hinnebusch A. G., Chen C., Fink G. R. Positive regulatory interactions of the HIS4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Jul;4(7):1326–1333. doi: 10.1128/mcb.4.7.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matts R. L., Levin D. H., London I. M. Effect of phosphorylation of the alpha-subunit of eukaryotic initiation factor 2 on the function of reversing factor in the initiation of protein synthesis. Proc Natl Acad Sci U S A. 1983 May;80(9):2559–2563. doi: 10.1073/pnas.80.9.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullin T. W., Haffter P., Fox T. D. A novel small-subunit ribosomal protein of yeast mitochondria that interacts functionally with an mRNA-specific translational activator. Mol Cell Biol. 1990 Sep;10(9):4590–4595. doi: 10.1128/mcb.10.9.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moehle C. M., Hinnebusch A. G. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991 May;11(5):2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldave K. Eukaryotic protein synthesis. Annu Rev Biochem. 1985;54:1109–1149. doi: 10.1146/annurev.bi.54.070185.005333. [DOI] [PubMed] [Google Scholar]
- Niederberger P., Aebi M., Hütter R. Identification and characterization of four new GCD genes in Saccharomyces cerevisiae. Curr Genet. 1986;10(9):657–664. doi: 10.1007/BF00410913. [DOI] [PubMed] [Google Scholar]
- Novick P., Osmond B. C., Botstein D. Suppressors of yeast actin mutations. Genetics. 1989 Apr;121(4):659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon C. J., Hannig E. M., Hinnebusch A. G. Amino acid sequence similarity between GCN3 and GCD2, positive and negative translational regulators of GCN4: evidence for antagonism by competition. Genetics. 1989 Jul;122(3):551–559. doi: 10.1093/genetics/122.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon C. J., Hinnebusch A. G. gcd12 mutations are gcn3-dependent alleles of GCD2, a negative regulator of GCN4 in the general amino acid control of Saccharomyces cerevisiae. Genetics. 1989 Jul;122(3):543–550. doi: 10.1093/genetics/122.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pain V. M. Initiation of protein synthesis in mammalian cells. Biochem J. 1986 May 1;235(3):625–637. doi: 10.1042/bj2350625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent S. A., Fenimore C. M., Bostian K. A. Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast. 1985 Dec;1(2):83–138. doi: 10.1002/yea.320010202. [DOI] [PubMed] [Google Scholar]
- Peterson C. L., Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992 Feb 7;68(3):573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Rowlands A. G., Panniers R., Henshaw E. C. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem. 1988 Apr 25;263(12):5526–5533. [PubMed] [Google Scholar]
- Siekierka J., Manne V., Ochoa S. Mechanism of translational control by partial phosphorylation of the alpha subunit of eukaryotic initiation factor 2. Proc Natl Acad Sci U S A. 1984 Jan;81(2):352–356. doi: 10.1073/pnas.81.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamkun J. W., Deuring R., Scott M. P., Kissinger M., Pattatucci A. M., Kaufman T. C., Kennison J. A. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell. 1992 Feb 7;68(3):561–572. doi: 10.1016/0092-8674(92)90191-e. [DOI] [PubMed] [Google Scholar]
- Thomas N. S., Matts R. L., Levin D. H., London I. M. The 60 S ribosomal subunit as a carrier of eukaryotic initiation factor 2 and the site of reversing factor activity during protein synthesis. J Biol Chem. 1985 Aug 15;260(17):9860–9866. [PubMed] [Google Scholar]
- Williams N. P., Hinnebusch A. G., Donahue T. F. Mutations in the structural genes for eukaryotic initiation factors 2 alpha and 2 beta of Saccharomyces cerevisiae disrupt translational control of GCN4 mRNA. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7515–7519. doi: 10.1073/pnas.86.19.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Chumley F., Fink G. R. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 1983;101:211–228. doi: 10.1016/0076-6879(83)01016-2. [DOI] [PubMed] [Google Scholar]