IL-27 promotes the differentiation of monocytes to HIV-resistant macrophages by down-regulating host factor SPTBN1.
Abstract
The susceptibility of macrophages to HIV-1 infection is modulated during monocyte differentiation. IL-27 is an anti-HIV cytokine that also modulates monocyte activation. In this study, we present new evidence that IL-27 promotes monocyte differentiation into macrophages that are nonpermissive for HIV-1 infection. Although IL-27 treatment does not affect expression of macrophage differentiation markers or macrophage biological functions, it confers HIV resistance by down-regulating spectrin β nonerythrocyte 1 (SPTBN1), a required host factor for HIV-1 infection. IL-27 down-regulates SPTBN1 through a TAK-1–mediated MAPK signaling pathway. Knockdown of SPTBN1 strongly inhibits HIV-1 infection of macrophages; conversely, overexpression of SPTBN1 markedly increases HIV susceptibility of IL-27–treated macrophages. Moreover, we demonstrate that SPTBN1 associates with HIV-1 gag proteins. Collectively, our results underscore the ability of IL-27 to protect macrophages from HIV-1 infection by down-regulating SPTBN1, thus indicating that SPTBN1 is an important host target to reduce HIV-1 replication in one major element of the viral reservoir.
Macrophages, as a major target of HIV-1, play an important role in HIV-1 infection. Macrophage infection is found extensively in body tissues and contributes to HIV-1 pathogenesis (Koenig et al., 1986; Salahuddin et al., 1986; Wang et al., 2001; Smith et al., 2003). Macrophage lineage cells are among the first cells to be infected because most viruses involved in the first round of infection use CCR5 as the co-receptor to initiate HIV-1 replication in vivo (Philpott, 2003). Once infected, macrophages have been shown to promote rapid virus dissemination by transmitting virus particles to CD4+ T cells via a transit “virological synapse” (Groot et al., 2008). Although most CD4+ T cells are eventually killed by HIV-1, infected macrophages survive longer and can harbor virus particles in intracellular compartments (Raposo et al., 2002; Pelchen-Matthews et al., 2003), thus maintaining a hidden HIV-1 reservoir for ongoing infection (Wahl et al., 1997; Lambotte et al., 2000; Zhu et al., 2002; Smith et al., 2003; Sharova et al., 2005). Collectively, macrophage infection is involved throughout the progression of disease. Therefore, restriction of macrophage infection may provide a key to eradication of HIV-1 infection.
HIV-1 infection is modulated by a variety of host cellular factors. HIV-1 has evolved to have specific viral proteins to counteract certain host restriction factors. Human HIV-1 restriction factors, including APOBEC3G and BST-2, have been reported (Neil et al., 2008; Sheehy et al., 2002) and models of how HIV-1 overcomes these restrictions have been described in reviews (Evans et al., 2010; Goila-Gaur and Strebel, 2008). More recently, SAMHD1, a restriction factor of myeloid cells, was found to limit HIV replication by depleting intracellular dNTPs, and it is largely opposed by Vpx (Hrecka et al., 2011; Laguette et al., 2011; Lahouassa et al., 2012). Release of these host restrictions, however, does not guarantee productive infection. HIV-1, with a limited genome of nine open reading frames, has to fully exploit an array of cellular proteins to facilitate its life cycle at almost every step (Goff, 2007). Genome-wide siRNA screens, using 293T or HeLa cells as HIV-1 targets, have revealed hundreds of potential supportive host factors (Brass et al., 2008; Zhou et al., 2008), only some of which have been validated in primary target cells. Regulation of host factors, both inhibitory and supportive, may offer great opportunities to prevent HIV-1 infection of macrophages.
Cytokine-mediated immunoregulation is an effective way to inhibit HIV-1 infection in cells of myeloid lineage (Kedzierska and Crowe, 2001). Our previous studies have demonstrated that IL-27 strongly inhibits HIV-1 replication in terminally differentiated monocyte-derived macrophages (MDMs) (Fakruddin et al., 2007). IL-27 is an IL-12 family cytokine mainly produced by dendritic cells and macrophages (Kastelein et al., 2007). It was originally characterized as a proinflammatory cytokines to induce Th1 responses in T cells (Pflanz et al., 2004; Villarino et al., 2004). However, the IL-27 receptor complex, consisting of WSX-1 and glycoprotein 130 (gp130), is also expressed on monocytes (Pflanz et al., 2004) and recent evidence has supported a role for IL-27 in monocyte activation (Kalliolias and Ivashkiv, 2008; Guzzo et al., 2010a). In the current study, we aim to investigate the role of IL-27 stimulation during monocyte differentiation in modulating macrophage susceptibility to HIV-1 infection, and our study will help to evaluate whether IL-27 can be used to prevent HIV-1 infection of macrophages.
RESULTS
IL-27 induces functional macrophages with HIV-1 resistance
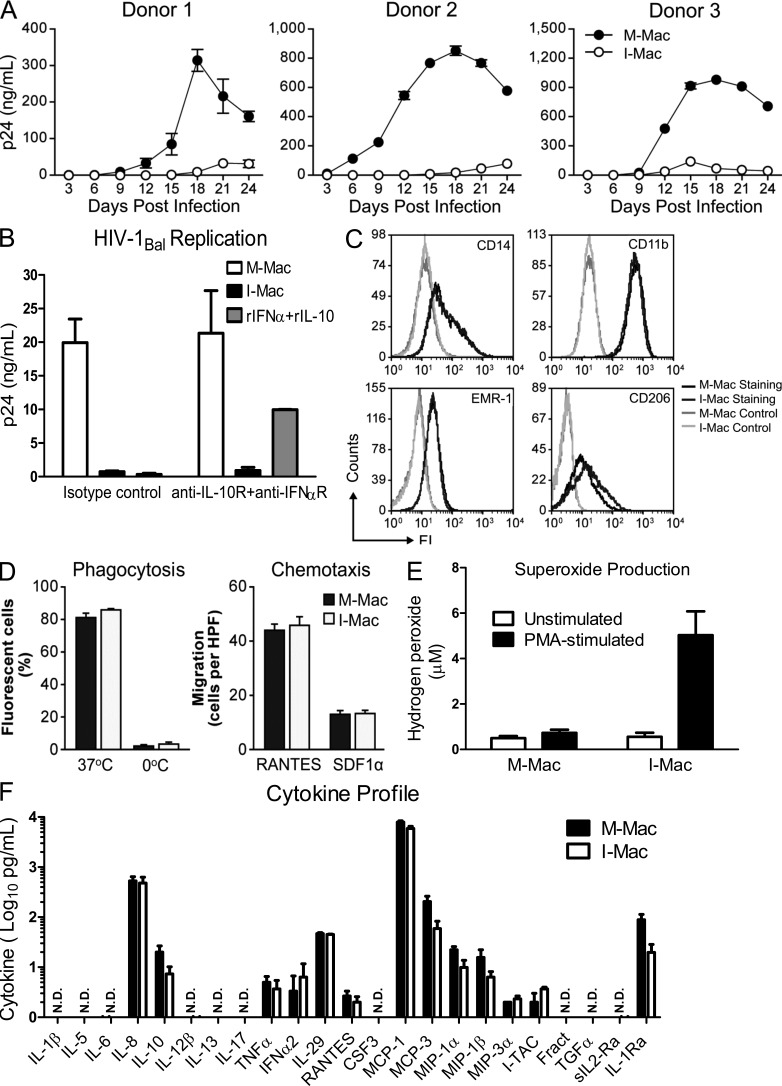
For the following experiments, we generated two types of MDMs in parallel for comparison: macrophages induced with M-CSF alone are termed “M-Mac” and macrophages induced with M-CSF combined with IL-27 are termed “I-Mac.” These two types of macrophages were infected with an R5 tropic HIV-1Bal virus strain and tested for their capacity to support HIV-1 replication. Although a robust spreading infection occurred in M-Mac, little replication was seen in I-Mac (Fig. 1 A). The inhibitory effect on the HIV-1 replication of I-Mac was not caused by cytotoxicity, as I-Mac and M-Mac were indistinguishable with respect to cell viability (unpublished data). Interestingly, blocking IFN-α and IL-10 receptors with neutralizing antibodies had no impact on the HIV-1 resistance of I-Mac (Fig. 1 B). Because susceptibility of macrophages to HIV-1 infection largely depends on the state of monocyte differentiation, we examined whether or not IL-27 treatment blocked macrophage differentiation. No significant difference was observed in the expression of macrophage differentiation markers such as CD14, CD11b, EMR1, or CD206. (Fig. 1 C). Similarly, I-Mac and M-Mac have indistinguishable phagocytosis and chemotaxis activities (Fig. 1 D). Moreover, I-Mac produced the same types of proinflammatory cytokines as M-Mac (Fig. 1 F), and I-Mac was able to produce high levels of superoxide upon stimulation with PMA (Fig. 1 E). Collectively, these results indicate that IL-27 promotes monocyte differentiation into HIV-resistant macrophages without compromising normal macrophage functions.
Figure 1.
IL-27 induces macrophages resistant to HIV-1 infection while retaining normal functions and differentiation markers. (A) M-Mac and I-Mac were infected with HIV-1BAL. Viral replication was monitored by measuring p24 antigen in culture supernatants. Results are shown for macrophages cultures obtained from three independent donors and data shown represent mean ± SD of triplicate infection samples. (B) Antibodies that neutralize human IFN-α receptor (10 µg/ml) and IL-10 receptors (10 µg/ml) were kept in culture during the 7-d differentiation of M-Mac and I-Mac. M-Mac and I-Mac differentiated in the presence of neutralizing antibodies were infected with HIV-1Bal, and p24 amount in culture supernatants was measured 14 d after infection. As an additional control to indicate the neutralizing effect, M-Mac differentiated in the presence of neutralizing antibodies were also incubated with recombinant human IFN-α (10 u/ml) and IL-10 (1 ng/ml) for 1 h before HIV-1 infection. Data shown represent means ± SE of three independent experiments. (C) M-Mac and I-Mac were analyzed for CD14, CD11b, EMR-1, and CD206 expression by FACS before infection. (D, left) Phagocytic activities of M-Mac and I-Mac were assessed using a pHrodo dye phagocytosis assay. As a negative control, macrophages were placed on ice to avoid phagocytosis. (right) the migration of M-Mac and I-Mac to RANTES (10 ng/ml) and SDF-1α (100 ng/ml) were assessed by counting cells migrating across filters of microchemotaxis chambers. Data shown represent mean ± SE of three independent experiments. (E) M-Mac and I-Mac were analyzed for superoxide production with or without PMA stimulation. Data shown represent means ± SE of three independent experiments. (F) Supernatants of M-Mac and I-Mac were analyzed for cytokine concentrations using the Multiplex Cytokine assay. Data are shown on a log scale. N.D., not detected. Data shown represent means ± SE of three independent experiments.
IL-27 induces a post-entry block to HIV-1 infection
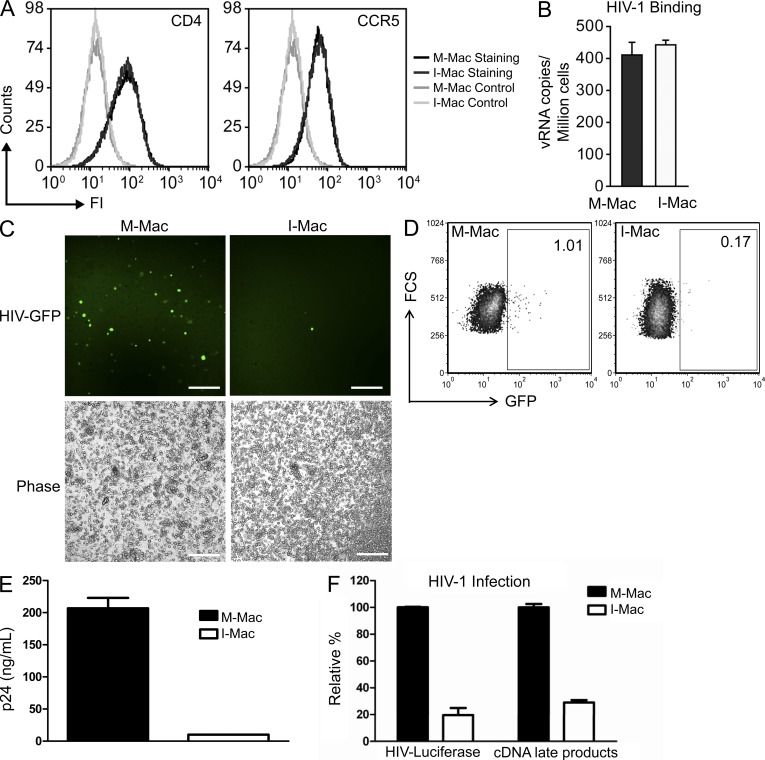
CD4 and CCR5 act as the receptor and co-receptor for HIV-1 entry into macrophages. FACS analysis showed that I-Mac and M-Mac expressed comparable levels of CD4 and CCR5 molecules (Fig. 2 A). Expression of these molecules was also sufficient to mediate efficient binding of comparable numbers of HIV particles to I-Mac (Fig. 2 B). Therefore, it appeared unlikely that HIV-1 infection of I-Mac was blocked at the level of virus entry. To further confirm this, we took advantage of vesicular stomatitis virus G glycoprotein (VSV-G)-pseudotyped HIV indicator viruses. The recombinant viruses, mutated in the Env gene and pseudotyped with VSV-G envelope, can enter macrophages through a CD4/CCR5-independent pathway and complete only a single round of infection. As such, the system allowed us to focus our study on the inhibition of HIV-1 infection in I-Mac at a post-entry level. The exposure of I-Mac to VSV-G–pseudotyped GFP encoding HIV-1 (HIV-EGFP-V) resulted in little infection, as observed by few green fluorescent cells in I-Mac (Fig. 2 C). A single round of infection led to 80 ± 3.2% (n = 6) less GFP-positive cells according to FACS analysis (Fig. 2 D) and a 95% reduction of soluble p24 antigen in I-Mac (Fig. 2 E). The pattern that I-Mac displayed significantly less (P < 0.05) GFP-positive cells was consistent in macrophage cultures prepared from an additional five independent donors (unpublished data). Likewise, when macrophages were infected with VSV-G-pseudotyped HIV-luciferase virus (HIV-LUC-V), HIV-1 infection of I-Mac was still inhibited, as indicated by 80% lower HIV-luciferase activity (Fig. 2 F). Importantly, we also compared the efficiency of proviral cDNA synthesis in M-Mac and I-Mac. M-Mac supported the synthesis of proviral cDNA as indicated by the amount of late products from viral cDNA synthesis. Infection of I-Mac by the same pseudotyped HIV luciferase virus led to 75% less proviral cDNA late products (Fig. 2 F). Thus, IL-27 appears to interfere with HIV-1 replication after viral entry and before reverse transcription.
Figure 2.
HIV-1 infection of I-Mac is blocked after entry and before reverse transcription of viral cDNA. (A) M-Mac and I-Mac were analyzed for CD4 and CCR5 expression by FACS. (B) M-Mac and I-Mac were incubated with HIV-1BAL on ice for 1 h to allow virus binding. Virus attachment was determined by measuring the copy number of HIV-1 RNA bound to the cells. Data shown represent means ± SE of three independent experiments. (C) M-Mac and I-Mac were infected with HIV-EGFP-V. (left) Infected cells were examined with fluorescent microscopy. Bar, 200 µm. (D) GFP+ cells were analyzed by FACS 4 d after infection. (E) p24 antigen amount was measured by ELISA. Data shown represent mean ± SD of triplicate infection samples. (F) M-Mac and I-Mac were infected with HIV-LUC-V. Viral cDNA copy number and luciferase activity was measured 48 h after infection. Data shown represent mean ± SE of three independent experiments.
I-Mac lacks a critical host factor to support HIV-1 infection
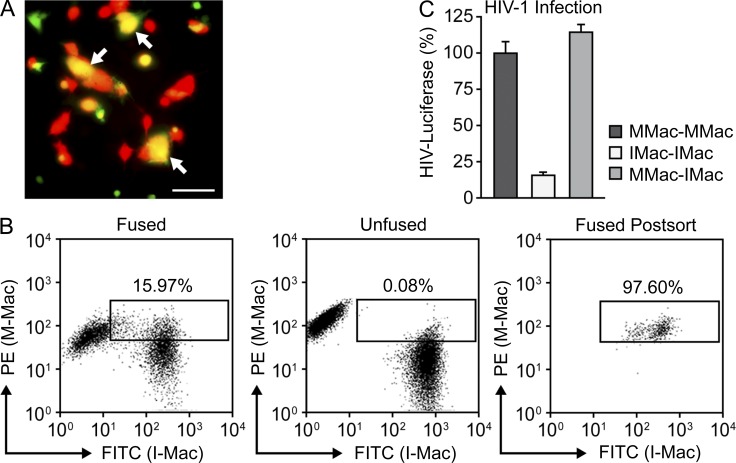
We have established that M-Mac and I-Mac display different susceptibility to HIV-1 infection. This indicates the existence of cellular factors, differentially expressed between M-Mac and I-Mac, which have the potential to impact HIV-1 infection. Gene expression profiling using the genome-wide Affymetrix GeneChip revealed that 178 genes were differentially expressed between the two cells with an absolute fold change greater than five (Table S1). Within this group, 60 genes showed decreased expression values, and 118 genes showed increased expression values in I-Mac. To determine whether the decreased HIV-1 infection was caused by lack of a required factor or increased expression of a restriction factor, we generated heterokaryons between M-Mac and I-Mac. M-Mac and I-Mac homokaryons were also generated as controls. Heterokaryon formation was confirmed with fluorescent microscopy as double-stained cells (Fig. 3 A). Fused cells were obtained with high purity by FACS sorting (Fig. 3 B). Susceptibility of the heterokaryons to HIV-LUC-V infection was compared with that of the homokaryons. Heterokaryons between M-Mac and I-Mac displayed full infection efficiency as M-Mac homokaryons (Fig. 3 C). As an additional control, we also tested mixed parental M-Mac and I-Mac cultures at a ratio of 1:1 without fusion. As expected, HIV-1 infection of this unfused control faithfully reflected the average level of HIV-luciferase between those of M-Mac and I-Mac (unpublished data). Because fusion with M-Mac largely rescued HIV-1 infection of I-Mac, this suggested that I-Mac is deficient in a host factor that is required for HIV-1 infection.
Figure 3.
I-Mac lacks a cellular host factor to support HIV-1 infection. (A) Heterokaryons were formed between M-Mac and I-Mac. M-Mac was stained with CellTracker Green and I-Mac was stained with CellTracker Red. Double-stained heterokaryons were yellow as indicated by arrows. Bar, 50 µm. (B, left) Double-stained heterokaryons between M-Mac and I-Mac were sorted by FACS with the indicated gate. (middle) M-Mac and I-Mac were mixed without fusion. (right) Heterokaryons were reanalyzed by FACS to confirm purity after sorting. (C) Sorted heterokaryons were infected with HIV-LUC-V. HIV-1 infection of heterokaryons between M-Mac and I-Mac was compared with infection levels of M-Mac or I-Mac homokaryons. Data shown represent mean ± SE of three independent experiments.
IL-27 down-regulates spectrin β nonerythrocyte 1 (SPTBN1) during monocyte differentiation
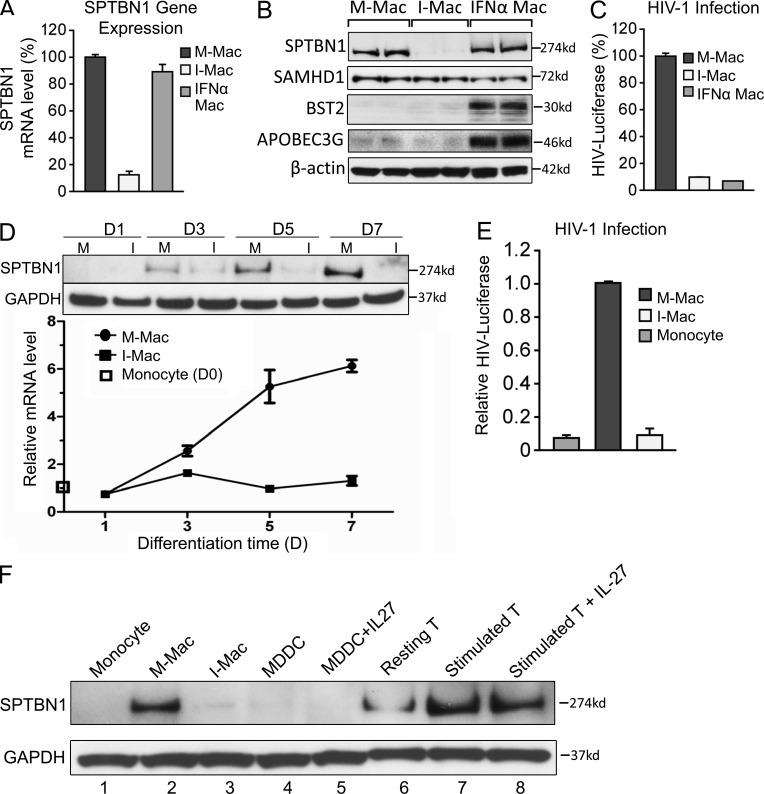
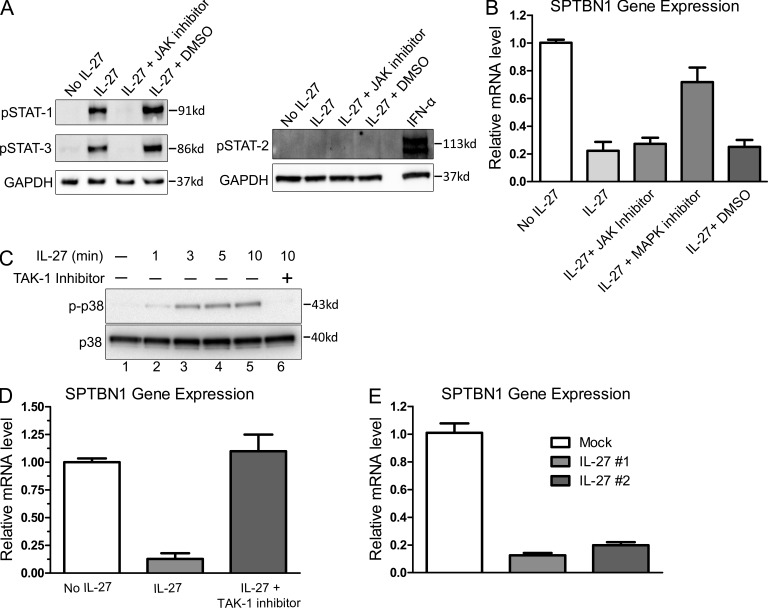
We next sought to identify the missing factor(s) for HIV-1 infection of macrophage from the 60 down-regulated candidate genes identified in Table S1, cross-referencing the 11 previously identified host factors predicted to facilitate the HIV life cycle after virus entry and before nuclear import (Brass et al., 2008). SPTBN1 was the only gene present in both groups. SPTBN1 belongs to the family of β spectrin and the gene encodes a 274-kD protein (Chang et al., 1993). We validated our findings by real-time PCR and Western blotting (Fig.4, A and B). We compared M-Mac, I-Mac, and macrophages treated with IFN-α for 24 h (IFN-α Mac). HIV-1 infection was highly inhibited in both I-Mac and IFN-α Mac, as expected (Fig. 4 C). A reduction of SPTBN1 mRNA level was only observed in I-Mac (Fig. 4 A). The specific reduction of SPTBN1 in I-Mac was also confirmed by Western blotting (Fig. 4 B). In agreement with the GeneChip microarray data, the expression of the IFN-inducible HIV-1 restriction factors APOBEC3G and BST-2 were not induced by IL-27, and the monocyte restriction factor SAMHD1 was not enhanced in I-Mac (Fig. 4 B). Thus, HIV-1 inhibition induced by IL-27 in macrophages seemed to be largely different from the anti-HIV events induced by IFN-α. We next examined whether or not SPTBN1 was expressed in primary monocytes. SPTBN1 expression was largely absent in monocytes and gradually became abundant in M-Mac along the 7-d differentiation (Fig. 4, D and F). HIV-1 transduction was undetectable in monocytes but was evident in M-Mac (Fig. 4 E). IL-27 efficiently down-regulated SPTBN1 of I-Mac during monocyte differentiation and led to lower susceptibility to HIV-1 infection (Fig. 4, D and E). We further compared the expression of SPTBN1 in monocytes, macrophages, monocyte-derived dendritic cells (MDDCs), and CD4+ T cells of the same donor (Fig. 4 F). SPTBN1 is highly expressed in differentiated macrophages and activated CD4+ T cells (Fig. 4 F, lane 2 and lane 7). In contrast, little SPTBN1 expression was found in monocytes or in MDDCs (Fig. 4 F, lanes 1 and 4). Notably, it seemed that IL-27 only strongly affected the SPTBN1 expression of macrophages (Fig.4F, lanes 2 and 3). In 293T and HeLa cell lines, IL-27 did not affect the gene expression of SPTBN1 (unpublished data). It is known that IL-27 activates STAT-1, -2, -3, and -5 in CD4+ T cells (Kamiya et al., 2004). In macrophages, however, IL-27 only activated STAT1 and STAT3, but not STAT2 (Fig. 5 A). A JAK-STAT inhibitor efficiently blocked the phosphorylation of STAT1 and STAT3 (Fig. 5 A), but had little impact on the down-regulation of SPTBN1 (Fig. 5 B). This result was surprising because JAK-STAT was considered as the classical apex of IL-27 signaling cascade. To exclude artificial effects from IL-27 preparation, we obtained another source of recombinant IL-27, which was produced by a distinct host and purified differently by another supplier. Because IL-27 from two different preparations similarly down-regulated SPTBN1 (Fig. 5 E), it was unlikely that the down-regulation of SPTBN1 resulted from artifacts of the IL-27 preparation. More importantly, we found that IL-27 also activated p38 MAPK in macrophages shortly after stimulation (Fig. 5 C, lanes 2–5), and that the down-regulation of SPTBN1 by IL-27 was sensitive to a p38 inhibitor, which inhibited p38 catalytic activity (Fig. 5 B), and also was restored by a TAK-1 inhibitor that blocked p38 phosphorylation (Fig. 5, C [lane 6] and D). Given that IL-6 activates TAK-1 through gp130 (Kojima et al., 2005), our results suggest IL-27 may down-regulate SPTBN1 through a TAK-1–mediated MAPK signaling pathway.
Figure 4.
IL-27 down-regulates SPTBN1 during monocyte differentiation. (A) M-Mac, I-Mac, and macrophages treated with IFN-α (1 ng/ml) for 24 h (IFN-α Mac) were analyzed for gene expression levels of SPTBN1 by RT-PCR. (B) Whole-cell lysates of M-Mac, I-Mac, and IFN-α Mac were used for Western blotting. Protein expression levels of SPTBN1, SAMHD1, BST-2, and APOBEC3G were examined with specific antibodies. Samples were loaded in duplicate and β-actin served as an internal loading control. (C) M-Mac, I-Mac, and IFN-α Mac were infected with HIV-LUC-V. Data shown represent mean ± SE of three independent experiments. (D) M-Mac and I-Mac were analyzed for SPTBN1 expression on day 1, 3, 5, and 7 during differentiation. (top) Whole-cell lysates were subjected to Western blotting to examine protein levels of SPTBN1. (bottom) Gene expression levels of SPTBN1 were examined by RT-PCR. Relative mRNA levels of M-Mac and I-Mac were compared with that of undifferentiated monocyte on day 0, which is equal to 1. (E) Monocytes, M-Mac, and I-Mac derived from the same donor were infected with HIV-LUC-V. Values of luciferase activity were expressed relative to those obtained for M-Mac. Data shown represent mean ± SE of three independent experiments. (F) Monocytes, macrophages, dendritic cells, and CD4+ T cells obtained from the same donor were analyzed for SPTBN1 expression by Western blotting. MDDCs were induced by G-MCSF (50 ng/ml) and IL-4 (50 ng/ml) with or without IL-27 (50 ng/ml). CD4+ T cells were stimulated with PHA (5 µg/ml) and IL-2 (20 U/ml) with or without IL-27 (50 ng/ml).
Figure 5.
IL-27 down-regulates SPTBN1 through a TAK-1–mediated p38 MAPK signaling pathway. On day 4 during differentiation, macrophages were stimulated with IL-27 (50 ng/ml), with or without the pretreatment of a JAK inhibitor or its solvent control DMSO. (A) Whole-cell lysates were harvested 15 min after IL-27 stimulation, and the phosphorylation of STAT1, STAT2, and STAT3 was examined by Western blotting using specific antibodies against p-STATs (Cell Signaling Technology). IFN-α–treated macrophage lysate was included to validate the antibody against phosphorylated STAT2. (B) Macrophages were pretreated with a JAK inhibitor (EMD), a p38 MAPK inhibitor (Cell Signaling Technology), or the solvent control DMSO. After 2 h, macrophages were stimulated with IL-27 (50 ng/ml). Gene expression of SPTBN1 was examined by RT-PCR 24h after IL-27 stimulation. Data shown represent means ± SE of three independent experiments. (C) Macrophages were pretreated with a TAK-1 inhibitor (Cell Signaling Technology) or mock treated for 2 h. Cells were then stimulated with IL-27 (50 ng/ml) and harvested at different time points as indicated. The phosphorylation of p38 MAPK was examined by Western blotting using an anti–p-p38 antibody (Cell Signaling Technology). The expression of unphosphorylated p38 was shown as an internal control. (D) Macrophages were pretreated with a TAK-1 inhibitor (Cell Signaling Technology) or mock treated. After 2 h, macrophages were stimulated with IL-27 (50 ng/ml). Gene expression of SPTBN1 was examined by RT-PCR 24h after IL-27 stimulation. Data shown represent means ± SE of three independent experiments. (E) Recombinant human IL-27 #1 and IL-27 #2 were obtained from R&D Systems and HumanZyme, respectively. On day 4 during differentiation, macrophages were stimulated with IL-27 #1 and IL-27 #2 (50 ng/ml). Gene expression of SPTBN1 was examined by RT-PCR 24 h after IL-27 stimulation. Data shown represent means ± SD of three independent samples.
SPTBN1 is required to support HIV-1 infection of macrophage
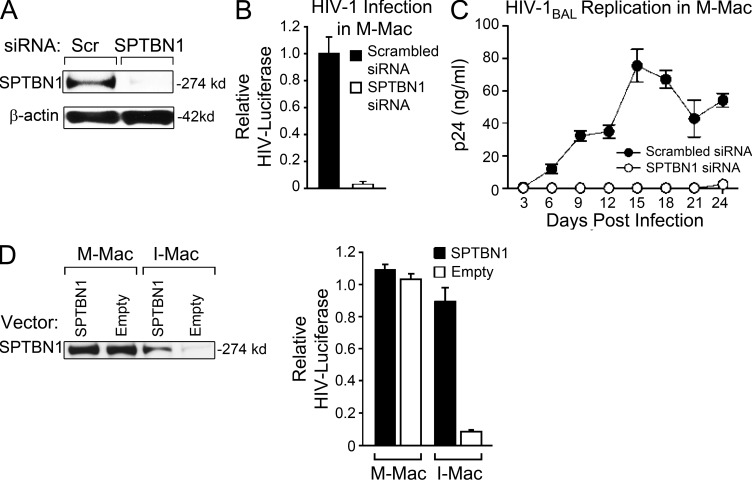
If a decrease in SPTBN1 is the key factor that determines the antiviral activity of IL-27, we would predict that knockdown of SPTBN1 in M-Mac or overexpression of SPTBN1 in I-Mac should reverse their phenotypes. To this end, we first determined the impact of SPTBN1 silencing on HIV-1 susceptibility of M-Mac. Transfection with a pool of three siRNAs targeting SPTBN1 resulted in complete knockdown of SPTBN1 in M-Mac (Fig. 6 A). Silencing SPTBN1 strikingly abrogated a single round of HIV-LUC-V infection as well as spreading infection of HIV-1Bal in M-Mac (Fig. 6, B and C). The transfection of SPTBN1 siRNA exerted no significant cytotoxic effect (unpublished data) and induction of IFN-α was not detected 6 or 24 h after transfection (unpublished data). To further exclude nonspecific effects of siRNA transfection, we used a neutralizing antibody against IFN-α/β receptor to blocks type I IFN responses, and the level of HIV-1 infection after the siRNA transfection of SPTBN1 was not affected (unpublished data). Additionally, we were able to silence the gene expression of SPTBN1 with the same siRNA in dendritic cells and CD4+ T cells. Knockdown of SPTBN1 in these cells, however, had no impact on the HIV-1 replication (unpublished data). Next, we compensated for the suppressed endogenous SPTBN1 of I-Mac by transfection with an SPTBN1 expression vector. With a fivefold increase of SPTBN1, HIV-1 infection of I-Mac was rescued to 83% that of M-Mac (Fig. 6 D). Collectively, these results demonstrate that SPTBN1 is a major host factor to support HIV-1 infection specifically in macrophages.
Figure 6.
SPTBN1 is required to support HIV-1 infection in primary macrophages. (A) M-Mac was mock transfected or transfected with siRNA targeting SPTBN1. 48 h after transfection, whole-cell lysates were used to examine SPTBN1 expression by Western blotting. (B) M-Mac was infected with HIV-LUC-V 48 h after siRNA transfection. Data shown represent mean ± SE of three independent experiments. (C) M-Mac was infected with HIV-1BAL. HIV-1 replication was monitored by measuring p24 antigen levels in culture supernatants for 24 d after infection. Data shown represent mean ± SD of triplicate infection samples. (D) M-Mac and I-Mac were transfected with an SPTBN1 expression vector or an empty vector for 48 h. (left) Whole-cell lysates were used to examine SPTBN1 expression by Western blotting. (right) Transfected cells were infected with HIV-LUC-V. Relative luciferase activity was shown, and data shown represent mean ± SE of three independent experiments.
SPTBN1 associates with HIV-1 gag proteins
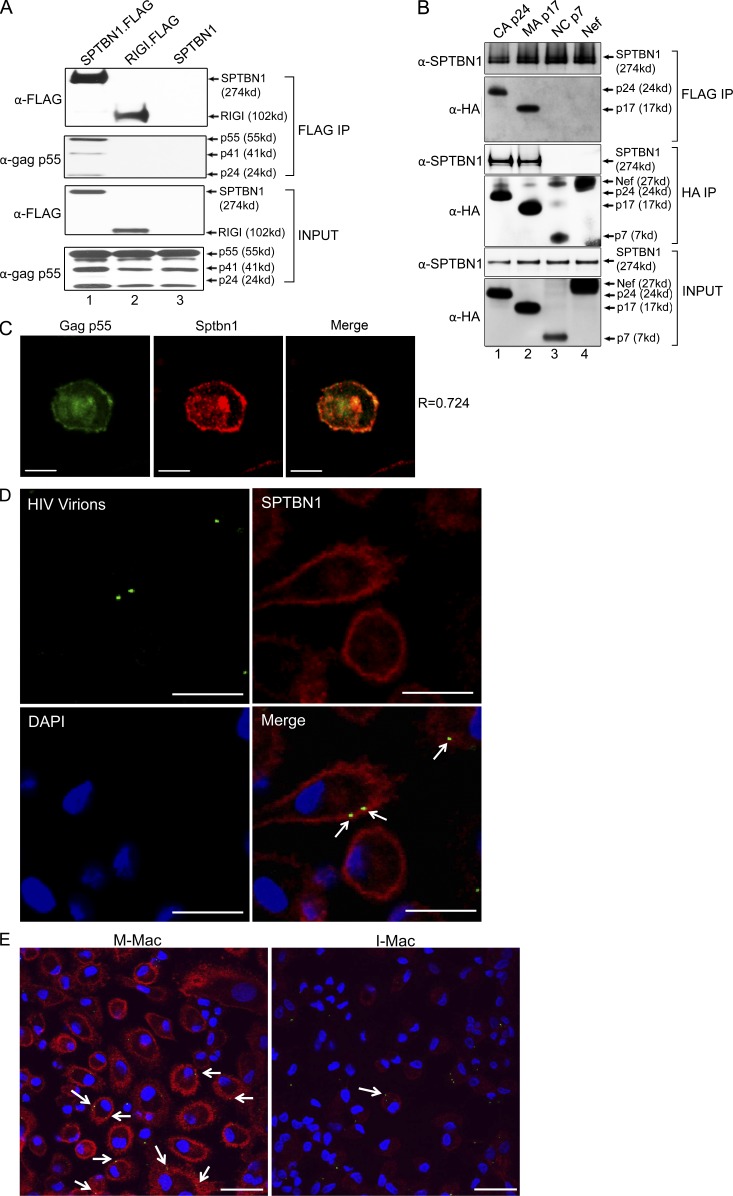
To investigate a potential protein–protein interaction that could link SPTBN1 and the early steps of the HIV-1 life cycle, we examined whether SPTBN1 associates with HIV-1 gag proteins. 293T cells were co-transfected with a pNL4.3 plasmid and a FLAG-tagged SPTBN1-expressing vector. Immunoblotting of the anti-FLAG immunoprecipitates revealed that SPTBN1 strongly binds to HIV-1 gag p55 (Fig. 7 A, lane 1), whereas no signal was detected with an unrelated FLAG-tagged protein RIG-I or untagged SPTBN1 control (Fig. 7 A, lanes 2 and 3). The antibody also detected gag p41 and p24, but not p17 or p7. In fact, HIV-1 gag p55 is the polyprotein precursor of capsid (CA) p24, matrix (MA) p17, and nucleocapsid (NC) p7. Because the interaction between SPTBN1 and gag p55 could be mediated by these smaller gag proteins, we performed coimmunoprecipitation to dissect the interaction of SPTBN1 with individual gag proteins. FLAG-tagged SPTBN1 was coexpressed with HA-tagged CA p24, MA p17, or NC p7. HA tagged-Nef was included as a control to rule out that SPTBN1 bound nonspecifically to HA tag in this assay. Immunoblotting of the anti-FLAG and anti-HA immunoprecipitates consistently showed that SPTBN1 associates with CA p24 and MA p17 (Fig. 7 B, lanes 1 and 2) but not with NC p7 or Nef (Fig. 7 B, lanes 3 and 4). These results demonstrate that SPTBN1 has a specific interaction with HIV-1 gag CA p24 and MA p17 proteins. We next sought to determine whether SPTBN1 colocalizes with HIV-1 gag in macrophages (Fig. 7 C). We transfected macrophages with a plasmid expressing GFP-tagged HIV-1 gag p55 (green) and labeled endogenous SPTBN1 with an Alexa Fluor 555–conjugated antibody (red). SPTBN1 displayed a localization pattern that largely overlapped with that of gag p55, as indicated with a Pearson correlation coefficient (R) value of 0.70 ± 0.05 (n = 5), and the colocalization (yellow) was observed both to the cell plasma membrane (PM) and to intracellular compartments (Fig. 7 C). To define the localization of gag in macrophages at early time points after infection, we infected macrophages with HIV-1 virus packaging Vpr.GFP fusion proteins (HIV-Vpr.GFP; Fig. 7 D). Because GFP fluorescence has been confirmed to highly associate with HIV-1 gag proteins CA p24 and MA p17 (McDonald et al., 2002), this system made it feasible for us to investigate the colocalization of SPTBN1 and HIV-1 virions in the context of viral infection. After macrophages were incubated with VSV-G–pseudotyped HIV-Vpr.GFP (green) for 20 min, the cells were extensively washed and fixed with formaldehyde, and then endogenous SPTBN1 was labeled with the Alexa Fluor 555–conjugated antibody (red). Colocalization of SPTBN1 and Vpr.GFP-labeled virions was observed on PM as well as close to the intracellular side of the PM (Fig. 7 D, bottom right). We observed in a total of 752 M-Mac cells that 75% of the virions displayed overlapped localization with SPTBN1, whereas only 13% did in a total of 825 I-Mac cells because of the substantial SPTBN1 reduction by IL-27 (Fig. 7 E). Additional statistical analysis confirmed that the possibility of the colocalization patterns of M-Mac and I-Mac being not significantly different was <10−11. Collectively, these results indicate that the lack of viral interaction with the host factor SPTBN1 may limit the completion of the HIV-1 life cycle.
Figure 7.
SPTBN1 associates with HIV-1 gag proteins. (A) 293T cells were transfected to express FLAG-tagged SPTBN1 and gag p55. Whole-cell lysates from transfected cells were subjected to a pull-down assay using anti-FLAG agarose. Unrelated FLAG-tagged RIG-I and untagged SPTBN1 were negative controls for nonspecific binding. Immunoprecipitates were analyzed by immunoblotting using anti-FLAG tag and anti-gag p55 antibodies. (B) 293T cells were transfected to express FLAG-tagged SPTBN1 and HA-tagged CA p24, MA p17, and NC p7. Whole-cell lysates from transfected cells were subjected to a pull-down assay using anti-FLAG agarose or anti-HA agarose. Immunoprecipitates were analyzed by immunoblotting using anti-SPTBN1 and anti-HA antibodies. Nef was a negative control for nonspecific binding. (C) Macrophages were transfected to express GFP-tagged Gag p55 (green). After 24 h, macrophages were fixed and labeled with antibody against SPTBN1 (red). A representative merged image of Gag p55 and SPTBN1 shown on the right (yellow) indicates the colocalization between Gag and SPTBN1. R, Pearson coefficient of correlation. Bars, 10 µm. (D) Macrophages were infected with VSV-G–pseudotyped Vpr-GFP-packaged HIV-1 virions (green). After 30-min incubation, cells were fixed and labeled with antibody against SPTBN1 (red) or DAPI (blue). White arrows on the merged image indicate the colocalization of SPTBN1 and incoming HIV-1 virions (yellow). Bars, 30 µm. (E) M-Mac and I-Mac were infected with VSV-G-pseudotyped Vpr-GFP-packaged HIV-1 virions (green). After 30-min incubation, cells were fixed and labeled with antibody against SPTBN1 (red) or DAPI (blue). White arrows on the merged image indicate the colocalization of SPTBN1 and incoming HIV-1 virions (yellow). Bars, 30 µm.
DISCUSSION
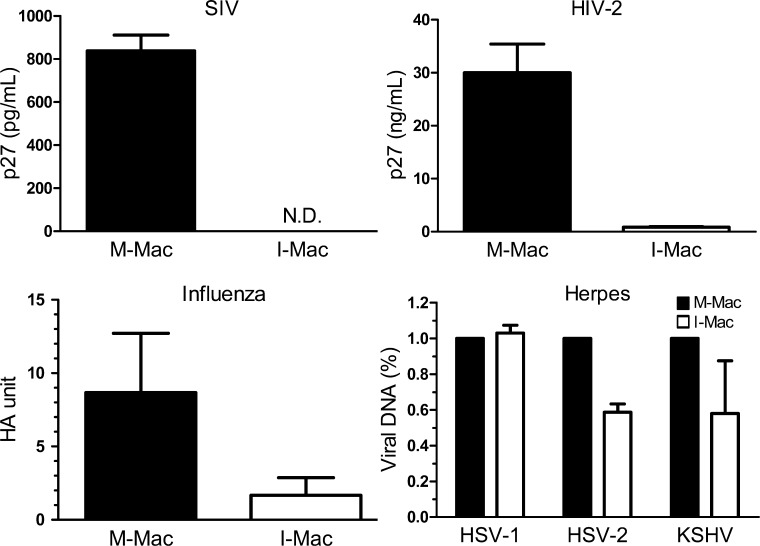
Our study has demonstrated that IL-27 promotes monocyte differentiation into HIV-1–resistant macrophages with no obvious impact on other biological functions. IL-27 induces a post-entry block to HIV-1 infection of macrophages by suppressing the host factor SPTBN1. Silencing of SPTBN1 in M-Mac strongly inhibits HIV-1 infection. Conversely, overexpression of SPTBN1 markedly increases the susceptibility of I-Mac to infection. Thus, our results suggest that modulating SPTBN1 level by IL-27 is an effective way to render human macrophages resistant to HIV-1 infection. IL-27 could be a promising therapeutic candidate, as the treatment of IL-27 has no significant impact on cell viability or biological functions of macrophages. Notably, the IL-27–induced viral resistance is not only against HIV-1 infection but also the infection of HCV (Frank et al., 2010), SIVmac239, HIV-2, influenza A, and some herpes viruses such as HSV-2 and KSHV (Fig. 8). Co-infection of other viruses with HIV may accelerate AIDS progression and increase the risk of death. Although IL-27 could be produced by dendritic cells and macrophages during viral infection, circulating HIV may suppress IL-27 production in infected patients (Guzzo et al., 2010b). Therefore, IL-27 treatment in HIV-1 patients could be beneficial to control systemic disease progression as a novel therapeutic candidate. Further studies are needed to first assess the antiviral effect of IL-27 in vivo using the SIV-infected monkey model.
Figure 8.
IL-27 inhibits various viral infections. M-Mac and I-Mac were infected with SIV, HIV-2, Influenza A, HSV-1, HSV-2, and KSHV. Infection of SIV and HIV-2 was monitored by measuring p27 antigen amount in supernatants. N.D., not detected. Detection limit of SIV p27 was 15.625 pg/ml. Infection of Influenza A was determined using HA assay. Infection of herpes viruses was gauged by measuring viral DNA copies with real-time PCR. Data shown represent means ± SE of three independent experiments.
Our group was the first to report the anti-HIV effect of IL-27 on terminally differentiated macrophages, and in those studies, IL-27 was always maintained in culture (Fakruddin et al., 2007; Imamichi et al., 2008). In the current study, we extend these studies and examine the impact of IL-27 on the primary monocytes. Our results demonstrate that IL-27 promotes the differentiation of monocytes into I-Mac which is nonpermissive to HIV-1 infection. Once I-Mac is established upon macrophage differentiation, IL-27 is no longer needed to maintain the HIV-1 resistance. Unlike the previous studies, IL-27 was completely removed after 7 d of macrophage differentiation and cells were challenged with HIV-1 in an IL-27–free environment. Thus, it is unlikely that IL-27 has a direct negative effect on the HIV-1 virus particles. Instead, IL-27 causes an intracellular block to HIV-1 infection.
The mechanism by which IL-27 inhibits HIV-1 replication in macrophages has been controversial: our previous results suggest the inhibition of HIV-1 by IL-27 is IFN-α–independent because neutralization of IFN-α has no impact on IL-27–mediated HIV inhibition (Imamichi et al., 2008), whereas another study has shown that type I IFNs are induced by IL-27 and subsequently induce APOBEC3G (Greenwell-Wild et al., 2009). This discrepancy could be attributed to the different methods used for macrophage differentiation. In the experiments presented here, the results of the microarray and Western blotting analysis display no differential expression of IFN-inducible genes such as APOBEC3G and BST-2 between M-Mac and I-Mac (Table S1 and Fig. 4 B). We have also examined the cytokine profiles of M-Mac and I-Mac (Fig. 1 F). I-Mac did not produce significantly more IFN-α2 or IL-10, which is another anti-HIV cytokine (Saville et al., 1994; Weissman et al., 1994), than M-Mac. Blocking IFN-α or IL-10 responses with neutralizing antibodies had no impact on the HIV-1 resistance of I-Mac (Fig. 1 B). Thus, our results indicate that IFN-α and IL-10 may only play a minimal role in the HIV-1 inhibition of I-Mac. Moreover, because co-culturing with I-Mac did not inhibit the HIV-1 infection of M-Mac (unpublished data), it seems unlikely that the HIV-1 resistance of I-Mac is mediated by a soluble factor in the supernatant. However, at present, we cannot fully exclude the possibility that other antiviral genes induced by IL-27 could also affect HIV replication of I-Mac, considering the known IFN-α–like and IFN-γ–like functions of IL-27 (Imamichi et al., 2008; Bender et al., 2009), and therefore further investigation will be needed to determine the individual role of any potential antiviral genes that could be induced by IL-27.
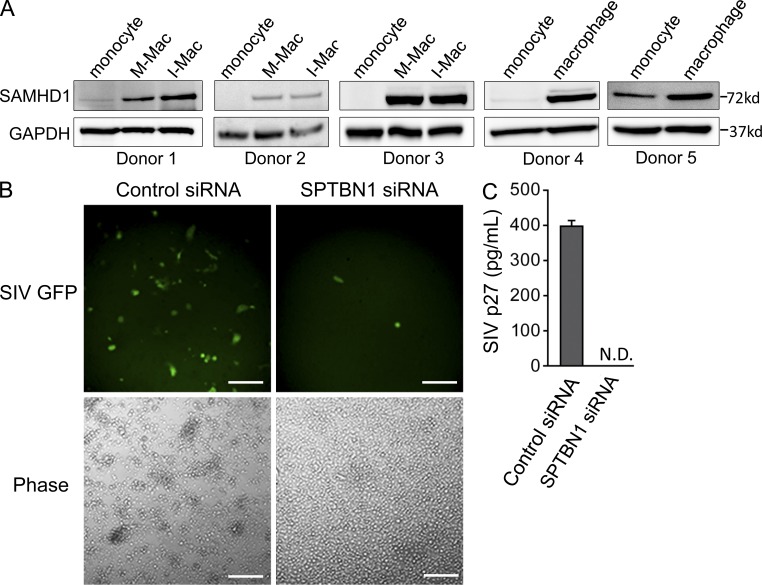
HIV-1 minimally infects peripheral blood monocytes in vitro because of a post-entry block (Sonza et al., 1996; Eisert et al., 2001; Neil et al., 2001). Susceptibility to HIV-1 is thought to be established once monocytes differentiate into macrophages (Di Marzio et al., 1998; Naif et al., 1998; Rich et al., 1992). The restriction of HIV-1 infection in monocytes seems to be the result of multiple limitations. Some studies have shown that monocytes express less CD4 and CCR5 receptors than macrophages (Di Marzio et al., 1998; Naif et al., 1998), although it does not explain why VSV-G–pseudotyped HIV-1 virus is still restricted. Other studies have shown that monocytes contain lower levels of dNTPs (O’Brien et al., 1994; Triques and Stevenson, 2004), and may have some anti-HIV miRNAs (Wang et al., 2009), which may be responsible for the lower infection of monocytes. Recently, SAMHD1 was identified as an important HIV-1 restriction factor of myeloid cells. The expression of SAMHD1 has been confirmed in dendritic cells, monocytes, and macrophages (Laguette et al., 2011). In this study, we have compared the expression of SAMHD1 in monocytes and monocyte-derived macrophages from the same donors. We found SAMHD1 expression was surprisingly enhanced after macrophage differentiation (Fig. 9 A). Thus, it appears that SAMHD1 is not the answer to explain why macrophages are more susceptible than monocytes. In fact, we found that SIVmac239, harboring Vpx to counteract SAMHD1, was still unable to replicate in macrophages when SPTBN1 was silenced (Fig. 9, B and C). The results in this study have identified SPTBN1 as a required host factor for HIV-1 infection of macrophages. The expression of SPTBN1 is missing in monocytes but significantly up-regulated upon macrophage differentiation, and suppression of SPTBN1 by IL-27 leads to impaired susceptibility (Fig. 4 C). Therefore, the absence of SPTBN1 in monocytes at least partially accounts for their relatively lower susceptibility to HIV-1 infection.
Figure 9.
SPTBN1 affects macrophage susceptibility to HIV-1 infection independently of SAMHD1. (A) Whole-cell lysates of monocytes, M-Mac, and I-Mac (donor 1–3) or whole-cell lysates of monocytes and macrophages (donor 4 and 5) were analyzed for protein expression levels of SPTBN1. GAPDH served as an internal loading control. (B) Macrophages were infected with SIV-GFP after transfection with control siRNA or SPTBN1 specific siRNA (50 pmol). A representative field of macrophages for SIV-GFP expression was shown. Bars, 200 µm. (C) SIV p27 level in culture supernatants was measured. Data shown represent means ± SD of triplicate infection samples.
Previous studies established that IL-27 mainly triggers a JAK-STAT signaling pathway in T cells (Takeda et al., 2003; Villarino et al., 2003). Owaki et al. (2006) reported that IL-27 induces Th1 differentiation through alternative MAPK/ERK-dependent pathways. Our results show that IL-27 activates p38, which confirms that the IL-27–mediated MAPK signaling pathway also exists in macrophages. Moreover, we found that IL-27–induced p38 activation is blocked by a TAK-1 inhibitor, indicating that TAK-1 is located upstream of p38 MAPK. Indeed, gp130, the common subunit of IL-27 and IL-6 receptor, has been shown to activate TAK-1 (Kojima et al., 2005). Collectively, our current study suggests that IL-27 activates an alternative MAPK signaling pathway via TAK-1 in macrophages.
SPTBN1 was previously implicated as a supportive host factor for HIV infection in the screen performed on HeLa-derived TZM-bl cells (Brass et al., 2008). Our results further demonstrate that SPTBN1 is required to support HIV-1 infection of macrophages. Moreover, SPTBN1 expression appears to correlate with the capacity of different primary cells to support HIV-1 infection (Fig. 4 F). This correlation, is not perfect, however, as exemplified by the fact that knock-down of SPTBN1 in CD4+ T cells does not restrict HIV-1 infection as it does in macrophages (unpublished data), and therefore the phenotype appears to be macrophage specific. Studies on the HIV-1 restriction factor SAMHD1 have demonstrated how a similar cell specificity issue is explained. SAMHD1 is highly expressed in restricting cells such as macrophages and dendritic cells, although it is largely missing in permissive cells such Jurkat and SupT1 cells (Laguette et al., 2011). This correlation is also not absolute. For example, 293T cells are permissive to HIV-1 infection but do express SAMHD1 (Hrecka et al., 2011). It is now understood that SAMHD1 restricts HIV reverse transcription by reducing intracellular dNTP pool (Lahouassa et al., 2012), and therefore this phenotype is only evident in the cells with relatively low concentrations of dNTPs such as monocytic cells. During the preparation of our manuscript, two groups reported that primary CD4+ T cells, both resting and activated, also express SAMHD1 and that differential dNTP concentrations may explain their contrast susceptibility to HIV-1 infection (Baldauf et al., 2012; Descours et al., 2012). Therefore, the presence or absence of a host factor alone is usually insufficient to perfectly explain its phenotype in different cells. In the case of SPTBN1, our current results indicate that SPTBN1 may facilitate a macrophage-specific post-entry viral activity that could be dispensable in CD4+ T cells, or alternatively, that in the absence of SPTBN1 HIV-1 can use redundant host factors to complete its life cycle in CD4+ T cells.
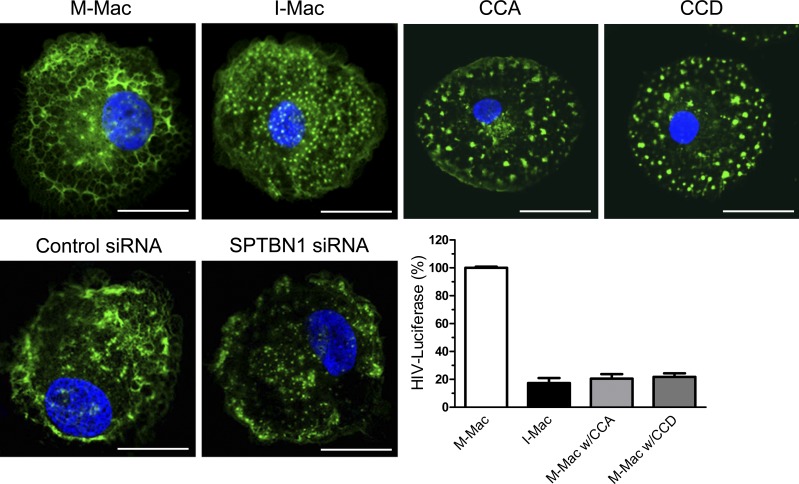
It is unclear by what exact mechanism SPTBN1 promotes the HIV-1 life cycle in macrophages. Nevertheless, our results indicate that a block to HIV-1 infection of I-Mac is present after entry and before the completion of reverse transcription, which puts SPTBN1 at a position to facilitate an early event of HIV-1 infection. Our results also indicate that SPTBN1 associates with HIV-1 gag CA p24 and MA p17. Mass spectrometry results from another group have shown that SPTBN1 also binds to Tat (Bushman et al., 2009). However, the interaction between SPTBN1 and Tat may be irrelevant to the block in I-Mac because Tat mainly enhances transcription of HIV-1 genome at a later stage. At present, we are not sure how SPTBN1 can bind to both p24 and p17, and we are constructing a series of mutants that will help us to determine the amino acid motif for the interactions in future studies, and will also help us understand the contribution of each interaction to HIV-1 infection. Although we currently have no direct evidence to support that these interactions have bona fide effects on the HIV-1 life cycle, it is tempting to speculate that such interactions may provide some possible mechanisms by which SPTBN1 may facilitate efficient reverse transcription. SPTBN1, with the N-terminal, actin-binding domain, has been implicated to serve as a scaffold protein for actin cytoskeleton (Hartwig, 1994; Davis et al., 2009). In fact, we have validated that SPTBN1 is required to maintain actin cytoskeletal structure in macrophages. IL-27 treatment or knockdown of SPTBN1 results in a severe damage to the cytoskeleton structure (Fig. 10). We quantitated a total of 50 I-Mac cells and found that 50% of the F-actin fibers were replaced with granular or punctate actin structures, an effect similar to the actin depolymerization caused by the treatment of cytochalasin A (CCA) and cytochalasin D (CCD). It should be also noted that the remaining 50% of the F-actin fibers, especially those close to the PM region, were still intact. Recent evidence has shown that phagocytic cups may still form, even with partially impaired F-actin structure (van Zon et al., 2009). This gives a possible explanation for the normal phagocytosis of I-Mac. Nevertheless, such actin disarrangement does lead to a dramatic reduction in HIV-1 infection (Fig. 10). The interaction between HIV-1 virus particles and the actin cytoskeleton has been reported to be important for virus entry, uncoating, and reverse transcription, as well as for virus trafficking to the nucleus (Bukrinskaya et al., 1998; Iyengar et al., 1998; McDonald et al., 2002; Viard et al., 2002; Arhel, 2006, 2010). SPTBN1, along with other spectrins, forms a lattice which supports a dense actin network beneath the cytoplasmic surface of the plasma membrane. In this study, SPTBN1 has been identified as a gag-interacting protein. In the context of macrophage infection, the colocalization of SPTBN1 and virus particles is also evident (Fig. 7). Thus, SPTBN1 may serve as an intracellular adaptor for HIV-1 viral particles after entry. The following uncoating process may be facilitated by association of the viral capsid core to the cytoskeleton through the interaction between SPTBN1 and CA. In this case, a simple cell-free system to study HIV uncoating will be of great help to initiate our further study (Shah and Aiken, 2011). Or alternatively, the gag MA within the reverse transcription complex (RTC) associates with SPTBN1 to locate the actin cytoskeleton to complete cDNA synthesis. In addition, if the preintegration complex (PIC) has to move along the cytoskeleton, from actin filaments to microtubules, to eventually reach the nucleus (Arhel et al., 2006; McDonald et al., 2002), disruption of the actin structure, due to the absence of SPTBN1, could make the HIV-1 PIC lost in the cytosol. Collectively, our current findings suggest a model that SPTBN1 mediates the interaction of virus particles and the actin cytoskeleton to facilitate an early step of the HIV-1 life cycle.
Figure 10.
Actin disarrangement impacts HIV-1 infection. Macrophages were treated with IL-27 (50 ng/ml), cytochalasin A (5 µM), or cytochalasin D (5 µM), or transfected with control or SPTBN1 siRNA (50 pmol). F-actin was labeled with fluorescein phalloidin and nuclei were stained with DAPI. Bars, 10 µm. Cells were infected with HIV-LUC-V and luciferase activity was measured 4 d after infection. Data shown represent means ± SE of three independent experiments.
MATERIALS AND METHODS
Plasmids.
pNL4-3.Luc were obtained from the AIDS reagent program of the National Institute of Allergy and Infectious Disease (NIAID), National Institutes of Health (NIH; Connor et al., 1995; He et al., 1995). It contains a luciferase reporter gene in the position of Nef and a frameshift mutation at 59 of Env. pNL4-3-deltaE-EGFP was obtained from the AIDS reagent program of NIAID (Zhang et al., 2004). It expresses truncated Env-EGFP fusion protein. pLTR-VSV, a VSV-G expression vector, was used to pseudotype envelope-deficient HIV-LUC and HIV-EGFP viruses. Construction of SPTBN1 expression vectors was conducted as follows: SPTBN1 cDNA was synthesized from 5 µg total macrophage RNA using Superscript First Stand Synthesis System for RT-PCR (Invitrogen). SPTBN1 cDNA was PCR amplified, and then cloned into pCMV5a and pCMV5b vectors (Sigma-Aldrich) to generate pSPTBN1 and pSPTBN1.FLAG, respectively. pNL4.3pb has been previously described (Imamichi et al., 2000). pNL4-3KFS and pVpr-GFP plasmids were obtained as a gift from E. Freed (Frederick National Laboratory for Cancer Research, Frederick, MD). pNL4-3KFS is an Env-HIV-1 NL4.3 clone with a frameshift mutation at the 5′ end of the env gene (Freed et al., 1992), and we further mutated the Vpr gene of this clone with a stop codon to generate pNL4-3KFS-Vpr-. pVpr-GFP expresses Vpr-GFP fusion proteins, which can be efficiently incorporated into HIV-1 virions (Campbell et al., 2007). pGag-EGFP was obtained from the NIAID AIDS reagent program (Schwartz et al., 1992; Hermida-Matsumoto and Resh, 2000; Lindwasser and Resh, 2002; Perlman and Resh, 2006). Codon-optimized CA, MA, or NC gene of HIV-1NL4.3 fused with a C-terminal HA tag was synthesized (Invitrogen) and subcloned into a pIRES2-ZsGreen1 expression vector (Takara Bio Inc.). The Nef expression vector has been previously described (Dai and Stevenson, 2010). RIG-I cDNA was PCR amplified, and then cloned into the pCMV5b vector (Sigma-Aldrich) to generate pRIG-I.FLAG.
Cells and viruses.
CD14+ monocytes were purified from PBMCs of healthy donors using CD14 MicroBeads (Miltenyi Biotec) according to the manufacturer’s instructions, as previously described (Fakruddin et al., 2007). To generate MDMs, isolated CD14+ monocytes were plated in 24-well plates at 0.7 × 106 cells/well. Monocytes were stimulated with 25 ng/ml M-CSF (R&D Systems) alone or in combination with 25–100 ng/ml IL-27 (R&D Systems) in macrophage serum-free medium (Invitrogen) for 7 d. MDMs were then maintained in DMEM (Invitrogen) containing 10% FBS (HyClone Laboratories), 25 mM Hepes (Quality Biology), and 5 µg/ml Gentamicin (Invitrogen) before use in experiments. 293T cells were obtained from H. Gottlinger (University of Massachussetts Medical School, Worcester, MA) and maintained in DMEM containing 10% FBS. HIV-1BAL virus stocks were obtained from Advanced Biotechnologies, Inc. HIV-LUC-V or HIV-EGFP-V were produced by co-transfecting 293T cells with pNL4-3.Luc or pNL4-3-deltaE-EGFP (10 µg) and pLTR-VSVG (1 µg) using a TransIT-LT1 transfection kit (Mirus). VSV-G–pseudotyped GFP-labeled HIV-1 virus HIV-Vpr.GFP-V was produced by co-transfecting 293T cells with pNL4-3KFS-Vpr- (10 µg), pVpr-GFP (3 µg), and pLTR-VSVG (1 µg) using the TransIT-LT1 transfection kit. Virus containing supernatants were harvested 48 h after transfection and filtered with 0.22-µm Steriflip Filter Units (Millipore). Virus particles were pelleted by ultracentrifugation through a 20% sucrose cushion and resuspended in PBS.
FACS analysis.
Expression of CD14, CD11b, EMR-1, CD206, CD4, and CCR5 on M-Mac or I-Mac was analyzed by flow cytometry. Cells were stained for 15 min at room temperature in PBS containing 2% BSA with the following antibodies: PE-conjugated anti-CD14 (BD), APC-conjugated anti-CD11b (BD), FITC-conjugated anti-F4/80 (Abcam), FITC-conjugated anti-CD206 (BD), FITC-conjugated anti-CD4 (BD), and PE-conjugated anti-CCR5 (BD). The cell preparations were analyzed with a FACSCalibur flow cytometer (BD). Positive and negative events were determined with matching isotype controls.
HIV binding assay.
Macrophages were incubated with virus at 4°C for 1 h to allow binding, but not membrane fusion, as previously described (McClure et al., 1992), and then washed to remove unbound virus. Total RNA was isolated and reverse transcription of viral RNA was performed as previously described (Brann et al., 2006). The levels of virus binding were estimated with copy numbers of viral RNA measured with quantitative RT-PCR.
HIV-1 replication assay.
To infect MDMs, 0.7 × 106 cells were incubated with 700 TCID50 HIV-1BAL for 2 h. Cells were washed and maintained in DMEM containing 10% FBS for 24 d. Half of the culture supernatants were replaced with fresh medium every 3 d. Viral replication was gauged from p24 levels in culture supernatants using an HIV-1 p24 ELISA kit (Perkin-Elmer).
HIV single-round infection.
MDMs were incubated with HIV-EGFP-V or HIV-LUC-V (1 µg/ml p24) for 2 h. Cells were washed and then cultured for 4 d. The EGFP expression of infected cells was analyzed with a FACSCalibur flow cytometer (BD). P24 antigen amount in supernatants was measured using an HIV-1 p24 ELISA kit (Perkin-Elmer). Luciferase activity was measured with Bright-Glo luciferase assay system (Promega). Quantitative analysis of viral cDNA late products was performed as previously described (Sharova et al., 2008). Copy number estimates of cDNA late product were determined by real-time PCR on an IQ5 RT-PCR detection system (Bio-Rad Laboratories). These copy numbers were normalized by the cell numbers determined by real-time PCR using CCR5-specific primers.
Cell fusion.
M-Mac was stained with CellTracker Red and I-Mac was stained with CellTracker Green (Invitrogen) following the manufacturer’s instructions. Cell fusion between M-Mac and I-Mac was achieved by using PEG-1450 as previously described (Sharova et al., 2008). Cells were then plated in a 100-mm culture dish (107 cells/dish) and cultured for 48 h. Double-stained cells were sorted with a FACSAria flow cytometer (BD).
Microarray analysis.
DNA microarray assay was performed using the Affymetrix GeneChip System (Affymetrix). The Affymetrix Human Exon 1.0 ST Array containing 1.4 million probe sets was used. Total cellular RNA was extracted from M-Mac and I-Mac with an RNeasy Isolation kit (QIAGEN) and quantitated following the manufacturer’s protocols (Affymetrix). Terminal labeling and hybridization, array wash, stain, and scan were processed according to the Affymetrix recommended standard protocol. Intensity data were processed and summarized to gene level with Partek (Partek). Differentially expressed gene candidates were selected for verification with an absolute fold change difference >5.0. The microarray data are available at the Gene Expression Omnibus under accession no. GSE43595
Quantitative RT-PCR.
Cells were washed using cold PBS, and RNA was isolated from the cells using the RNeasy Isolation kit (QIAGEN). Total cDNA was synthesized using TaqMan reverse transcription reagents (Applied Biosystems). SPTBN1 expression levels were measured using quantitative RT-PCR on CFX96 Real-Time system (Bio-Rad Laboratories). Copy numbers of SPTBN1 were normalized by GAPDH. Probes specific for SPTBN1 (Hs00162271_m1) and GAPDH (Hs99999905_m1) were purchased from Applied Biosystems.
siRNA Silencing of SPTBN1 in Macrophages.
SPTBN1 specific siRNA (SR304571) and scrambled control siRNA (SR30004) were purchased from OriGene. Macrophages were transfected with siRNA using Lipofectamine 2000 (Invitrogen). In brief, macrophages (0.7 × 106 /well) were seeded in a 24-well plate. 50 pmol siRNA and 3 µl Lipofectamine 2000 were mixed well in 350 µl plain DMEM. The transfection mixture was incubated at room temperature for 30 min. Macrophages were washed with plain DMEM once and incubated with the transfection mixture at 37°C. After 2 h, the transfection mixture was removed and replaced with DMEM containing 10% FBS. SPTBN1 knockdown was assessed by RT-PCR and Western blotting 48 h after transfection.
DNA transfection of macrophages.
DNA transfection of macrophages was performed using a P3 Primary Cell kit (V4SP-3096) in 96-well Shuttle Nucleofector system (Lonza). To transfect macrophages, 105/well cells were mixed with 0.4 µg plasmid DNA in 20 µl P3 primary cell solution, and program 96-DP-148 was applied for nucleofection. SPTBN1 expression was assessed by Western blotting 48 h after transfection.
Western blotting.
Whole-cell lysates were used to detect SPTBN1 expression by Western blotting. Samples were loaded on 3–8% Tris-Acetate NuPAGE gels (Invitrogen) and analyzed by immunoblotting with a rabbit anti-SPTBN1 antibody (Abcam). Other antibodies used in this study include anti-SAMHD1 (Abcam), anti-APOBEC3G (Abcam), anti-BST2 serum (AIDS Reagent Program) (Miyagi et al., 2009), anti–β-actin (Santa Cruz Biotechnology, Inc.), and anti-GAPDH (Abcam). Intensity of Western blot bands was analyzed with the ImageJ (NIH).
Coimmunoprecipitation.
293T cells were transfected with a FLAG-tagged SPTBN1 expression vector along with vectors expressing gag p55, HA-tagged CA p24, MA p17, NC p7, or Nef by using TransIT-LT1 transfection reagents (Mirus). The transfected cells were lysed after 48 h in Pierce IP Lysis Buffer (Thermo Fisher Scientific) with 10% glycerol. The lysates were centrifuged at 18,000× g for 10 min to remove artifacts caused by incomplete solubilization. The supernatants were immunoprecipitated with anti-FLAG agarose (Sigma-Aldrich) or Protein G agarose (Sigma-Aldrich) coated with anti-HA antibody (Covance). Immunoprecipitates were analyzed by immunoblotting with anti-FLAG antibody (Cell Signaling Technology), anti-HIV-1 gag p55 (Abcam), or anti-HA antibody (Covance).
Confocal microscopy.
To monitor the colocalization of Gag p55 and SPTBN1, macrophages were transfected with pGag-EGFP using a P3 Primary Cell kit (V4SP-3096; Lonza) in 96-well Shuttle Nucleofector system (Lonza). In brief, 0.106/well cells were mixed with 0.4 µg DNA plasmid in 20 µl of P3 primary cell solution and program DP-148 was applied for nucleofection. Transfected macrophages were transferred into glass-bottomed 24-well plates (MatTek) and cultured in DMEM-10% FCS medium for 48 h. To monitor the colocalization of incoming HIV-1 viral particles and SPTBN1, 0.7 × 106 macrophages per well were cultured in glass-bottomed 24-well plates (MatTek), incubated with HIV-Vpr.GFP-V for 30 min, and washed extensively to remove free virions. To label SPTBN1, cells were incubated with 0.4 ml Fc Blocker (Innovex) for 1 h at room temperature and fixed using 3.7% methanol-free formaldehyde in PBS for 15 min at room temperature. The cells were then permeabilized and blocked with 0.2% Triton X-100 and 1% BSA in PBS for 30 min, incubated with the anti-SPTBN1 antibody for 1 h, washed, and incubated with Alexa Fluor 555–conjugated anti–rabbit IgG (Cell Signaling Technology) for 30 min at room temperature. Samples were air dried and maintained in a ProLong Gold with DAPI reagent (Invitrogen). The cells were imaged using a FluoView FV1000 Confocal Microscope (Olympus). Photographs were obtained with a PLAPON objective (60 × 1.42 NA) under oil immersion. To quantitate the extent of colocalization, we calculated the Pearson correlation coefficient (R) value, a standard measurement of colocalization (Manders et al., 1993). The R value was calculated using the JAcoP plug-in of the ImageJ software (NIH).
Phagocytosis assay.
Phagocytosis assay was performed using a pHrodo E. coli BioParticles Phagocytosis kit (Invitrogen). The E. coli particles are nonfluorescent at neutral pH but emit strong fluorescence in an acidic environment after phagocytosis. 106 macrophages were incubated with 20 µl particles at 37°C for 30 min. A negative control was set on ice to prevent phagocytosis. Cells were washed and analyzed by flow cytometry.
Chemotaxis assay.
Migration of macrophages to SDF-1α (R&D System) or RANTES (R&D System) was assessed using a 48-well microchemotaxis chamber technique as previously described (Falk et al., 1980). The results are presented as number of cells per high power field.
Multiplex cytokine assay.
The concentrations of multiple proinflammatory cytokines in the culture supernatants of M-Mac and I-Mac were analyzed with a Milliplex assay kit (Millipore). Data were collected using Luminex100 instrumentation (Luminex). The sample concentrations were determined using a Logistic-5PL regression method with the Bio-Plex manager 5.0 software (Bio-Rad Laboratories) following the manufacture’s protocol. The limits of detection for undetected cytokines are as follows: IL-1β, 0.7 pg/ml; IL-5, 0.1 pg/ml; IL-6, 0.4 pg/ml; IL-12β, 12.3 pg/ml; IL-13, 0.3 pg/ml; IL-17, 0.4 pg/ml; CSF3, 3.9 pg/ml; Fractalkine, 7.6 pg/ml; TGFα, 1.4 pg/ml; sIL-2Rα, 7.5 pg/ml.
Superoxide production assay.
M-Mac and I-Mac were seeded into 48-well plates (0.4 × 106 cells/well) in 100 µl Krebs–Ringer phosphate buffer (pH 7.4) containing 5.5 mM glucose. Cells were then incubated with the buffer alone or with 100 ng/ml of PMA (Sigma Aldrich) for 30 min at 37°C. Production of hydrogen peroxide (H2O2) was quantitated using Amplex Red Hydrogen Peroxide/Peroxidase Assay kit (Invitrogen) following the manufacture’s protocol.
SIV infection assay.
SIV-GFP was constructed by inserting GFP in the position of Nef. SIV-GFP virus stocks were produced by transfecting 293T cells with SIV-GFP, and were normalized by SIV p27 protein level. To infect MDMs, cells were incubated with SIV-GFP (125 ng p27) for 2 h. Cells were washed and maintained in DMEM containing 10% FBS for 7 d. Half of the culture supernatants were replaced with fresh medium on Day 4. GFP expression was visualized with fluorescent microscopy. Viral replication was gauged from p27 levels in culture supernatants using an SIV p27 ELISA kit (ZeptoMetrix).
HIV-2 infection assay.
HIV-2 virus was isolated from culture supernatants of an HIV-2–infected H9 cell line (MVP-11971). Virus particles were pelleted by ultracentrifugation through a 20% sucrose cushion and resuspended in PBS. Virus stocks were semiquantitated using a p27 ELISA kit (ZeptoMetrix). M-Mac and I-Mac (1.2 × 105 cells/well) were seeded in 96-well plates and inoculated with HIV-2 (0.5 ng p27 /well) for 2 h at 37°C. HIV-2 replication was determined by measuring the amount of p27 antigen in the culture supernatants 2 wk after infection.
Influenza infection and hemagglutination (HA) assay.
Viral stock of influenza A/Victoria/210/2009 H3N2 strain was produced in chicken eggs and purified from allantoic fluid. M-Mac and I-Mac (3.5 × 106/well) were seeded in 6-well plates and washed twice with PBS to remove residual FBS before infection. Viral stocks prepared at 10-, 100-, and 1,000-fold dilution in serum-free medium were used to inoculate macrophages for 1 h. Cells were then washed with PBS and cultured in DMEM containing 2% FBS and 2 µg/ml TCPK-treated trypsin. After 48 h, culture supernatants were collected for the following HA assay. HA assay was performed in 96-well V-bottom microtiter plates. Virus containing cell culture supernatant was serially diluted from 1:2 to 1:2,048. 0.5% Turkey erythrocytes (Lampire Biological Laboratories) were then added at an equal volume. After 30 min, HA titer was determined and the highest dilution of the virus that cause complete hemagglutination is considered the HA titration end point.
HSV-1, HSV-2, and KSHV infection assay.
HSV-1 and HSV-2 were obtained from Advanced Biotechnologies Inc. and virus titer was determined by plaque-forming assay using Vero cells (American Type Culture Collection [ATCC]; Andrei et al., 2000). KSHV was prepared from BCLB-1 cells (ATCC) stimulated with 20 ng/ml PMA (Sigma Aldrich) and virus stocks were quantitated by viral DNA copy numbers measured by real-time PCR (Wu et al., 2006). Macrophages (1.5 × 106 cells/well) were seeded in 6-well plates and incubated with HSV-1 (MOI = 1), HSV-2 (MOI = 1), or KSHV (50 copies /cell) for 1 h. Heat-inactivated viruses were used as a negative control. Infected cells were cultured for 24 h and genomic DNA was extracted using QIAamp DNA Blood Mini kit (QIAGEN). Copy number estimates of viral DNA were determined by real-time PCR on a CFX96 RT-PCR detection system (Bio-Rad Laboratories). Copy numbers were normalized by the cell numbers determined by real-time PCR using RNaseP-specific primers.
Statistical analysis and image processing.
Non-image statistical analysis was performed using GraphPad Prism 5 software. Error bars were SD or SE from means as indicated. An unpaired Student’s t test was used and P values lower than 0.05 were considered significant where indicated. To analyze microscope images, 10 images of M-Mac and I-Mac were processed as follows: the SPTBN1 channel was segmented into background and cellular regions using K-means classification with three output classes and treating the lowest intensity class as background. The mean background intensity was calculated and subtracted from pixel intensities in the cell regions. The pixel intensities in cell regions were rescaled such that the mean pixel intensity in cell regions was 1.0. The GFP channel was similarly segmented with the highest class representing the HIV particles. The rescaled SPTBN1 intensity at the location of each detected HIV particle was recorded. The difference in the distribution of SPTBN1 intensities at the locations of HIV particles for M-MAC and I-MAC was determined by the Kolmogorov-Smirnov test.
Online supplemental material.
Table S1 shows differential gene expression profile of I-Mac. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120572/DC1.
Supplementary Material
Acknowledgments
We thank Eric Freed for providing plasmids and helpful discussions; Heinrich Gottlinger for providing virus-producing cells; Hongyan Sui and De Yang for suggestions on the experiments; Hiromi Imamichi and Doug Kuhns for technical support; Alicia Troxell for assistance with experimental supplies; and Howard Young, Sanjay Swaminathan, and Raphael Oguariri for critical review of the manuscript. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pNL4-3.Luc.R–E– from Dr. Nathaniel Landau; pNL4-3-deltaE-EGFP (Cat# 11100) from Drs. Haili Zhang, Yan Zhou, and Robert Siliciano; Anti-Bst-2 (Cat# 11722) from Drs. Klaus Strebel and Amy Andrew.
This project has been funded in whole or in part with federal funds from the NCI, NIH, under contract no. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by NIAID.
All authors declare no competing interests.
Footnotes
Abbreviations used:
- CA
- capsid
- gp130
- glycoprotein 130
- HIV-EGFP-V
- VSV-G-pseudotyped GFP-encoding HIV-1
- HIV-LUC-V
- VSV-G-pseudotyped HIV-luciferase virus
- I-Mac
- macrophage induced with M-CSF and IL-27
- MA
- matrix
- MDDC
- monocyte-derived dendritic cell
- MDM
- monocyte-derived macrophage
- M-Mac
- macrophages induced with M-CSF alone
- NC
- nucleocapsid
- PM
- plasma membrane
- SPTBN1
- spectrin β nonerythrocyte 1
- VSV-G
- vesicular stomatitis virus G glycoprotein
References
- Andrei G., Fiten P., De Clercq E., Snoeck R., Opdenakker G. 2000. Monitoring drug resistance for herpesviruses. Methods Mol. Med. 24:151–169 [DOI] [PubMed] [Google Scholar]
- Arhel N. 2010. Revisiting HIV-1 uncoating. Retrovirology. 7:96 10.1186/1742-4690-7-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhel N., Genovesio A., Kim K.A., Miko S., Perret E., Olivo-Marin J.C., Shorte S., Charneau P. 2006. Quantitative four-dimensional tracking of cytoplasmic and nuclear HIV-1 complexes. Nat. Methods. 3:817–824 10.1038/nmeth928 [DOI] [PubMed] [Google Scholar]
- Baldauf H.M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., et al. 2012. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat. Med. 18:1682–1687 10.1038/nm.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender H., Wiesinger M.Y., Nordhoff C., Schoenherr C., Haan C., Ludwig S., Weiskirchen R., Kato N., Heinrich P.C., Haan S. 2009. Interleukin-27 displays interferon-gamma-like functions in human hepatoma cells and hepatocytes. Hepatology. 50:585–591 10.1002/hep.22988 [DOI] [PubMed] [Google Scholar]
- Brann T.W., Dewar R.L., Jiang M.K., Shah A., Nagashima K., Metcalf J.A., Falloon J., Lane H.C., Imamichi T. 2006. Functional correlation between a novel amino acid insertion at codon 19 in the protease of human immunodeficiency virus type 1 and polymorphism in the p1/p6 Gag cleavage site in drug resistance and replication fitness. J. Virol. 80:6136–6145 10.1128/JVI.02212-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L., Dykxhoorn D.M., Benita Y., Yan N., Engelman A., Xavier R.J., Lieberman J., Elledge S.J. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 319:921–926 10.1126/science.1152725 [DOI] [PubMed] [Google Scholar]
- Bukrinskaya A., Brichacek B., Mann A., Stevenson M. 1998. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J. Exp. Med. 188:2113–2125 10.1084/jem.188.11.2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F.D., Malani N., Fernandes J., D’Orso I., Cagney G., Diamond T.L., Zhou H., Hazuda D.J., Espeseth A.S., König R., et al. 2009. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 5:e1000437 10.1371/journal.ppat.1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell E.M., Perez O., Melar M., Hope T.J. 2007. Labeling HIV-1 virions with two fluorescent proteins allows identification of virions that have productively entered the target cell. Virology. 360:286–293 10.1016/j.virol.2006.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.G., Scarpa A., Eddy R.L., Byers M.G., Harris A.S., Morrow J.S., Watkins P., Shows T.B., Forget B.G. 1993. Cloning of a portion of the chromosomal gene and cDNA for human beta-fodrin, the nonerythroid form of beta-spectrin. Genomics. 17:287–293 10.1006/geno.1993.1323 [DOI] [PubMed] [Google Scholar]
- Connor R.I., Chen B.K., Choe S., Landau N.R. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 206:935–944 10.1006/viro.1995.1016 [DOI] [PubMed] [Google Scholar]
- Dai L., Stevenson M. 2010. A novel motif in HIV-1 Nef that regulates MIP-1beta chemokine release in macrophages. J. Virol. 84:8327–8331 10.1128/JVI.00741-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L., Abdi K., Machius M., Brautigam C., Tomchick D.R., Bennett V., Michaely P. 2009. Localization and structure of the ankyrin-binding site on beta2-spectrin. J. Biol. Chem. 284:6982–6987 10.1074/jbc.M809245200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descours B., Cribier A., Chable-Bessia C., Ayinde D., Rice G., Crow Y., Yatim A., Schwartz O., Laguette N., Benkirane M. 2012. SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology. 9:87 10.1186/1742-4690-9-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marzio P., Tse J., Landau N.R. 1998. Chemokine receptor regulation and HIV type 1 tropism in monocyte-macrophages. AIDS Res. Hum. Retroviruses. 14:129–138 10.1089/aid.1998.14.129 [DOI] [PubMed] [Google Scholar]
- Eisert V., Kreutz M., Becker K., Königs C., Alex U., Rübsamen-Waigmann H., Andreesen R., von Briesen H. 2001. Analysis of cellular factors influencing the replication of human immunodeficiency virus type I in human macrophages derived from blood of different healthy donors. Virology. 286:31–44 10.1006/viro.2001.0940 [DOI] [PubMed] [Google Scholar]
- Evans D.T., Serra-Moreno R., Singh R.K., Guatelli J.C. 2010. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18:388–396 10.1016/j.tim.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakruddin J.M., Lempicki R.A., Gorelick R.J., Yang J., Adelsberger J.W., Garcia-Pineres A.J., Pinto L.A., Lane H.C., Imamichi T. 2007. Noninfectious papilloma virus-like particles inhibit HIV-1 replication: implications for immune control of HIV-1 infection by IL-27. Blood. 109:1841–1849 10.1182/blood-2006-02-001578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk W., Goodwin R.H., Jr, Leonard E.J. 1980. A 48-well micro chemotaxis assembly for rapid and accurate measurement of leukocyte migration. J. Immunol. Methods. 33:239–247 [DOI] [PubMed] [Google Scholar]
- Frank A.C., Zhang X., Katsounas A., Bharucha J.P., Kottilil S., Imamichi T. 2010. Interleukin-27, an anti-HIV-1 cytokine, inhibits replication of hepatitis C virus. J. Interferon Cytokine Res. 30:427–431 10.1089/jir.2009.0093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed E.O., Delwart E.L., Buchschacher G.L., Jr, Panganiban A.T. 1992. A mutation in the human immunodeficiency virus type 1 transmembrane glycoprotein gp41 dominantly interferes with fusion and infectivity. Proc. Natl. Acad. Sci. USA. 89:70–74 10.1073/pnas.89.1.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.P. 2007. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 5:253–263 10.1038/nrmicro1541 [DOI] [PubMed] [Google Scholar]
- Goila-Gaur R., Strebel K. 2008. HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology. 5:51 10.1186/1742-4690-5-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell-Wild T., Vázquez N., Jin W., Rangel Z., Munson P.J., Wahl S.M. 2009. Interleukin-27 inhibition of HIV-1 involves an intermediate induction of type I interferon. Blood. 114:1864–1874 10.1182/blood-2009-03-211540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot F., Welsch S., Sattentau Q.J. 2008. Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood. 111:4660–4663 10.1182/blood-2007-12-130070 [DOI] [PubMed] [Google Scholar]
- Guzzo C., Che Mat N.F., Gee K. 2010a. Interleukin-27 induces a STAT1/3- and NF-kappaB-dependent proinflammatory cytokine profile in human monocytes. J. Biol. Chem. 285:24404–24411 10.1074/jbc.M110.112599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo C., Hopman W.M., Che Mat N.F., Wobeser W., Gee K. 2010b. Impact of HIV infection, highly active antiretroviral therapy, and hepatitis C coinfection on serum interleukin-27. AIDS. 24:1371–1374 [DOI] [PubMed] [Google Scholar]
- Hartwig J.H. 1994. Actin-binding proteins 1: spectrin superfamily. Protein Profile. 1:706–778 [PubMed] [Google Scholar]
- He J., Choe S., Walker R., Di Marzio P., Morgan D.O., Landau N.R. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermida-Matsumoto L., Resh M.D. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J. Virol. 74:8670–8679 10.1128/JVI.74.18.8670-8679.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K., Hao C., Gierszewska M., Swanson S.K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M.P., Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 474:658–661 10.1038/nature10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamichi T., Berg S.C., Imamichi H., Lopez J.C., Metcalf J.A., Falloon J., Lane H.C. 2000. Relative replication fitness of a high-level 3′-azido-3′-deoxythymidine-resistant variant of human immunodeficiency virus type 1 possessing an amino acid deletion at codon 67 and a novel substitution (Thr—>Gly) at codon 69. J. Virol. 74:10958–10964 10.1128/JVI.74.23.10958-10964.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamichi T., Yang J., Huang D.W., Brann T.W., Fullmer B.A., Adelsberger J.W., Lempicki R.A., Baseler M.W., Lane H.C. 2008. IL-27, a novel anti-HIV cytokine, activates multiple interferon-inducible genes in macrophages. AIDS. 22:39–45 10.1097/QAD.0b013e3282f3356c [DOI] [PubMed] [Google Scholar]
- Iyengar S., Hildreth J.E., Schwartz D.H. 1998. Actin-dependent receptor colocalization required for human immunodeficiency virus entry into host cells. J. Virol. 72:5251–5255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliolias G.D., Ivashkiv L.B. 2008. IL-27 activates human monocytes via STAT1 and suppresses IL-10 production but the inflammatory functions of IL-27 are abrogated by TLRs and p38. J. Immunol. 180:6325–6333 [DOI] [PubMed] [Google Scholar]
- Kamiya S., Owaki T., Morishima N., Fukai F., Mizuguchi J., Yoshimoto T. 2004. An indispensable role for STAT1 in IL-27-induced T-bet expression but not proliferation of naive CD4+ T cells. J. Immunol. 173:3871–3877 [DOI] [PubMed] [Google Scholar]
- Kastelein R.A., Hunter C.A., Cua D.J. 2007. Discovery and biology of IL-23 and IL-27: related but functionally distinct regulators of inflammation. Annu. Rev. Immunol. 25:221–242 10.1146/annurev.immunol.22.012703.104758 [DOI] [PubMed] [Google Scholar]
- Kedzierska K., Crowe S.M. 2001. Cytokines and HIV-1: interactions and clinical implications. Antivir. Chem. Chemother. 12:133–150 [DOI] [PubMed] [Google Scholar]
- Koenig S., Gendelman H.E., Orenstein J.M., Dal Canto M.C., Pezeshkpour G.H., Yungbluth M., Janotta F., Aksamit A., Martin M.A., Fauci A.S. 1986. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 233:1089–1093 10.1126/science.3016903 [DOI] [PubMed] [Google Scholar]
- Kojima H., Sasaki T., Ishitani T., Iemura S., Zhao H., Kaneko S., Kunimoto H., Natsume T., Matsumoto K., Nakajima K. 2005. STAT3 regulates Nemo-like kinase by mediating its interaction with IL-6-stimulated TGFbeta-activated kinase 1 for STAT3 Ser-727 phosphorylation. Proc. Natl. Acad. Sci. USA. 102:4524–4529 10.1073/pnas.0500679102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 474:654–657 10.1038/nature10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahouassa H., Daddacha W., Hofmann H., Ayinde D., Logue E.C., Dragin L., Bloch N., Maudet C., Bertrand M., Gramberg T., et al. 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat. Immunol. 13:223–228 10.1038/ni.2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambotte O., Taoufik Y., de Goër M.G., Wallon C., Goujard C., Delfraissy J.F. 2000. Detection of infectious HIV in circulating monocytes from patients on prolonged highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 23:114–119 [DOI] [PubMed] [Google Scholar]
- Lindwasser O.W., Resh M.D. 2002. Myristoylation as a target for inhibiting HIV assembly: unsaturated fatty acids block viral budding. Proc. Natl. Acad. Sci. USA. 99:13037–13042 10.1073/pnas.212409999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders E.M.M., Verbeek F.J., Aten J.A. 1993. Measurement of Colocalization of Objects in Dual-Color Confocal Images. J. Microsc. 169:375–382 10.1111/j.1365-2818.1993.tb03313.x [DOI] [PubMed] [Google Scholar]
- McClure M.O., Moore J.P., Blanc D.F., Scotting P., Cook G.M., Keynes R.J., Weber J.N., Davies D., Weiss R.A. 1992. Investigations into the mechanism by which sulfated polysaccharides inhibit HIV infection in vitro. AIDS Res. Hum. Retroviruses. 8:19–26 10.1089/aid.1992.8.19 [DOI] [PubMed] [Google Scholar]
- McDonald D., Vodicka M.A., Lucero G., Svitkina T.M., Borisy G.G., Emerman M., Hope T.J. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441–452 10.1083/jcb.200203150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi E., Andrew A.J., Kao S., Strebel K. 2009. Vpu enhances HIV-1 virus release in the absence of Bst-2 cell surface down-modulation and intracellular depletion. Proc. Natl. Acad. Sci. USA. 106:2868–2873 10.1073/pnas.0813223106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naif H.M., Li S., Alali M., Sloane A., Wu L., Kelly M., Lynch G., Lloyd A., Cunningham A.L. 1998. CCR5 expression correlates with susceptibility of maturing monocytes to human immunodeficiency virus type 1 infection. J. Virol. 72:830–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil S., Martin F., Ikeda Y., Collins M. 2001. Postentry restriction to human immunodeficiency virus-based vector transduction in human monocytes. J. Virol. 75:5448–5456 10.1128/JVI.75.12.5448-5456.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil S.J., Zang T., Bieniasz P.D. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 451:425–430 10.1038/nature06553 [DOI] [PubMed] [Google Scholar]
- O’Brien W.A., Namazi A., Kalhor H., Mao S.H., Zack J.A., Chen I.S. 1994. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleotide precursors. J. Virol. 68:1258–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owaki T., Asakawa M., Fukai F., Mizuguchi J., Yoshimoto T. 2006. IL-27 induces Th1 differentiation via p38 MAPK/T-bet- and intercellular adhesion molecule-1/LFA-1/ERK1/2-dependent pathways. J. Immunol. 177:7579–7587 [DOI] [PubMed] [Google Scholar]
- Pelchen-Matthews A., Kramer B., Marsh M. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443–455 10.1083/jcb.200304008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman M., Resh M.D. 2006. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic. 7:731–745 10.1111/j.1398-9219.2006.00428.x [DOI] [PubMed] [Google Scholar]
- Pflanz S., Hibbert L., Mattson J., Rosales R., Vaisberg E., Bazan J.F., Phillips J.H., McClanahan T.K., de Waal Malefyt R., Kastelein R.A. 2004. WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J. Immunol. 172:2225–2231 [DOI] [PubMed] [Google Scholar]
- Philpott S.M. 2003. HIV-1 coreceptor usage, transmission, and disease progression. Curr. HIV Res. 1:217–227 10.2174/1570162033485357 [DOI] [PubMed] [Google Scholar]
- Raposo G., Moore M., Innes D., Leijendekker R., Leigh-Brown A., Benaroch P., Geuze H. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 3:718–729 10.1034/j.1600-0854.2002.31004.x [DOI] [PubMed] [Google Scholar]
- Rich E.A., Chen I.S., Zack J.A., Leonard M.L., O’Brien W.A. 1992. Increased susceptibility of differentiated mononuclear phagocytes to productive infection with human immunodeficiency virus-1 (HIV-1). J. Clin. Invest. 89:176–183 10.1172/JCI115559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahuddin S.Z., Rose R.M., Groopman J.E., Markham P.D., Gallo R.C. 1986. Human T lymphotropic virus type III infection of human alveolar macrophages. Blood. 68:281–284 [PubMed] [Google Scholar]
- Saville M.W., Taga K., Foli A., Broder S., Tosato G., Yarchoan R. 1994. Interleukin-10 suppresses human immunodeficiency virus-1 replication in vitro in cells of the monocyte/macrophage lineage. Blood. 83:3591–3599 [PubMed] [Google Scholar]
- Schwartz S., Campbell M., Nasioulas G., Harrison J., Felber B.K., Pavlakis G.N. 1992. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent gag expression. J. Virol. 66:7176–7182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V.B., Aiken C. 2011. In vitro uncoating of HIV-1 cores. J. Vis. Exp. 57:3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova N., Swingler C., Sharkey M., Stevenson M. 2005. Macrophages archive HIV-1 virions for dissemination in trans. EMBO J. 24:2481–2489 10.1038/sj.emboj.7600707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova N., Wu Y., Zhu X., Stranska R., Kaushik R., Sharkey M., Stevenson M. 2008. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 4:e1000057 10.1371/journal.ppat.1000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 418:646–650 10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- Smith P.D., Meng G., Salazar-Gonzalez J.F., Shaw G.M. 2003. Macrophage HIV-1 infection and the gastrointestinal tract reservoir. J. Leukoc. Biol. 74:642–649 10.1189/jlb.0503219 [DOI] [PubMed] [Google Scholar]
- Sonza S., Maerz A., Deacon N., Meanger J., Mills J., Crowe S. 1996. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J. Virol. 70:3863–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A., Hamano S., Yamanaka A., Hanada T., Ishibashi T., Mak T.W., Yoshimura A., Yoshida H. 2003. Cutting edge: role of IL-27/WSX-1 signaling for induction of T-bet through activation of STAT1 during initial Th1 commitment. J. Immunol. 170:4886–4890 [DOI] [PubMed] [Google Scholar]
- Triques K., Stevenson M. 2004. Characterization of restrictions to human immunodeficiency virus type 1 infection of monocytes. J. Virol. 78:5523–5527 10.1128/JVI.78.10.5523-5527.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zon J.S., Tzircotis G., Caron E., Howard M. 2009. A mechanical bottleneck explains the variation in cup growth during FcgammaR phagocytosis. Mol. Syst. Biol. 5:298 10.1038/msb.2009.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viard M., Parolini I., Sargiacomo M., Fecchi K., Ramoni C., Ablan S., Ruscetti F.W., Wang J.M., Blumenthal R. 2002. Role of cholesterol in human immunodeficiency virus type 1 envelope protein-mediated fusion with host cells. J. Virol. 76:11584–11595 10.1128/JVI.76.22.11584-11595.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino A., Hibbert L., Lieberman L., Wilson E., Mak T., Yoshida H., Kastelein R.A., Saris C., Hunter C.A. 2003. The IL-27R (WSX-1) is required to suppress T cell hyperactivity during infection. Immunity. 19:645–655 10.1016/S1074-7613(03)00300-5 [DOI] [PubMed] [Google Scholar]
- Villarino A.V., Huang E., Hunter C.A. 2004. Understanding the pro- and anti-inflammatory properties of IL-27. J. Immunol. 173:715–720 [DOI] [PubMed] [Google Scholar]
- Wahl R.C., Costigan V.J., Batac J.P., Chen K., Cam L., Courchesne P.L., Patterson S.D., Zhang K., Pacifici R.E. 1997. Mutation of Cys672 allows recombinant expression of activatible macrophage-stimulating protein. J. Biol. Chem. 272:15053–15056 10.1074/jbc.272.24.15053 [DOI] [PubMed] [Google Scholar]
- Wang T.H., Donaldson Y.K., Brettle R.P., Bell J.E., Simmonds P. 2001. Identification of shared populations of human immunodeficiency virus type 1 infecting microglia and tissue macrophages outside the central nervous system. J. Virol. 75:11686–11699 10.1128/JVI.75.23.11686-11699.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ye L., Hou W., Zhou Y., Wang Y.J., Metzger D.S., Ho W.Z. 2009. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 113:671–674 10.1182/blood-2008-09-175000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Poli G., Fauci A.S. 1994. Interleukin 10 blocks HIV replication in macrophages by inhibiting the autocrine loop of tumor necrosis factor alpha and interleukin 6 induction of virus. AIDS Res. Hum. Retroviruses. 10:1199–1206 10.1089/aid.1994.10.1199 [DOI] [PubMed] [Google Scholar]
- Wu W., Vieira J., Fiore N., Banerjee P., Sieburg M., Rochford R., Harrington W., Jr, Feuer G. 2006. KSHV/HHV-8 infection of human hematopoietic progenitor (CD34+) cells: persistence of infection during hematopoiesis in vitro and in vivo. Blood. 108:141–151 10.1182/blood-2005-04-1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhou Y., Alcock C., Kiefer T., Monie D., Siliciano J., Li Q., Pham P., Cofrancesco J., Persaud D., Siliciano R.F. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 78:1718–1729 10.1128/JVI.78.4.1718-1729.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Xu M., Huang Q., Gates A.T., Zhang X.D., Castle J.C., Stec E., Ferrer M., Strulovici B., Hazuda D.J., Espeseth A.S. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 4:495–504 10.1016/j.chom.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Zhu T., Muthui D., Holte S., Nickle D., Feng F., Brodie S., Hwangbo Y., Mullins J.I., Corey L. 2002. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J. Virol. 76:707–716 10.1128/JVI.76.2.707-716.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.