Abstract
Background & Aims
Genomic instability participates in the pathogenesis of hepatocellular carcinoma (HCC). Apurinic/apyrimidinic endonuclease 1 (APEX1) participates in the base excision repair of premutagenic apurinic/apyrimidinic (AP) sites. Mice deficient in methionine adenosyltransferase 1a (Mat1a KO) have chronic hepatic deficiency of S-adenosylmethionine (SAMe) and increased oxidative stress, and develop HCC. We examined livers of Mat1a KO mice for genomic instability and dysregulation of APEX1.
Methods
Studies were conducted using Mat1a KO mice livers and cultured mouse and human hepatocytes.
Results
Genomic instability increased in the livers of 1-month-old Mat1a KO mice, compared with wild-type mice, whereas Apex1 mRNA and protein levels were reduced by 20% and 50%, respectively, in Mat1a KO mice of all ages. These changes correlated with increased numbers of AP sites and reduced expression of Bax, Fas, and p21 (all APEX targets). When human and mouse hepatocytes were placed in culture, transcription of MAT1A mRNA decreased whereas that of APEX1 and c-MYC increased. However, the protein levels of APEX1 decreased to 60% of baseline. Addition of 2 mmol/L SAMe prevented increases in APEX1 and c-MYC mRNA levels, as well as decreases in MAT1A expression and cytosolic and nuclear APEX1 protein levels.
Conclusions
By 1 month of age, genomic instability increases in livers of Mat1a KO mice, possibly due to reduced APEX1 levels. Although SAMe inhibits APEX1 transcription, it stabilizes the APEX1 protein. This novel aspect of SAMe on APEX1 regulation might explain the chemopreventive action of SAMe and the reason that chronic SAMe deficiency predisposes to HCC.
Neoplastic cells typically possess numerous genomic mutations and chromosomal aberrations, including point mutations, gene amplifications and deletions, and replication errors. Acquisition of such genomic instability represents an early step in the process of carcinogenesis. 1 Proteins involved in DNA replication, DNA repair, cell cycle progression, and others are all components of complex overlapping biochemical pathways that function to maintain cellular homeostasis. Therefore, mutational alteration of genes encoding proteins involved in these cellular processes can contribute to genomic instability.1
Genomic instability can arise when there is inefficient or defective repair of the normally occurring damages that appear in the genome.2 Damage repair deals with single events through base excision repair, nucleotide excision repair, mismatch repair, or homologous recombination repair systems.2 Specialized genes exist for various forms of repair optimized for the particular damage. 3 Apurinic/apyrimidinic (AP) sites are the most frequent DNA lesions in cells.4 These lesions are generated by spontaneous hydrolysis after exposure to ionizing radiation or as an intermediate of DNA repair by N-glycosylases acting on modified DNA base.4 Base modifications occur endogenously or are the result of environmental mutagens, in particular alkylating compounds and reactive oxygen species (ROS). In concert with several other DNA repair proteins, AP endonuclease redox effector (APE1/REF-1/APEX1) participates in base excision repair.5 APEX1 is a protein involved both in the base excision repair pathways of DNA lesions and in the regulation of gene expression as a redox coactivator of different transcription factors, such as early growth response protein-1, p53, and AP-1.6 Apex1 is well known to be induced by ROS at the transcriptional level,7 which is part of the defense mechanism against genomic instability. 8
Chronic hepatic S-adenosylmethionine (SAMe) deficiency generates oxidative stress and predisposes to injury and malignant transformation.9 SAMe is the principal biological methyl donor and a precursor for glutathione and polyamines.10 It is generated in the first reaction in the metabolism of methionine, catalyzed by methionine adenosyltransferase (MAT). In mammals two MAT genes, MAT1A and MAT2A, encode for two catalytic subunits of MAT, α1 and α2, respectively. MAT1A is largely expressed in normal liver, whereas MAT2A is widely expressed.10 Patients with chronic liver disease have impaired hepatic SAMe biosynthesis because of decreased MAT1A mRNA levels and posttranslational inhibition of the MAT1Aencoded isoenzymes.10 Chronic hepatic SAMe deficiency occurs in Mat1a knockout (KO) mice, which exhibit an increased propensity to choline-deficient, diet-induced fatty liver, a higher level of lipid peroxidation, and the spontaneous development of steatohepatitis and hepatocellular carcinoma (HCC).9,11 In this study, we investigated the hypothesis that increased genomic instability occurs in this model, and uncovered the unexpected finding that the APEX1 protein level is markedly down-regulated in the Mat1a KO mice livers early on. Our investigation of the molecular mechanisms revealed novel regulation of Apex1 expression by SAMe, which may be one of the mechanisms of HCC formation in this model.
Materials and Methods
Primary Cultures and Reagents
Mouse hepatocytes from 2- to 3-month-old male C57/B6 mice were isolated by collagenase perfusion as described.12 Primary human hepatocytes were obtained in suspension culture in cold preservation medium (24 hours after the livers were harvested) from CellzDirect (Pittsboro, NC). Cells were plated at a density of 0.5 × 106 cells per well in 6-well plates, unless otherwise mentioned. Cultures were maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum, 2 mmol/L glutamine, 50 mmol/L penicillin, and 50 mg/mL streptomycin sulfate. After 2 hours, the medium was changed to contain 2.5% fetal calf serum.13 SAMe in the form of disulfate p-toluenesulfonate dried powder was generously provided by Gnosis SRL (Cairate, Italy). All other reagents were of analytical grade and were obtained from commercial sources.
Cell Treatments
Hepatocytes were treated with SAMe (2 mmol/L), methylthioadenosine (MTA, 1 mmol/L), or MTA plus cycloleucine (20 mmol/L pretreatment for 1 hour) for different time points. When indicated, cells were pretreated with actinomycin D (5 μg/mL) for 5 minutes or cycloheximide (5 μg/mL) for 5 minutes. Treatments with actinomycin D and cycloheximide at these doses did not cause any toxicity as evaluated by trypan blue exclusion for up to 24 hours.14
Primary mouse hepatocytes were transiently transfected with c-Myc small interfering RNA (siRNA) duplex (Qiagen) as previously described.15 Briefly, c-Myc or scrambled siRNA duplexes were mixed with 4 μL of Lipofactamine RNAiMAX Reagents (Invitrogen, Carlsbad, CA) in the OPTI medium (Invitrogen) and then added to hepatocytes. Six hours later, the media were replaced with fresh Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum, and the cells were treated with SAMe (2 mmol/L) or vehicle and cultured for another 18 and 30 hours.
Mat1a KO Mice
Male Mat1a KO and wild-type (WT) littermates were previously described.11 They were fed ad libitum a standard diet (Harland Teklad irradiated mouse diet 7912, Madison, WI) and housed in a temperature-controlled animal facility with 12-hour light-dark cycles. Mice were killed and liver tissues removed, snap frozen in liquid nitrogen, and stored at −80°C until analysis. Animals were treated humanely, and all procedures were in compliance with our institution’s guidelines for the use of laboratory animals.
DNA Extraction and Random Amplified Polymorphic DNA Analysis
DNA was extracted from mouse liver tissues using the phenol/chloroform extraction and ethanol precipitation method.16 Eighteen island GpC-rich arbitrary primers were used to score genomic alterations in Mat1a KO and WT liver samples as described.17 The polymerase chain reaction (PCR) mixture (12 μL) consisted of 2 units of AmpliTaq DNA polymerase, 1.6 mmol/L deoxynucleoside triphosphate, 24 pmol/μL of random primer, and 5 or 10 ng of genomic DNA. The final reaction mixture was placed in a DNA thermal cycler (Perkin Elmer 9700, Waltham, MA). The PCR program included an initial denaturation step at 98°C for 2 minutes followed by 45 cycles at 95°C for 45 seconds for DNA denaturation, annealing step at 33°C for 45 seconds, extension at 72°C for 1 minute, and final extension at 72°C for 10 minutes. The amplified DNA fragments were separated on 2% agarose gel and stained with ethidium bromide. The amplified pattern was visualized on an ultraviolet trans-illuminator and photographed. The banding profile was analyzed on the basis of change of the intensity of the band, a missing band, or the appearance of a new band in the Mat1a KO liver samples compared with WT. The value in terms of percentage of genomic instability was calculated as previously described.17
AP Site Counting in Genomic DNA
Genomic DNA was purified from mouse liver samples using a DNA isolation kit (Dojindo Molecular Technologies, Inc, Gaithersburg), and AP site number was determined as described previously,18 using a DNA Damage Quantification Kit (Dojindo Molecular Technologies, Inc) according to the manufacturer’s instructions.
8-Hydroxy-2′-deoxyguanosine Measurement
8-Hydroxy-2′-deoxyguanosine (8-OHdG) levels in mouse livers were measured using a commercially available enzyme-linked immunosorbent assay (Cell Biolabs, Inc, San Diego, CA) following the instructions of the manufacturer.
Quantitative PCR Analysis
Total RNA isolated from hepatocytes and livers as described19 was subjected to reverse transcription by using M-MLV Reverse Transcriptase (Invitrogen). One microliter of reverse transcription product was subjected to quantitative real-time PCR analysis. The primers and TaqMan probes for Apex1/APEX1, c-Myc/c-MYC, Mat1a/MAT1A, p21 and Fas, and Universal PCR Master Mix were purchased from ABI (Foster City, CA). 18SrRNA was used as a housekeeping gene as described.20 The delta Ct (ΔCt) obtained was used to find the relative expression of genes according to the formula: relative expression Δ 2−ΔΔCt, where ΔΔCt Δ ΔCt of respective genes in experimental groups - ΔCt of the same genes in control group.
Protein Extraction and Western Blot Analysis
Protein extracts from primary hepatocytes and liver samples were prepared as described12 and resolved by electrophoresis on 12%–13% sodium dodecyl sulfate– polyacrylamide gel. Western blotting was performed following standard protocols (Amersham BioSciences, Piscataway, NJ) using primary antibodies for APEX1 (Novus, Littleton, CO), BAX (Cell Signaling, Danvers, MA), and β-actin (Santa Cruz Biotechnologies, Santa Cruz, CA).
Immunohistochemistry
Hepatocytes were plated at a density of 0.2 × 106 cells in 6-well plate and treated with SAMe (2 mmol/L) for 30 hours. The cells were fixed, made permeable, and processed for the direct immunofluorescence microscopy by the ICC Abcam protocol (Abcam, Inc, Cambridge, MA). Cells were blocked in 5% bovine serum albumin in phosphate-buffered saline for 1 hour and incubated with a 1:500 dilution of a monoclonal anti-APEX1 antibody (Novus, Littleton, CO), and 1 hour with rabbit polyclonal to chicken IgY (FITC) (Abcam, Inc). The slides were washed and mounted with Dako fluorescent mounting medium (Dako Co, Carpinteria, CA) and examined using the Nikon Eclipse TE300 confocal microscope (Nikon Instruments Inc, Mellville, NY).
Determination of SAMe Levels
Hepatocytes were plated at a density of 1.5 × 106 cells in 100-mm dishes and treated with SAMe (2 mmol/L) for different time points. Cellular SAMe levels were measured by high-performance liquid chromatography as described.12
Statistical Analysis
Data are given as mean ± SEM. Statistical analysis was performed using analysis of variance and Fisher’s test. For mRNA and protein levels, ratios of genes and proteins to respective housekeeping densitometric values were compared. Significance was defined by P < .05.
Results
Histologic Changes in Mat1a KO Mice
One-month-old KO mice did not have any histologic abnormality, but by 4–6 months, there is some fatty liver change in the liver, but no dysplasia or HCC is evident11; at 19–25 months, steatosis was more severe. Although 62.5% of the KO mice developed HCC by 18 months,9 the animals included in this study had not developed overt HCC.
Increased Genomic Instability in Mat1a KO Mice
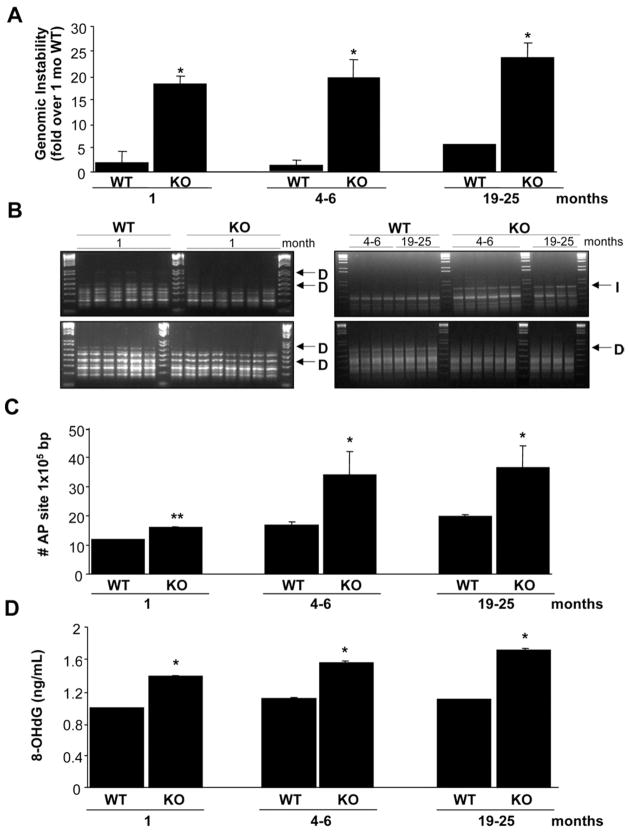
DNA from livers of Mat1a KO mice showed an increased level of genomic instability compared with WT controls (Figure 1A). This increased genomic instability was found as early as 1 month of age in Mat1a KO mice. Spontaneous oxidative stress occurs in livers of 3-monthold Mat1a KO mice.9 Oxidative DNA damage might lead to the release of free bases from DNA, generating strand breaks with various sugar modifications and AP sites, which could account for the genomic instability.4 To test this hypothesis, AP sites in genomic DNA from livers of Mat1a KO mice were quantified. We found that the AP site number was increased in the Mat1a KO mice at all age groups (Figure 1B and C). Another measure of oxidative DNA damage is 8-OHdG.21 Consistent with the increase in genomic instability, there is also a significant increase in the level of 8-OHdG in all age groups in the Mat1a KO mice (Figure 1D).
Figure 1.
Increased genomic instability in Mat1a KO mice. (A) The percentage of genomic instability in liver was calculated in different age groups of Mat1a KO and WT mice as described in the Materials and Methods section. Results are expressed as fold over results from 1-monthold WT mice (mean ± SEM) from 3 to 6 mice per group. *P < .001 vs respective WT. (B) The RAPDPCR analysis of KO mouse livers (KO) and corresponding normal liver tissue DNA from WT mice (WT) with same primers. Each lane represents sample from one mouse. Genetic alterations are indicated by arrows: D, deletion, I, insertion. The 1.5-kilobase (kb) ladder was used as a control marker. (C) The number of AP sites per 105 base pairs was detected in DNA obtained from livers of different age groups of Mat1a KO and WT mice. Results are expressed as absolute number of AP sites per 105 base pairs (mean ± SEM) from 3 mice per group. *P < .001, **P < .05 vs respective WT. (D) Amount of oxidative DNA damage was assessed in each age group by measuring 8-OHdG as described in the Materials and Methods section. *P < .001 vs respective WT.
Decreased Apex1 Expression in Mat1a KO Mice
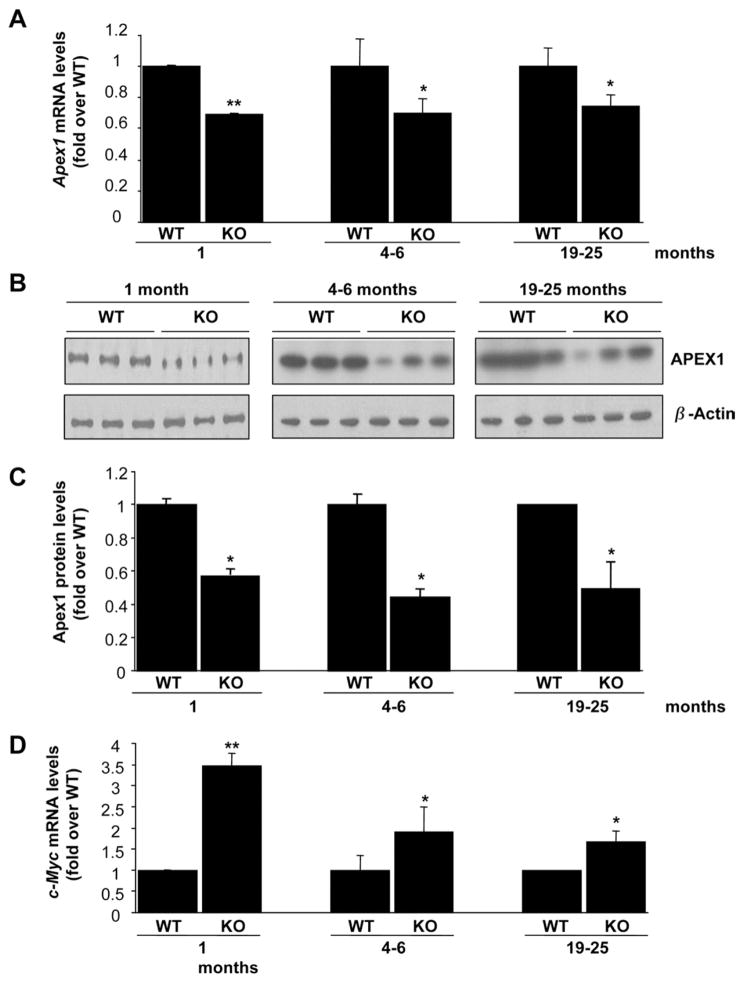
APEX1 plays an essential role in the repair of AP sites of DNA damage.22 To see whether APEX1 may be dysregulated in the Mat1a KO mice livers, we analyzed the expression of Apex1 in liver samples of Mat1a KO mice (Figure 2). Apex1 mRNA levels decreased 20% in Mat1a KO mice in all age groups (Figure 2A). We further evaluated whether this change in mRNA levels translated to a change in APEX1 protein levels (Figure 2B and C). Interestingly, APEX1 protein levels were reduced even more in Mat1a KO mice in all age groups (Figure 2B and C). A known transcriptional activator of APEX1 is c-MYC.18 However, c-Myc mRNA levels were increased in the Mat1a KO mice by about 2.5-fold at 1 month, and about 1-fold in older mice (Figure 2D).
Figure 2.
Apex1 and c-Myc expression in Mat1a KO mice. (A and D) RNA from livers of different age groups of Mat1a KO and WT mice was subjected to quantitative real-time PCR analysis for Apex1 (A) and c-Myc (D) as described in the Materials and Methods section. Results are expressed as fold over respective WT mice (mean ± SEM) from 3 to 5 mice per group. *P < .05, **P < .001 vs respective WT. (B) Western blot analyses for APEX1 and β-actin were done using 10 μg of tissue extracts from livers of different age groups of Mat1a KO and WT mice. (C) Results are expressed as fold over respective WT mice (mean ± SEM) from 3 to 4 mice per group. *P < .002 vs respective WT.
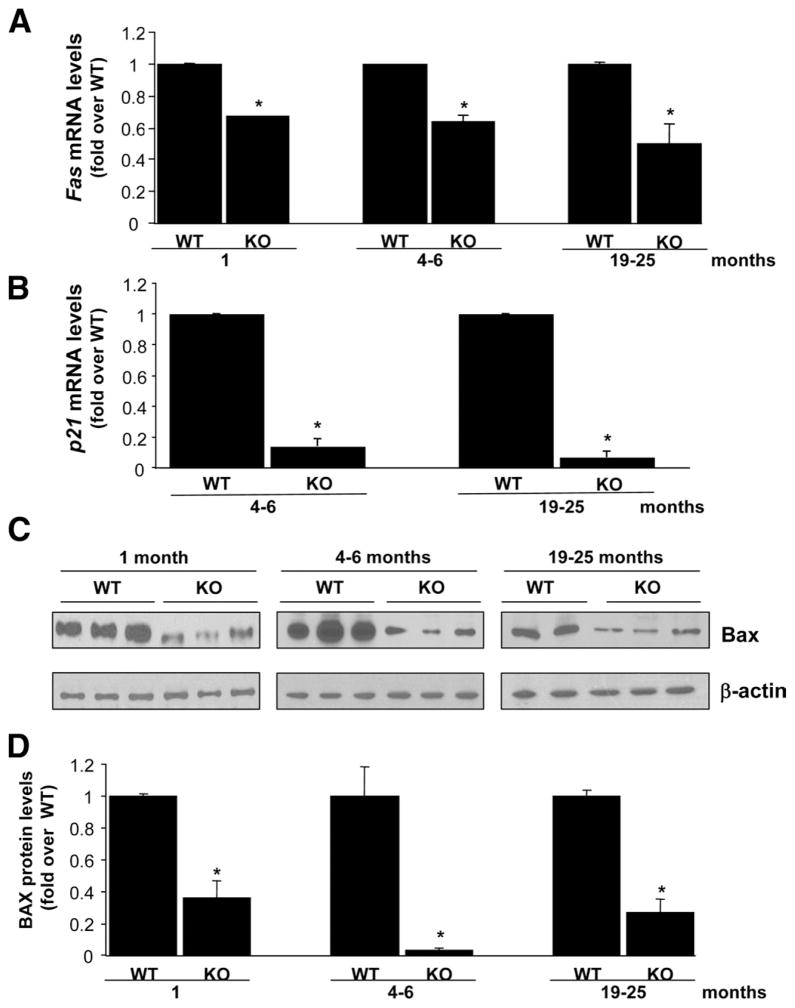
APEX1 is known to regulate the transactivation and proapoptotic functions of p53.23 To analyze whether the reduction in APEX1 protein found in the Mat1a KO mice would cause a reduction in p53-dependent transcriptional activity, we examined the expression of Fas, p21, and Bax (Figure 3), known transcriptional targets of p53.24 We found that Fas and p21 mRNA levels and BAX protein levels were notably reduced in Mat1a KO mice livers (Figure 3).
Figure 3.
Fas, p21, and BAX expression are reduced in Mat1a KO mice. (A and B) Fas (A) and p21 (B) mRNA levels were measured by real-time PCR in different age groups of KO and WT mice. (C) Western blot analysis for BAX and β-actin were done using 10 μg of tissue extracts from livers of different age groups of Mat1a KO and WT mice. (D) Results are expressed as fold over respective WT mice (mean ± SEM) from 3 to 5 mice per group. *P < .001 vs respective WT.
Effects of SAMe on Apex1 Expression in Cultured Hepatocytes
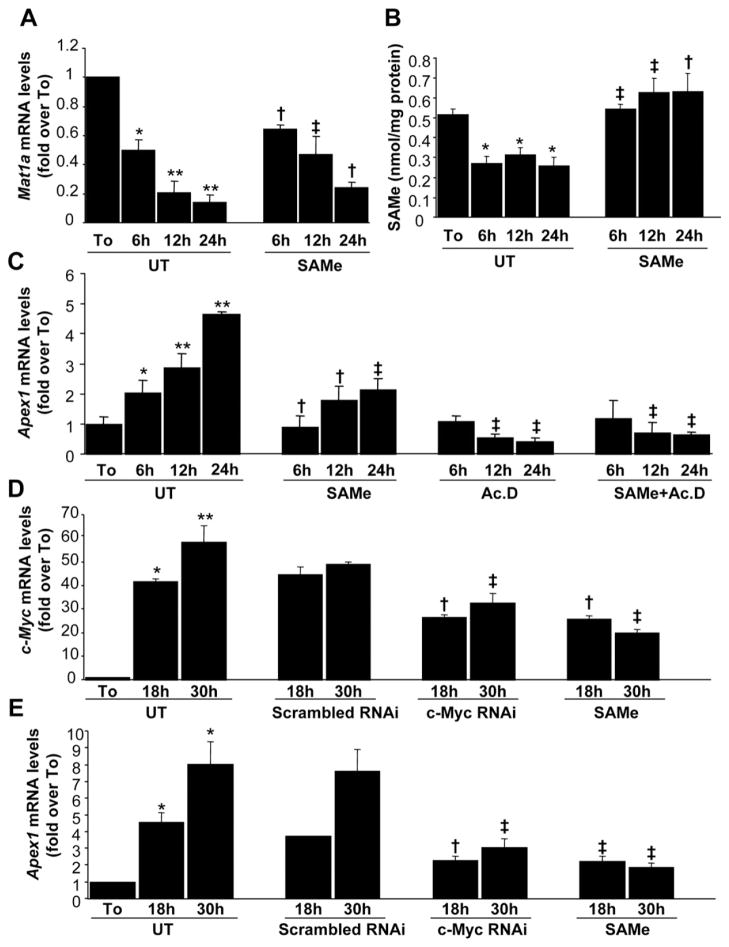
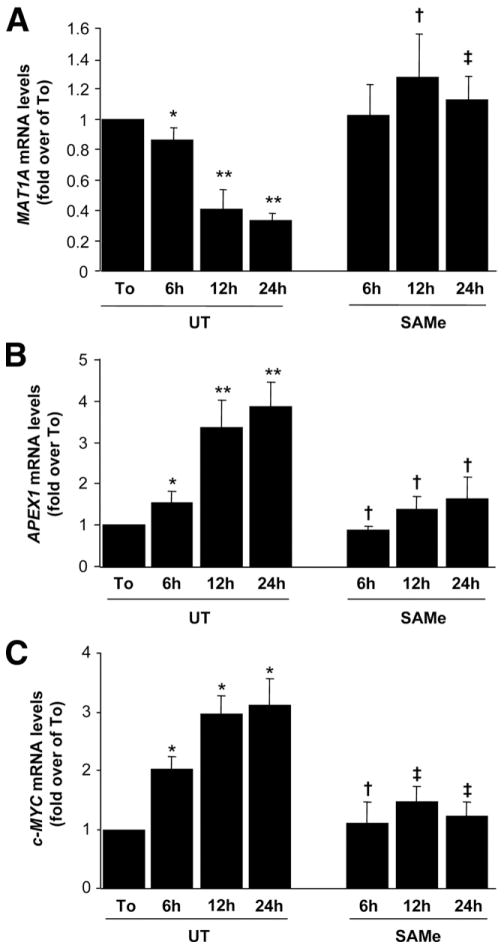
To investigate the mechanisms of the interplay between Mat1a expression, SAMe, and Apex1 expression, we performed a series of analyses in cultured mouse hepatocytes (Figure 4). It has been reported that Mat1a expression and SAMe levels decrease in rat hepatocytes after 12 hours of culture.13 This characteristic mimics the scenario found in Mat1a KO mice.11 In mouse hepatocytes, Mat1a mRNA levels and SAMe levels decreased during acute culture (Figure 4A and B). Treatment with 2 mmol/L SAMe raised intracellular SAMe levels (Figure 4B) and ameliorated the drop in Mat1a expression (Figure 4A), confirming a previous report.13 Apex1 mRNA levels, on the other hand, increased in a time-dependent fashion, whereas SAMe treatment partially prevented it (Figure 4C). We also examined c-Myc expression, as Apex1 is a well-known downstream target of c-MYC.18 c-Myc mRNA levels increased sharply to more than 45-fold of baseline, and this was also partially prevented by SAMe treatment (Figure 4D). To delineate the mechanism of the change in mRNA levels, we found that actinomycin D treatment prevented Apex1 upregulation (Figure 4C). Cotreatment with SAMe had the same effect as actinomycin D treatment alone (Figure 4C), suggesting that the changes in Apex1 mRNA levels in the cultured hepatocytes were at the transcriptional level. To further prove a causal role of c-Myc in the upregulation of Apex1, we blunted the increase in c-Myc expression with RNA interference (RNAi) to a similar degree as SAMe treatment (Figure 4D). This significantly prevented the increase in Apex1 mRNA levels (Figure 4E).
Figure 4.
Effect of SAMe treatment on Mat1a, Apex1, and c-Myc mRNA levels in mouse hepatocytes. Mouse hepatocytes were treated with SAMe (2 mmol/L) for different time points. When indicated, cells were pretreated with actinomycin D (5 μg/mL) for 5 minutes. At the indicated time points, RNA from hepatocyte suspension right after isolation (To) or cultured hepatocytes were extracted for real-time PCR. (A) Effect of SAMe treatment on Mat1a mRNA levels. Results are expressed as fold over To cells (mean ± SEM) from 3 independent experiments. *P < .005 vs To, **P < .0001 vs To, †P < .05 vs untreated (UT) at respective time points, ‡P < .01 vs UT at respective time points. (B) Effect of SAMe treatment on intracellular SAMe levels. Hepatocytes were treated with SAMe (2 mmol/L) for different time points. Cellular SAMe levels were measured by high-performance liquid chromatography. The results are expressed as nanomoles of SAMe per milligrams of protein (mean ± SEM) from 3 independent experiments. *P < .005 vs To, †P < .05 vs UT at respective time points, ‡P < .001 vs UT at respective time points. (C) Effect of SAMe treatment on Apex1 mRNA levels as determined by real-time PCR. Results are expressed as fold over To cells (mean ± SEM) from 3 independent experiments. *P < .04 vs To, **P < .001 vs To, †P < .04 vs UT at respective time points, ‡P < .001 vs UT at respective time points. (D) Effect of c-Myc RNAi and SAMe treatments on c-Myc mRNA levels. Hepatocytes were treated with scrambled or c-Myc RNAi as described in the Materials and Methods section or with 2 mmol/L SAMe for indicated times. RNA was subjected to quantitative real-time PCR analysis. Results are expressed as fold over To cells (mean ± SEM) from 4 independent experiments. *P < .004 vs To, **P < .0001 vs To, †P < .05, ††P < .0001 vs UT at respective time points. (E) Effect of c-Myc RNAi and SAMe treatments on Apex1 mRNA levels. The same samples from D were subjected to real-time PCR for Apex1 mRNA measurement. *P < .0002 vs To, †P < .004, ††P < .002 vs UT at respective time points.
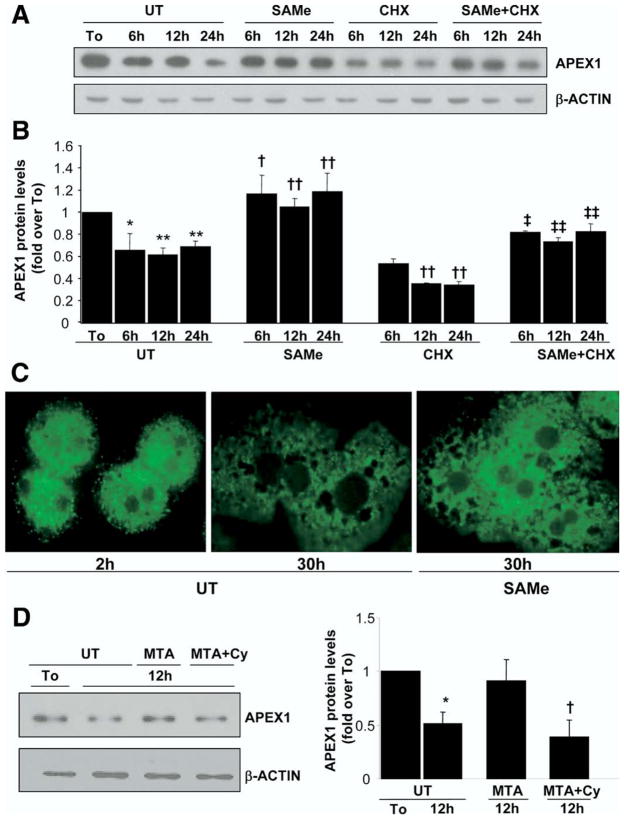
Interestingly, we found the opposite scenario when we checked APEX1 protein levels in mouse cultured hepatocytes (Figure 5); that is, APEX1 protein levels decreased by 40% despite the increase in mRNA levels (Figures 4C and 5B), and treatment with 2 mmol/L SAMe was able to completely prevent it (Figure 5A and B). To see whether the effect of SAMe on APEX1 protein was due to decreased protein degradation versus enhanced protein translation, we compared the effect of SAMe in the presence of cycloheximide, an inhibitor of de novo protein synthesis. Cycloheximide treatment further decreased APEX1 protein levels as compared with untreated mouse hepatocytes, and importantly, SAMe cotreatment was able to increase APEX1 protein levels as compared with cycloheximide treatment alone (Figure 5A and B), suggesting a new role for SAMe in the stabilization of the APEX1 protein. Lastly, confocal microscopy showed that APEX1 is largely localized to the cytosol in mouse hepatocytes, with a small amount in the nucleus at 2 hours after culturing (Figure 5C). APEX1 content in both compartments decreased significantly by 30 hours in culture, and the addition of SAMe to the media prevented this decrease (Figure 5C). Since MTA is a metabolite of SAMe that can mimic many of the actions of SAMe,12 we also tested its effect. MTA also prevented the decrease in APEX1 protein level (Figure 5D). Importantly, cylcoleucine pretreatment completely abolished the effect of MTA (Figure 5D).
Figure 5.
Effect of SAMe and MTA treatment on APEX1 protein levels in mouse cultured hepatocytes. Mouse hepatocytes were treated with SAMe (2 mmol/L) or MTA (1 mmol/L) for different time points. When indicated, cells were pretreated with cycloheximide (CHX, 5 μg/mL) for 5 minutes or cycloleucine (Cy, 20 mmol/L) for 1 hour. (A) At the indicated times, Western blot analysis was done for APEX1 or β-actin. (B) Results are expressed as fold over To cells (mean ± SEM) from 3 to 5 independent experiments. *P < .005 vs To, **P < .001 vs To, †P < .05 vs UT at respective time points, ††P < .02 vs UT at respective time points, ‡P < .05 vs CHX at respective time points, ‡‡P < .02 vs CHX at respective time points. (C) Subcellular localization of APEX1 was evaluated by confocal microscopy as described in the Materials and Methods section. APEX1 protein appears green on fluorescent microscopy. (D) Representative Western blots are shown from 3 independent experiments showing the effect of MTA with or without Cy pretreatment. Densitometric changes are shown and expressed as fold over To in the graph on right. *P < .002 vs UT, †P < .002 vs MTA.
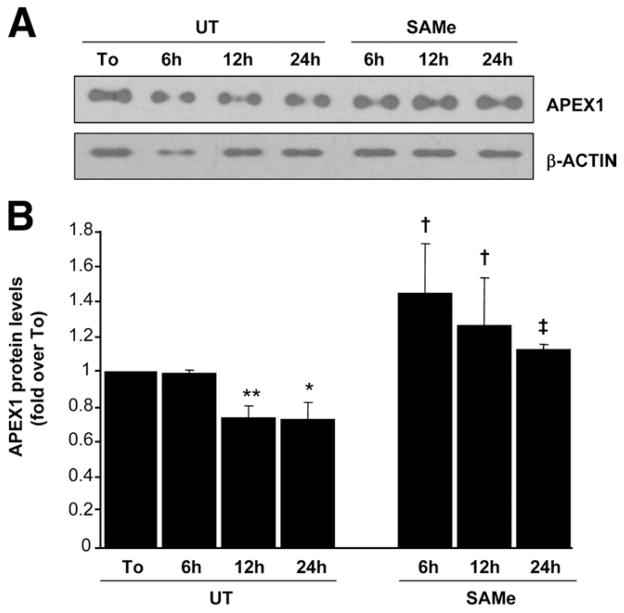
To make sure that our observations in mouse hepatocytes also occur in human hepatocytes, we examined the same variables, and Figure 6 shows that in human hepatocytes, APEX1 and c-MYC mRNA levels also increased while MAT1A mRNA levels decreased after culture, and these changes were prevented by SAMe treatment. Similarly, APEX1 protein levels decreased despite the increase in mRNA level, and this was prevented by SAMe (Figure 7).
Figure 6.
Effect of SAMe treatment on MAT1A, APEX1, and c-MYC mRNA levels in human cultured hepatocytes. Human hepatocytes were treated with SAMe (2 mmol/L) for different time points. At the indicated time points, RNA from hepatocytes received in suspension (To) or cultured hepatocytes were extracted for real-time PCR. (A) Effect of SAMe treatment on MAT1A mRNA levels. Results are expressed as fold over To cells (mean ± SEM) from 3 independent donors. *P < .03 vs To, **P < .001 vs To, †P < .004 vs UT at respective time points, ‡P < .001 vs UT at respective time points. (B) Effect of SAMe treatment on APEX1 mRNA levels. Results are expressed as fold over To cells (mean ± SEM) from 3 independent donors. *P < .02 vs To, **P < .004 vs To, †P < .02 vs UT at respective time points. (C) Effect of SAMe treatment on c-MYC mRNA levels. Results are expressed as fold over To cells (mean ± SEM) from 3 independent donors. *P < .001 vs To, †P < .02 vs UT at respective time points, ‡P < .001 vs UT at respective time points.
Figure 7.
Effect of SAMe treatment on APEX1 protein levels in human cultured hepatocytes. Human hepatocytes were treated with SAMe (2 mmol/L) for different time points. Protein from hepatocytes received in suspension (To) or cultured hepatocytes were extracted. (A) Western blot analysis using 2 μg of protein was done for APEX1 or β-actin. (B) Results are expressed as fold over To cells (mean ± SEM) from 3 independent donors. *P < .004 vs To, **P < .0001 vs To, †P < .004 vs UT at respective time points, ‡P < .002 vs UT at respective time points.
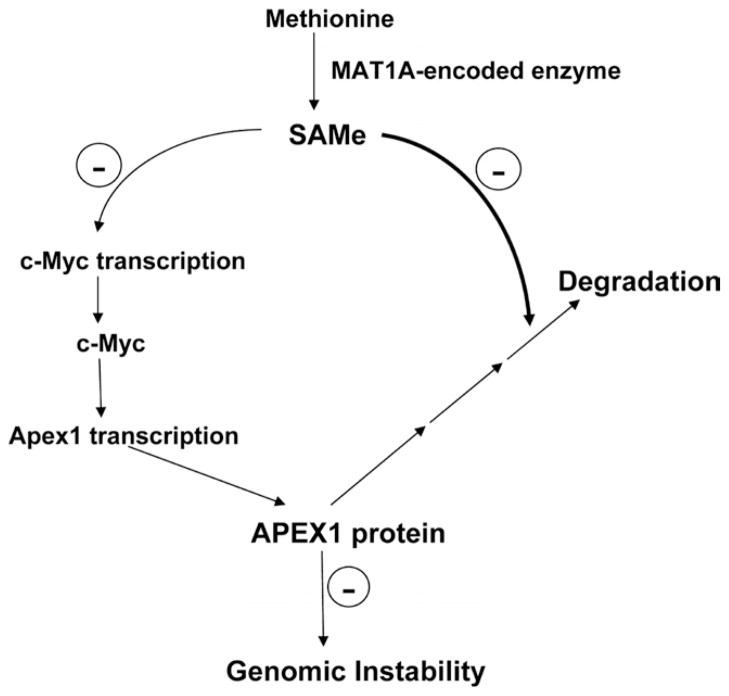
Figure 8 summarizes the results from our study and suggests a model for involvement of SAMe in genomic instability.
Figure 8.
Summary of findings and proposed model of SAMe regulation of APEX1 and genomic instability in hepatocytes. SAMe inhibits Apex1 transcription indirectly via c-Myc but increases APEX1 protein level by blocking its degradation. Thus, SAMe depletion results in lower APEX1 protein level and contributes to genomic instability.
Discussion
Aggressive tumors are characterized by genomic instability, which is thought to favor cancer progression and adaptation.25 As in other human malignancies, HCC demonstrates a high incidence of genomic instability, and the level of genomic instability correlates with tumor stage.25,26 The defense against genomic instability relies, among others, on APEX1, which is a multifunctional protein possessing both DNA repair and redox regulatory activities.22 ROS are implicated in the oxidative damage of main cellular components, such as DNA, in cancer,27 and Apex1 is well known to be induced by oxidative stress,7 which is part of the defense mechanism against genomic instability.8
Since livers of Mat1a KO mice exhibit increased oxidative stress and malignant transformation,9 we examined the degree of genomic instability in this model and found that it was increased as early as 1 month of age. One would have expected Apex1 to be induced in the Mat1a KO mice, since these mice have increased oxidative stress. To our surprise, Apex1 was not only not induced, but it was reduced in this model. Mat1a KO mice show chronic hepatic SAMe deficiency.11 This prompted us to examine how SAMe might regulate Apex1, using the cultured hepatocytes model. Using this model, we were able to demonstrate that SAMe regulates Apex1 both transcriptionally and posttranslationally.
Apex1 is regulated in a complex manner. At the transcriptional level, Apex1 is activated by oxidative stress.7 It has also been suggested that Apex1 is able to regulate its own expression.5 In addition, cell cycle–dependent expression of Apex1 has been reported.28 Finally, c-MYC has been shown to be a transcriptional activator of Apex1.18 Our results show that SAMe regulates Apex1 at the transcriptional level (Figure 4). SAMe treatment did not enhance mRNA degradation when de novo mRNA synthesis was blocked by actinomycin D (Figure 4C), suggesting that the inhibition is solely at the level of transcription. However, the effect of SAMe on Apex1 transcription is likely to be an indirect effect via c-Myc, which is rapidly induced during culture. Supporting this is the fact that blunting c-Myc upregulation by RNAi to a similar degree as SAMe blocked the increase in Apex1 mRNA level by a similar magnitude (Figure 4D and E). The observation that SAMe can prevent c-Myc induction is consistent with studies from Garcea et al29 showing c-Myc induction during hepatocarcinogenesis can be blocked by exogenous SAMe. However, the molecular mechanism of the inhibitory effect of SAMe on c-Myc expression remains to be fully elucidated.
Less is known about the regulation of APEX1 protein stability. One report showed that simultaneous overexpression of ubiquitin 9 and APEX1 in HeLa cells dramatically lowered APEX1 protein level, suggesting that ubiquitin 9 is involved in the APEX1 protein degradation.30 In this aspect, our data suggest a new role for SAMe in the regulation of APEX1 protein stability. SAMe has been shown to stabilize other proteins implicated in the redox regulation—for example, cystathionine β-synthase (CBS).31 A previous report had suggested that the carboxy-terminal sequence between residues 414 and 551 of CBS is essential for SAMe activation of the enzyme.32 SAMe binds to the regulatory domain of CBS, which contains a tandem repeat of “CBS domains,” a secondary structure motif that binds various adenine nucleotides and is believed to play a role in energy sensing.33 However, computerized studies of protein domain homology failed to detect the SAMe-binding domain in APEX1 protein (data not shown). This suggests that SAMe might regulate APEX1 stability directly, but through a different and unknown SAMe-binding domain, or indirectly. With respect to the indirect regulation, we speculate the possibility that SAMe might prevent the proteosomal degradation of APEX1, but more work will be required to elucidate the molecular mechanisms of how SAMe regulates APEX1 protein stability.
We investigated whether our observations in mouse hepatocytes also occur in human hepatocytes to further strengthen the relevance of our data. In agreement with the findings in mouse hepatocytes, we observed that in human hepatocytes, APEX1 and c-MYC mRNA levels also increased while MAT1A mRNA levels decreased after culture, and these changes were prevented by SAMe treatment. Similarly, APEX1 protein levels decreased despite the increase in mRNA level, and this was prevented by SAMe.
We have previously shown that when SAMe is used exogenously, much of its effect may be mediated by MTA.12 This is because SAMe is unstable and readily converts to MTA, which is membrane permeant.12 However, MTA can be converted back to SAMe via the methionine salvage pathway.10 Interestingly, we found that MTA also stabilized the APEX1 protein level in cultured mouse hepatocytes, but this effect was blocked if cells were pretreated with cycloleucine, an inhibitor of MAT that would have prevented the conversion of MTA to SAMe.34 Thus, in the case of APEX1 stabilization, SAMe is likely to be the major mediator.
There is an apparent discrepancy between the data from mouse hepatocytes in culture and the Mat1a KO livers where Apex1 mRNA levels are slightly lower despite lower hepatic SAMe levels. This occurred despite an increase in c-Myc mRNA levels in the Mat1a KO livers. In both models, the loss of Mat1a results in SAMe depletion, and the APEX1 protein level decreases. The Apex1 mRNA level, which increases in the culture model, is slightly lower in the KO model. Regulation of APEX1 at the mRNA level is complex and involves more than c-Myc. In addition, while the c-Myc induction in the culture model is more than 45-fold, c-Myc is only induced by 2.5-fold in the young KO mice. We can only conclude that with an acute decrease in SAMe, c-Myc is rapidly upregulated, and this led to APEX1 induction at the mRNA level. Chronically other adaptive responses are likely to set in so that we don’t see this in the KO.
A recent study by Di Maso et al35 reported an increased cytoplasmic APEX level in a large cohort of human HCC and found this localization correlated with worse prognosis. Given that APEX serves to defend against genomic instability, this would seem to be a paradox. One plausible explanation is that despite a higher cytoplasmic APEX level, the nuclear APEX level may be lower in cancerous cells. This remains to be further investigated. In addition, given that genomic instability is already present in 1-month-old KO mice but HCC develops much later (18 months), other factors are likely to take over at the later stages of hepatocarcinogenesis.
The importance of APEX1 in mutagenesis has been demonstrated in Apex1+/− mice.36 Spontaneous mutant frequencies were elevated by 2-fold in livers of 3-monthold heterozygous mice. Centrilobular fatty changes in the livers of several 9-month-old Apex1+/− mice were noted, but there was no mention of HCC up to 9 months of age. It would be of interest to see whether these animals develop HCC at a later age.
In summary, we have uncovered a novel mechanism for SAMe in the regulation of Apex1 expression. A decrease in SAMe level can lead to increased Apex1 gene transcription, but more importantly, it destabilizes APEX1 protein. This novel aspect of SAMe on Apex1 regulation may explain the chemopreventive action of SAMe and how chronic SAMe deficiency predisposes to HCC formation.
Acknowledgments
M. L. Tomasi and A. Iglesias-Ara share first authorship.
Funding
Supported by National Institutes of Health grants DK51719 (S.C.L.), AA13847 (S.C.L. and J.M.M.), and AT1576 (S.C.L., M.L.M.-C., and J.M.M.), Plan Nacional of I+D SAF 2005-00855, HEPADIP-EULSHM-CT-205, and ETORTEK-2005 (M.L.M.-C. and J.M.M.), and Associazione Italiana per la Ricerca sul Cancro, Regional Grant (F.F. and M.R.P.). Primary hepatocytes were provided by the Cell Culture Core and the confocal microscope was provided by the Cell Imaging Core of the USC Research Center for Liver Diseases (DK48522). Ciberehd is funded by the Instituto de Salud Carlos III. Maria Lauda Tomasi is a PhD student funded by the University of Turin in collaboration with the University of Sassari, Italy, Ainhoa Iglesias-Ara is a recipient of the Postdoctoral Fellowship supported by the CIC bioGUNE, Centro de Investigación Cooperativa en Biociencías, and Komal Ramani is a recipient of the Postdoctoral Fellowship of the Training Program in Alcoholic Liver and Pancreatic Diseases (T32 AA07578).
Abbreviations used in this paper
- AP
apurinic/apyrimidinic
- Apex1
apurinic/apyrimidinic endonuclease 1
- CBS
cystathionine β-synthase
- 8-OHdG
8-hydroxy-2′-deoxyguanosine
- HCC
hepatocellular carcinoma
- KO
knockout
- MAT
methionine adenosyltransferase
- MTA
methylthioadenosine
- PCR
polymerase chain reaction
- RAPD
random amplified polymorphic DNA analysis
- RNAi
RNA interference
- ROS
reactive oxygen species
- SAMe
S-adenosylmethionine
- siRNA
small interfering RNA
- WT
wild type
Footnotes
Conflict of interest
The authors disclose no conflicts.
References
- 1.Coleman WB, Tsongalis GJ. Multiple mechanisms account for genomic instability and molecular mutation in neoplastic transformation. Clin Chem. 1995;41:644–657. [PubMed] [Google Scholar]
- 2.Lindahl T, Wood RD. Quality control by DNA repair. Science. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 3.Anderson GR. Genomic instability in cancer. Curr Sci. 2001;81:501–507. [Google Scholar]
- 4.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair (Amst) 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 5.Izumi T, Brown DB, Naidu CV, et al. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci U S A. 2005;102:5739–5743. doi: 10.1073/pnas.0500986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tell G, Damante G, Caldwell D, et al. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- 7.Ramana CV, Boldogh I, Izumi T, et al. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc Natl Acad Sci U S A. 1998;95:5061–5066. doi: 10.1073/pnas.95.9.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grösch S, Fritz G, Kaina B. Apurinic endonuclease (Ref-1) is induced in mammalian cells by oxidative stress and involved in clastogenic adaptation. Cancer Res. 1998;58:4410–4416. [PubMed] [Google Scholar]
- 9.Martínez-Chantar ML, Corrales FJ, Martínez-Cruz LA, et al. Spontaneous oxidative stress and liver tumors in mice lacking methionine adenosyltransferase 1A. FASEB J. 2002;16:1292–1294. doi: 10.1096/fj.02-0078fje. [DOI] [PubMed] [Google Scholar]
- 10.Mato JM, Lu SC. Role of S-adenosyl-L-methionine in liver health and injury. Hepatology. 2007;45:1306–1312. doi: 10.1002/hep.21650. [DOI] [PubMed] [Google Scholar]
- 11.Lu SC, Alvarez L, Huang ZZ, et al. Methionine adenosyltransferase 1A knockout mice are predisposed to liver injury and exhibit increased expression of genes involved in proliferation. Proc Natl Acad Sci U S A. 2001;98:5560–5565. doi: 10.1073/pnas.091016398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramani K, Yang H, Xia M, et al. Leptin’s mitogenic effect in human liver cancer cells requires induction of both methionine adenosyltransferase 2A and 2beta. Hepatology. 2008;47:521–531. doi: 10.1002/hep.22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Trevijano ER, Latasa MU, Carretero MV, et al. S-adenosylmethionine regulates MAT1A and MAT2A gene expression in cultured rat hepatocytes: a new role for S-adenosylmethionine in the maintenance of the differentiated status of the liver. FASEB J. 2000;14:2511–2518. doi: 10.1096/fj.00-0121com. [DOI] [PubMed] [Google Scholar]
- 14.Freshney R. Culture of animal cells: a manual of basic technique. NewYork: Alan R. Liss; 1987. [Google Scholar]
- 15.Collins QF, Xiong Y, Lupo EG, et al. p38 mitogen-activated protein kinase mediates free fatty acid-induced gluconeogenesis in hepatocytes. J Biol Chem. 2006;281:24336–24344. doi: 10.1074/jbc.M602177200. [DOI] [PubMed] [Google Scholar]
- 16.Sood AK, Buller RE. Genomic instability in ovarian cancer: a reassessment using an arbitrarily primed polymerase chain reaction. Oncogene. 1996;13:2499–2504. [PubMed] [Google Scholar]
- 17.Calvisi DF, Factor VM, Ladu S, et al. Disruption of beta-catenin pathway or genomic instability define two distinct categories of liver cancer in transgenic mice. Gastroenterology. 2004;126:1374–1386. doi: 10.1053/j.gastro.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Jin Z, May WS, Gao F, et al. Bcl2 suppresses DNA repair by enhancing c-Myc transcriptional activity. J Biol Chem. 2006;281:14446–14456. doi: 10.1074/jbc.M511914200. [DOI] [PubMed] [Google Scholar]
- 19.Yang H, Ara AI, Magilnick N, et al. Expression pattern, regulation, and functions of methionine adenosyltransferase 2beta splicing variants in hepatoma cells. Gastroenterology. 2008;134:281–291. doi: 10.1053/j.gastro.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veal N, Hsieh CL, Xiong S, et al. Inhibition of lipopolysaccharide-stimulated TNF-alpha promoter activity by S-adenosylmethionine and 5′-methylthioadenosine. Am J Physiol Gastrointest Liver Physiol. 2004;287:G352–G362. doi: 10.1152/ajpgi.00316.2003. [DOI] [PubMed] [Google Scholar]
- 21.Patel PR, Bevan RJ, Mistry N, et al. Evidence of oligonucleotides containing 8-hydroxy-2′-deoxyguanosine in human urine. Free Radic Biol Med. 2007;42:552–558. doi: 10.1016/j.freeradbiomed.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 22.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat Res. 2000;460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 23.Gaiddon C, Moorthy NC, Prives C. Ref-1 regulates the transactivation and pro-apoptotic functions of p53 in vivo. EMBO J. 1999;18:5609–5621. doi: 10.1093/emboj/18.20.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miled C, Pontoglio M, Garbay S, et al. A genomic map of p53 binding sites identifies novel p53 targets involved in an apoptotic network. Cancer Res. 2005;65:5096–5104. doi: 10.1158/0008-5472.CAN-04-4232. [DOI] [PubMed] [Google Scholar]
- 25.Mann CD, Neal CP, Garcea G, et al. Prognostic molecular markers in hepatocellular carcinoma: a systematic review. Eur J Cancer. 2007;43:979–992. doi: 10.1016/j.ejca.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Kusano N, Okita K, Shirahashi H, et al. Chromosomal imbalances detected by comparative genomic hybridization are associated with outcome of patients with hepatocellular carcinoma. Cancer. 2002;94:746–751. doi: 10.1002/cncr.10254. [DOI] [PubMed] [Google Scholar]
- 27.Trush MA, Kensler TW. An overview of the relationship between oxidative stress and chemical carcinogenesis. Free Radic Biol Med. 1991;10:201–209. doi: 10.1016/0891-5849(91)90077-g. [DOI] [PubMed] [Google Scholar]
- 28.Fung H, Bennett RA, Demple B. Key role of a downstream specificity protein 1 site in cell cycle-regulated transcription of the AP endonuclease gene APE1/APEX in NIH3T3 cells. J Biol Chem. 2001;276:42011–42017. doi: 10.1074/jbc.M106423200. [DOI] [PubMed] [Google Scholar]
- 29.Garcea R, Daino L, Pascale R, et al. Protooncogene methylation and expression in regenerating liver and preneoplastic liver nodules induced in the rat by diethylnitrosamine: effect of variations of S-adenosylmethionine:S-adenosylhomocysteine ratio. Carcinogenesis. 1989;10:1183–1192. doi: 10.1093/carcin/10.7.1183. [DOI] [PubMed] [Google Scholar]
- 30.Yan MD, Xu WJ, Lu LR, et al. Ubiquitin conjugating enzyme Ubc9 is involved in protein degradation of redox factor-1 (Ref-1) Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 2000;32:63–68. [PubMed] [Google Scholar]
- 31.Prudova A, Bauman Z, Braun A, et al. S-adenosylmethionine stabilizes cystathionine beta synthase and modulates redox capacity. Proc Natl Acad Sci U S A. 2006;103:6489–6494. doi: 10.1073/pnas.0509531103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kery V, Poneleit L, Kraus JP. Trypsin cleavage of human cystathionine beta-synthase into an evolutionarily conserved active core: structural and functional consequences. Arch Biochem Biophys. 1998;355:222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- 33.Scott JW, Hawley SA, Green KA, et al. CBS domains form energysensing modules whose binding of adenosine ligands is disrupted by disease mutations. J Clin Invest. 2004;113:274–284. doi: 10.1172/JCI19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ou XP, Yang HP, Ramani K, et al. Inhibition of human betaine homocysteine methyltransferase expression by S-adenosylmethionine and methylthioadenosine. Biochem J. 2007;401:87–96. doi: 10.1042/BJ20061119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Maso V, Avellini C, Crocè LS, et al. Subcellular localization of APE1/Ref-1 in human hepatocellular carcinoma: possible prognostic significance. Mol Med. 2007;13:89–96. doi: 10.2119/2006-00084.DiMaso. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huamani J, McMahan CA, Herbert DC, et al. Spontaneous mutagenesis is enhanced in Apex heterozygous mice. Mol Cell Biol. 2004;24:8145–8153. doi: 10.1128/MCB.24.18.8145-8153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]