Abstract
Background: Efficient screening for detection of colorectal cancer (CRC) at earlier stages reduces its mortality. The purpose of this study was to investigate expression of carcinoembryonic antigen (CEA) and human telomerase reverse transcriptase (hTERT) mRNA in peripheral blood of CRC patients and to present strategies for early detection screen test. Methods: Twenty seven patients in non-metastatic stage and 27 healthy individuals were studied. Expression of CEA, hTERT mRNA and 18srRNA (18s subunit of ribosomal RNA, as reference gene) were determined based on real-time RT-PCR on 3 µg of total RNA from blood in 3 separate vials (1 µg per vial). Results: Positive expression rate of CEA mRNA (78%) and hTERT mRNA (81%) were higher in patient group (P<0.001). These rates were meaningfully higher than the results of individual vials containing only 1 µg of total RNA. Difference between Ct values of markers with 18srRNA (ΔCt) was higher in healthy group than patient one. Therefore, a ΔCt cut-off value was determined for distinguishing between true- and false-positive results. Concurrent expression of both markers was found in 67% of the patients, which was higher than healthy cases (11%). Combination of concurrent marker expression with cut-off point strategy increased specificity to 100%. Conclusion: These results showed that concurrent evaluation of marker expression and performing the test on 3 µg of samples in 3 separate vials may increase specificity and sensitivity of real-time RT-PCR for early detection of non-metastatic CRC. However, more investigations with larger numbers of samples are needed to verify these results.
Key Words: Carcinoembryonic antigen, Biomarker, Colorectal cancer
Introduction
colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and the second in females, with over 1.2 million new cases and 608,700 deaths estimated to have occurred in 2008 [1]. Patient prognosis depends on stage of CRC at the time of diagnosis. Range of 5-year survival of patients varies from 90% for localized cancer [2] to 68% and 10% for regional and metastatic cancer, respectively [2, 3]. As indicated in a study by Japanese Society for Cancer of the Colon and Rectum following a curative surgery, overall 5-year survival of patients is nearly 81% in stages 0 to III [4]. Since more than 60% of CRC is diagnosed at the symptomatic stages with lower rate of long-term survival [5], efficient screening for detection of the disease at earlier asymptomatic stages is very important. CRC mortality can be reduced by screening all men and women aged 50 years and older for CRC [6]. Although common endoscopic methods for CRC screening find adenomas in precancerous stage [2], peoples are unwilling to them because of invasive nature and serious complications. Therefore, non-invasive tests with high sensitivity and specificity must be designed for screening the disease. Detection of CRC blood markers is one of the most interesting ideas. An important source of blood biomarkers in cancer is circulating cancer cells (CTC). By determination of mRNA expression of specific tumor markers, real-time RT PCR could indirectly detect cancer cells in peripheral blood of cancer patients compared with healthy subjects [7]. The main advantages of real-time RT-PCR are high sensitivity, reliability and specificity [8]. Statistical characteristics of the assay, which might be affected by tumor cell heterogeneity, could be improved by multiple molecular marker analyses [9, 10].
Carcinoembryonic antigen (CEA) is one of the reliable target genes for detection of CTC [10]. CEA mRNA can be detected in the peripheral blood of patients with colorectal carcinoma by means of RT-PCR [11]. CEA mRNA in combination with other RNA markers has been detected in peripheral blood of patients with postoperative relapse of disease before CEA antigen raising [12]. A wide range of sensitivity and specificity of CEA mRNA detection tests have been reported in several studies [13, 14].
Telomerase activation has been detected during development of colorectal adenomas from low- to high-grade and eventually carcinoma [15]. Expression of human telomerase reverse transcriptase (hTERT) mRNA in the peripheral blood of CRC patients has been also investigated in several studies. Result of several studies showed that levels of hTERT mRNA expression were increased in CRC [16-18].
The 18srRNA (18s subunit of ribosomal RNA) is a reliable house-keeping gene which has been used in a similar previous study [17] and therefore was chosen as reference gene of this study.
The main purpose of the present study was to detect blood CEA mRNA and hTERT mRNA by real-time RT-PCR method and compare their single and combined sensitivity and specificity in CRC patients. As a secondary goal, using simple and practical plans for increasing sensitivity and specificity of these markers was also considered. Our findings may enable this method to be used for early detection of the non-metastatic disease in the future if more widespread studies with enough sample sizes verify the results. The term "early" used above as concept of the study does not exclusively mean the short time after beginning of tumor formation, but it might be considered as having enough time for complete cure of disease [19].
MATERIALS AND METHODS
Study group . The study group consisted of 27 patients (13 males and 14 Females) with colorectal carcinoma from stages I, II and III (9, 10 and 8 patients, respectively), who were admitted at Gastrointestinal Department of Beheshti Hospital (Hamadan) and General Surgery Department of Imam Khomeini Hospital (Tehran). The control group consisted of 27 healthy volunteers (13 males and 14 females), who were referred to Colonoscopy Unit of Beheshti Hospital of Hamadan and the results of the exams for the disease were negative. The patients were diagnosed by pathologic examination of specimens after colonoscopy from June 2011 until August 2012. Exclusion criteria of the patients were: 1) undergoing curative surgery, chemotherapy and/or radiotherapy before blood sampling and 2) presence of known second neoplastic disease. The protocol was approved by ethical committee of Hamadan University of Medical Sciences prior to the study. The research was carried out according to the principles set out in the Declaration of Helsinki 1964 and all subsequent revisions.
Blood sampling. Following a brief explanation of study purpose and taking an informed consent, 12 ml peripheral venous blood was obtained at the time of admission. The first 2 ml of blood was discarded to avoid false-positive results due to contamination with with skin epithelial cells. Afterward, 10 ml blood was collected in tubes containing sodium EDTA, kept on ice, transferred to the laboratory and processed within 1 hour after collection.
Lysis of red blood cells . Erythrocyte lysis buffer (Tris-HCl + saccharose + MgCl2 + Triton X-100) was made and 40 ml solution was added to 10 ml blood sample, held on ice for 30 min and then centrifuged at 3000 ×g for 20 minutes. Precipitated WBC pellet was washed by PBS solution. The second step of lysis (with 20 milliliters of the buffer) and wash by PBS solution was applied for complete elimination of hemoglobin.
RNA extraction. The RNeasy Midi Kit (Qiagen, Hilden, Germany) was used for RNA extraction according to the manufacturer's instructions. The entire isolated RNA was dissolved in 300 μl RNase-free water. Integrity of RNA was assessed by 1% agarose gel electrophoresis. RNA purity and concentration were evaluated by optical density measurement applying a Nano-Drop spectrophotometer (Bio-TeK, USA) and 3 µg of RNA in 3 aliquots (1 µg per vial) underwent reverse transcription.
Reverse transcription. QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany) was used for reverse transcription. Integrity of produced cDNA was confirmed by 2.5% agarose gel electrophoresis. Purity and concentration of cDNA were evaluated by optical density measurement as mentioned above. Quality of cDNA as template of real-time RT-PCR was confirmed by detection of reference gene (18s rRNA) expression.
Table1.
Properties and amounts of primers used in real-time RT-PCR assays of marker and reference genes.
| Property | CEA | hTERT | 18srRNA |
|---|---|---|---|
| NCBI accession number | M29540 | NM_198253 | X03205 |
| Forward primer | accctggatgtcctctatgg | tgtcacagcctgtttctgga | gtaacccgttgaaccccatt |
| (primer length) | (20) | (20) | (20) |
| (amount of use) | (10 pmol) | (15 pmol) | (10 pmol) |
| Reverse primer | caggcataggtcccgttatta | gttcttggctttcaggatgg | ccatccaatcggtagtagcg |
| (primer length) | (21) | (20) | (20) |
| (amount of use) | (10 pmol) | (15 pmol) | (10 pmol) |
| Amplicon length | 209 | 210 | 152 |
| Optimized annealing temperature | 51.2˚C | 48˚C | 53.5˚C |
CEA, carcinoembryonic antigen; hTERT, human telomerase reverse transcriptase; 18srRNA, 18s subunit of ribosomal RNA
Primers. Design of primers was done by AlleleID 7 software (Premier Biosoft Corporation, USA). Primer properties have been shown in Table 1. Primer efficacy was checked by preliminary tests on positive and negative controls.
Real-time qRT-PCR assay. To determine CEA and hTERT mRNA, real-time qRT-PCR assays were constructed using the QuantiTect® SYBR® Green PCR Kit (Qiagen, Hilden, Germany) in a CFX96 real-time PCR detection system (BioRad, USA). Positive and negative controls were used as quality controls of the process. Assessment of gene expression markers were made in 3 separate vials. It seems probable that the number of circulating tumor cells is very low, particularly in earlier stages of the disease. On the other hand, expression of gene markers is also low for the same reason; therefore, detection of these mRNA markers could be influenced by the mentioned important factors. So, the possibility of markers’ detection may be increased by making more separate vials of cDNA as template of real-time qRT-PCR assay for each sample.
Statistical analysis. The sample size was calculated based on difference between proportions of positive ratios in two groups as reported in previous similar studies [7, 20, 21]. All statistics were calculated using the SPSS software (version 10.0). Student's t-test was applied for comparison of two means. Two-sample binomial test was used to compare the positivity rate between 2 study groups. A P value <0.05 was considered significant. For calculation of marker sensitivity and specificity, results of pathological examination of sample tissues were considered as a gold standard.
Results
In total, 48% of the patients were men and 52% were women. The mean of age in patient and healthy groups were respectively 61.6 (range: 26-87 years, standard deviation: 17.68) and 62.1 years (range 25-88: years, standard deviation: 18.76), with no significant statistical difference (P = 0.911). The same results were found when this variable was compared between men and women in patient (P = 0.772) and healthy (P = 0.143) groups. Therefore, both patient and healthy groups were similar in age and sex variables and none of the factors were confounding. Location of tumor was colon (17 patients, 63%) and rectum (10 patients, 37%). Ten out of 13 male patients and 7 out of 14 female patients suffered from colon cancer and the others had rectal cancer.
Expression levels of reference gene. In order to evaluate the reference gene expression in patient and healthy groups, Ct value of the marker was determined in each sample in triplicate assays and an average of 3 values was considered as 18srRNA Ct value, which could be an index of 18srRNA gene expression. The average of calculated 18srRNA Ct values was 22.7 ± 2.85 in patient and 21.9 ± 4.61 in healthy groups. There was no significant statistical difference between the two study groups in 18s gene expression (P = 0.479). The same results were found when this index was compared between men and women and also among individuals who were older and younger than median age (64 years) in both main groups of the study. Therefore, this marker could be assumed as a reference for normalization of our biomarker expression in blood samples.
Expression of carcinoembryonic antigen and hTERT mRNA in peripheral blood of colorectal cancer patients and healthy volunteers and combined marker analysis. CEA and hTERT mRNA expressions were studied in 3 separated vials and the results were considered as positive if at least one of three vials showed typical signals. According to the findings, sensitivity of the each marker was calculated. Final results of statistical parameters were calculated for two markers (including combined analysis) compared with pathologic findings as a gold standard test (Tables 2 and 3). Statistical analysis showed the significant differences between positive ratios of patient and healthy groups in CEA, hTERT and combination markers (P<0.001). However, no significant differences were found between sensitivity of combination marker assay and single CEA (P = 0.139) or hTERT (P = 0.226) assay. Comparison of single and combined markers positive ratios between male and female patients, between older (age > 64) and younger (age ≤ 64) patients, between colon and rectal cancer patients and between different stages of disease have been presented in Table 4.
Table 2.
Results of real-time RT-PCR assay of carcino-embryonic antigen (CEA), human telomerase reverse transcriptase (hTERT) mRNA and combination markers in peripheral blood of 27 CRC patients and 27 healthy volunteers in comparison to pathologic results as gold standard diagnostic test.
| Marker results |
Gold standard
|
||
|---|---|---|---|
| Positive | Negative | ||
| CEA mRNA | positive | 21* | 3** |
| negative | 6*** | 24**** | |
| hTERT mRNA | positive | 22* | 9** |
| negative | 5*** | 18**** | |
| CEA + hTERT mRNA | positive | 25* | 9** |
| negative | 2*** | 18**** | |
Numbers of *true positives; **false positives; ***false negatives; ****true negatives
Table 3.
Calculated statistical parameters of carcino-embryonic antigen (CEA) mRNA, human telomerase reverse transcriptase (hTERT) mRNA and combination markers tests according to the data of Table 2.
| Marker | Sensitivity (%) | Specificity (%) |
|---|---|---|
| CEA mRNA | 78 | 89 |
| hTERT mRNA | 81 | 67 |
| CEA or hTERT mRNA | 93 | 67 |
Comparison of positive rate of carcinoembryonic antigen and hTERT mRNA in each vial with calculated sensitivity of the markers. To analyze the initial theory, comparison of positive rate of CEA and hTERT mRNA in each vial with total calculated sensitivity of the markers was performed. As shown in Table 5, significant statistical differences were revealed in patient group.
Evaluation of expression levels for the markers between patient and healthy volunteers. To evaluate the expression levels of markers in patients and healthy volunteers whose assay results were positive, a simple and practical plan was applied. The plan was based on results of a previous study [22] which showed the ratio of marker/reference genes was significantly different between true-positive results of the assay in patients and false-positive results due to background expression of marker in healthy individuals. Briefly, we calculated ΔCt of both CEA and hTERT using the following parameters: [Ct value of maker - Ct value of reference] and then, mean of this value was calculated in two study groups. It was found that means of ΔCt were 8.79 ± 3.2 (range: 3.78-13.23) for CEA and 7.30 ± 2.8 (range: 2.02-11.30) for hTERT in patient group. In healthy group, these parameters were 16.85 ± 0.28 (range: 16.52-17.02) and 12.27 ± 1.5 (range: 9.46-13.58) for CEA and hTERT, respectively. Then, a range of mean of ΔCt for both markers was calculated in healthy volunteers with 95% of confidence interval. Lower limit of this range for each marker was determined in healthy group and considered as a cut-off point. Values lower and higher than the cut-off points were assumed as true and background-induced positive results, respectively. According to these findings, all of three false-positive cases in CEA mRNA assay and 7 out of 9 false-positive cases in hTERT mRNA were distinguished in healthy group.
Table 4.
Positive ratios of markers in patients according to sex, age and tumor location category. In each fraction, denominator and numerator stand for total individuals in each subgroup and number of persons with positive results, respectively
| Clinical feature |
CEA
positivity (%) |
hTERT
positivity (%) |
CEA or hTERT
Positivity (%) |
|---|---|---|---|
| Sex | |||
| Male | 11/13 (85) | 13/13 (100) | 13/13 (100) |
| Female | 10/14 (71) | 9/14 (64) | 12/14 (86) |
| Age | |||
| >64 | 12/13 (92) | 11/13 (85) | 13/13 (100) |
| ≤64 | 9/14 (64) | 11/14 (79) | 12/14 (86) |
| Location | |||
| Colon | 11/17 (65) | 14/17 (82) | 15/17 (88) |
| Rectum | 10/10 (100) | 8/10 (80) | 10/10 (100) |
| Stage of disease | |||
| І | 6/9 (67) | 6/9 (82) | 8/9 (89) |
| II | 8/10 (80) | 9/10 (90) | 9/10 (90) |
| III | 7/8 (88) | 7/8 (78) | 8/8 (100) |
Table 5.
Comparison of positive rate of carcinoembryonic antigen (CEA) and human telomerase reverse transcriptase (hTERT) mRNA in each vial with calculated sensitivity of the markers in patient group.
| Vial |
CEA mRNA
positive rate (%) |
CEA mRNA
sensitivity (%) |
P
value |
hTERT mRNA
positive rate (%) |
hTERT mRNA
sensitivity (%) |
P
value |
|
|---|---|---|---|---|---|---|---|
| 1 | 33 | 78 | <0.001 | 41 | 81 | 0.002 | |
| 2 | 37 | 0.003 | 33 | <0.001 | |||
| 3 | 26 | <0.001 | 22 | <0.001 |
Based on these results, specificity of CEA mRNA, hTERT mRNA and combination markers were increased to 100%, 93% and 93%, respectively. Compared to primary calculations, specificity of CEA mRNA was not meaningfully elevated (P = 0.077) but in case of hTERT mRNA and combination markers, specificity was meaningfully raised (P = 0.018).
Discussion
CTC markers might be considered as potential biomarkers for detection of colorectal carcinoma [23]. Shedding of CTC from primary cancer into the circulation begins at early stage of cancer development process [24]. Therefore, finding tumor cells by detection of their specific markers may result in early diagnosis of CRC. Use of appropriate markers is very critical, because tracking of rare tumor cells (about 1 tumor cell among more than 106 WBC of peripheral blood) is the main challenge [25]. A few mRNA markers have been studied as blood markers for early detection of colorectal carcinoma [26]. Several studies with different methods of sampling, assay protocols and interpretation of results have been also performed [11, 13, 21].
In this study, real-time PCR was applied to detect CEA and hTERT mRNA in peripheral blood of CRC patients. Sensitivity and specificity of CEA and hTERT mRNA have been reported in a wide range in different investigations [11, 13, 17, 27].
Increasing marker sensitivity and specificity have been achieved by means of two essential strategies: CTC enrichment techniques [7, 28, 29] and multi-sampling methods [27, 30]. Since these two strategies require high cost and long period of time, we attempted to increase our detection possibility by producing of cDNA from 3 μg of total mRNA in 3 separate vials. Results of our study showed that this strategy might be considered as a tool for increasing sensitivity. Although sensitivity of marker combination (93%) is not meaningfully higher than single marker sensitivity, it may be presumed that similar studies with larger sample size may result in considerable higher differences. The calculated sensitivity of CEA mRNA in the present study was similar to the results of a study presented a 3-time sampling method and reported CEA mRNA sensitivity equal to 74% [27]. Results of other studies on CRC patients from all stages of disease presented 36% [20] and 68% [31] of CEA mRNA sensitivity. All of these rates were meaningfully lower than our results.
False-positive results of tumor marker expression in healthy group may be due to illegitimate transcription or background expression [32] and result in a decrease in specificity of marker test. To increase the specificity of markers, we designed the mentioned cut-off point strategy, which revealed false-positive results. Based on this strategy, specificity of the method was raised to 93%, which was similar to the previous studies [20, 31].
By comparison with colonoscopy whose sensitivity (95%) and specificity (100%) are more than the other routine screening procedures of CRC [33, 34], our primary findings showed relatively optimistic results. Therefore, it may be considered as a tool for detection of non-metastatic disease in people with no tendency to invasive screening procedures; however, it should not be considered as a substitution method for colono-scopy.
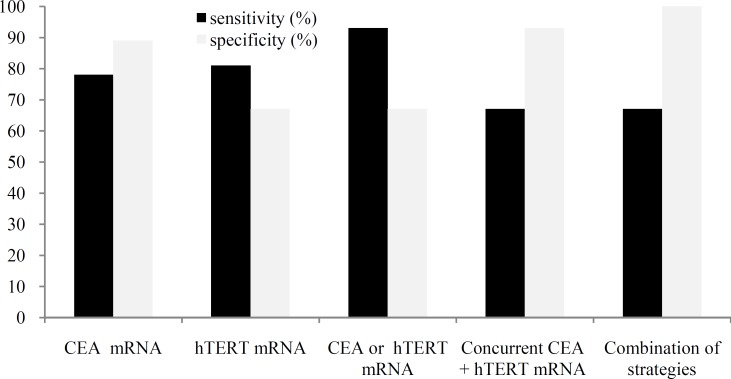
An investigation has suggested a combined panel of marker evaluation for finding of CTC in earlier stages of CRC [7]. According to the previous studies, to detect only a single marker gene in peripheral blood indicates the presence of tumor cells in peripheral blood of samples (of course under definite conditions which distinguishes between true- and false-positive results). This concept usually leads to more sensitivity and perhaps less specificity in comparison to single marker assays. The combined marker expression may be judged as positive only if results of at least a couple of markers were positive concurrently. Did this assumption overcome the problem of background expression of markers and decrease false-positive findings of CTC detection? What about decreasing the sensitivity? To answer the questions, we analyzed our findings. Interestingly, we found that 18 out of 27 patients and 2 out of 27 healthy volunteers showed concurrent expression of CEA and hTERT. Therefore, sensitivity and specificity were calculated as 67% and 93%, respectively. Finally, if we combine concurrent marker expression theory with cut-off point strategy, sensitivity will be calculated as 67%. This finding is similar to the reported values of studies which have used CTC enrichment techniques [7] and multi-sampling methods [27]; however, specificity will increase to 100%. Summary of different calculated sensitivities and specificities of our study has been presented in Figure 1. To increase the sensitivity of concurrent combination biomarker test, we suggest that more than 3 vials to be considered as templates of real-time RT-PCR in future studies. The rate of gene expression of markers among the patient’s clinical features implies that screening test for primary detection of CRC could be designed based on tracking of these markers in peripheral blood. Such a screening test might be applied for this purpose regardless of subjects' age, sex, tumor location and disease stage. However, similar studies with enough sample sizes should be done to confirm the statistical equality of these probable confounding factors.
Fig. 1.
Comparison of calculated sensitivity and specificity according to single marker and different protocols of combined markers evaluation. CEA, carcino-embryonic antigen; hTERT, human telomerase reverse transcriptase
In conclusion, the results of this study suggest that mRNA markers of peripheral blood may be considered as useful tools to find non-metastatic CRC by real-time RT-PCR. To improve the sensitivity and specificity of the assay, we suggest performing combination marker assays on at least 3 separate vials and interpretation of results according to the mentioned protocols. However, more widespread studies are required to confirm our findings.
Acknowledgment
This work was supported by Research and Technology Deputy of Hamadan University of Medical Sciences as a Ph.D. thesis. We thank staffs of Beheshti (Hamadan) and Imam Khomeini Hospitals (Tehran).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011 Feb;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Levin B, Lieberman DA, McFarland B, Smith RA, Brooks D, Andrews KS, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US multi-society task force on colorectal cancer, and the American College of Radiology. CA Cancer J Clin. 2008 May-Jun;58(3):130–60. doi: 10.3322/CA.2007.0018. [DOI] [PubMed] [Google Scholar]
- 3.Davila RE, Rajan E, Baron TH, Adler DG, Egan JV, Faigel DO, et al. ASGE guideline: colorectal cancer screening and surveillance. Gastrointest Endosc. 2006 Apr;63(4):546–57. doi: 10.1016/j.gie.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe T, Itabashi M, Shimada Y, Tanaka S, Ito Y, Ajioka Y, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2010 for the treatment of colorectal cancer. Int J Clin Oncol. 2012 Feb;17(1):1–29. doi: 10.1007/s10147-011-0315-2. [DOI] [PubMed] [Google Scholar]
- 5.Sun L, Wu H, Guan YS. Colonography by CT, MRI and PET/CT combined with conventional colonoscopy in colorectal cancer screening and staging. World J Gastroentro. 2008 Feb;14(6):853–63. doi: 10.3748/wjg.14.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh JM, Terdiman JP. Colorectal cancer screening: scientific review. Jama. 2003 Mar;289(10):1288–96. doi: 10.1001/jama.289.10.1288. [DOI] [PubMed] [Google Scholar]
- 7.Shen C, Hu L, Xia L, Li Y. Quantitative real-time RT-PCR detection for survivin, CK20 and CEA in peripheral blood of colorectal cancer patients. Jpn J Clin Oncol. 2008 Nov;38(11):770–6. doi: 10.1093/jjco/hyn105. [DOI] [PubMed] [Google Scholar]
- 8.Zieglschmid V, Hollmann C, Bocher O. Detection of disseminated tumor cells in peripheral blood. Crit Rev Clin Lab Sci. 2005;42(2):155–96. doi: 10.1080/10408360590913696. [DOI] [PubMed] [Google Scholar]
- 9.Guller U, Zajac P, Schnider A, Bosch B, Vorburger S, Zuber M, et al. Disseminated single tumor cells as detected by real-time quantitative polymerase chain reaction represent a prognostic factor in patients undergoing surgery for colorectal cancer. Ann surg. 2002 Dec;236(6):768–75. doi: 10.1097/00000658-200212000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mocellin S, Rossi CR, Pilati P, Nitti D, Marincola FM. Quantitative real-time PCR: a powerful ally in cancer research. Trends Mol Med. 2003 May;9(5):189–95. doi: 10.1016/s1471-4914(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 11.Castells A, Boix L, Bessa X, Gargallo L, Pique JM. Detection of colonic cells in peripheral blood of colorectal cancer patients by means of reverse transcriptase and polymerase chain reaction. Brit J Cancer. 1998 Nov;78(10):1368–72. doi: 10.1038/bjc.1998.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang JY, Lin SR, Wu DC, Lu CY, Yu FJ, Hsieh JS, et al. Multiple molecular markers as predictors of colorectal cancer in patients with normal perioperative serum carcinoembryonic antigen levels. Clin Cancer Res. 2007 Apr;13(8):2406–13. doi: 10.1158/1078-0432.CCR-06-2054. [DOI] [PubMed] [Google Scholar]
- 13.Guadagni F, Kantor J, Aloe S, Carone MD, Spila A, D'Alessandro R, et al. Detection of blood-borne cells in colorectal cancer patients by nested reverse transcription-polymerase chain reaction for carcinoembryonic antigen messenger RNA: longitudinal analyses and demonstration of its potential importance as an adjunct to multiple serum markers. Cancer Res. 2001 Mar;61(6):2523–32. [PubMed] [Google Scholar]
- 14.Miura M, Ichikawa Y, Tanaka K, Kamiyama M, Hamaguchi Y, Ishikawa T, et al. Real-time PCR (TaqMan PCR) quantification of carcinoembryonic antigen (CEA) mRNA in the peripheral blood of colorectal cancer patients. Anticancer Res. 2003 Mar-Apr;23(2B):1271–6. [PubMed] [Google Scholar]
- 15.Kanamaru T, Tanaka K, Kotani J, Ueno K, Yamamoto M, Idei Y, et al. Telomerase activity and hTERT mRNA in development and progression of adenoma to colorectal cancer. Int J Mol Med. 2002 Aug;10(2):205–10. [PubMed] [Google Scholar]
- 16.Niiyama H, Mizumoto K, Sato N, Nagai E, Mibu R, Fukui T, et al. Quantitative analysis of hTERT mRNA expression in colorectal cancer. Am J Gastroenterol. 2001 Jun;96(6):1895–900. doi: 10.1111/j.1572-0241.2001.03890.x. [DOI] [PubMed] [Google Scholar]
- 17.Lledo SM, Garcia-Granero E, Dasi F, Ripoli R, Garcia SA, Cervantes A, et al. Real time quantification in plasma of human telomerase reverse transcriptase (hTERT) mRNA in patients with colorectal cancer. Colorectal Dis. 2004 Jul;6(4):236–42. doi: 10.1111/j.1463-1318.2004.00627.x. [DOI] [PubMed] [Google Scholar]
- 18.Terrin L, Rampazzo E, Pucciarelli S, Agostini M, Bertorelle R, Esposito G, et al. Relationship between tumor and plasma levels of hTERT mRNA in patients with colorectal cancer: implications for monitoring of neoplastic disease. Clin Cancer Res. 2008 Nov;14(22):7444–51. doi: 10.1158/1078-0432.CCR-08-0478. [DOI] [PubMed] [Google Scholar]
- 19.Kashida H, Kudo SE. Early colorectal cancer: concept, diagnosis, and management. Intl J Clin Oncol. 2006 Feb;11(1):1–8. doi: 10.1007/s10147-005-0550-5. [DOI] [PubMed] [Google Scholar]
- 20.Xu D, Li XF, Zheng S, Jiang WZ. Quantitative real-time RT-PCR detection for CEA, CK20 and CK19 mRNA in peripheral blood of colorectal cancer patients. J Zhejiang Univ Sci B. 2006 Jun;7(6):445–51. doi: 10.1631/jzus.2006.B0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadahiro S, Suzuki T, Tokunaga N, Yurimoto S, Yasuda S, Tajima T, et al. Detection of tumor cells in the portal and peripheral blood of patients with colorectal carcinoma using competitive reverse transcriptase-polymerase chain reaction. Cancer. 2001 Sep;92(5):1251–8. doi: 10.1002/1097-0142(20010901)92:5<1251::aid-cncr1445>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 22.Schuster R, Max N, Mann B, Heufelder K, Thilo F, Grone J, et al. Quantitative real-time RT-PCR for detection of disseminated tumor cells in peripheral blood of patients with colorectal cancer using different mRNA markers. Int J Cancer. 2004 Jan;108(2):219–27. doi: 10.1002/ijc.11547. [DOI] [PubMed] [Google Scholar]
- 23.Tsouma A, Aggeli C, Pissimissis N, Lembessis P, Zografos GN, Koutsilieris M. Circulating tumor cells in colorectal cancer: detection methods and clinical significance. Anticancer Res. 2008 Nov-Dec;28(6B):3945–60. [PubMed] [Google Scholar]
- 24.Wang JY, Yeh CS, Chen YF, Wu CH, Hsieh JS, Huang TJ, et al. Development and evaluation of a colorimetric membrane-array method for the detection of circulating tumor cells in the peripheral blood of Taiwanese patients with colorectal cancer. Int J Mol Med. 2006 May;17(5):737–47. [PubMed] [Google Scholar]
- 25.Xi L, Nicastri DG, El-Hefnawy T, Hughes SJ, Luketich JD, Godfrey TE. Optimal markers for real-time quantitative reverse transcription PCR detection of circulating tumor cells from melanoma, breast, colon, esophageal, head and neck, and lung cancers. Clin Chem. 2007 Jul;53(7):1206–15. doi: 10.1373/clinchem.2006.081828. [DOI] [PubMed] [Google Scholar]
- 26.Hundt S, Haug U, Brenner H. Blood markers for early detection of colorectal cancer: a systematic review. Cancer Epidemiol Biomarkers Prev. 2007 Oct;16(10):1935–53. doi: 10.1158/1055-9965.EPI-06-0994. [DOI] [PubMed] [Google Scholar]
- 27.Wharton RQ, Jonas SK, Glover C, Khan ZA, Klokouzas A, Quinn H, et al. Increased detection of circulating tumor cells in the blood of colorectal carcinoma patients using two reverse transcription-PCR assays and multiple blood samples. Clin Cancer Res. 1999 Dec;5(12):4158–63. [PubMed] [Google Scholar]
- 28.Khair G, Monson JR, Greenman J. Epithelial molecular markers in the peripheral blood of patients with colorectal cancer. Dis Colon Recumt. 2007 Aug;50(8):1188–1203. doi: 10.1007/s10350-006-0875-9. [DOI] [PubMed] [Google Scholar]
- 29.Zieglschmid V, Hollmann C, Gutierrez B, Albert W, Strothoff D, Gross E, et al. Combination of immunomagnetic enrichment with multiplex RT-PCR analysis for the detection of disseminated tumor cells. Anticancer Res. 2005 May-Jun;25(3A):1803–10. [PubMed] [Google Scholar]
- 30.Zhang XW, Yang HY, Fan P, Yang L, Chen GY. Detection of micrometastasis in peripheral blood by multi-sampling in patients with colorectal cancer. World J Gastroentrol. 2005 Jan;11(3):436–8. doi: 10.3748/wjg.v11.i3.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yun S, Zhen-Jun , W , Bin W, et al. The clinical value of CEA mRNA, CEA protein of peripheral blood in the prediction of colorectal cancer. J Chin Phys. 2005:S1. [Google Scholar]
- 32.Yadegarazari R, Saidijam M. Using RT-PCR and qRT-PCR to assay RNA markers in detection of peripheral colorectal circulating cells: A systemic review. Clin Biochem. 2011;44(13):196s. [Google Scholar]
- 33.Rex DK, Rahmani EY, Haseman JH, Lemmel GT, Kaster S, Buckley JS. Relative sensitivity of colonoscopy and barium enema for detection of colorectal cancer in clinical practice. Gastroenterology. 1997 Jan;112(1):17–23. doi: 10.1016/s0016-5085(97)70213-0. [DOI] [PubMed] [Google Scholar]
- 34.Allameh Z, Davari M, Emami MH. Cost-effectiveness analysis of colorectal cancer screening methods in Iran. Arch Iran Med. 2011 Mar;14(2):110–4. [PubMed] [Google Scholar]