Abstract
Aberrant methylation of gene promoters and corresponding loss of gene expression plays a critical role in the initiation and progression of colorectal cancer. An IL-6-type cytokine receptor, leukemia inhibitory factor receptor (LIFR), is a component of cell-surface receptor complexes for multi-functional cytokines such as LIF. Herein, we report that LIFR is methylated in human colon cancer. LIFR promoter was methylated in primary tumor tissues with high frequency (65%, 52/80). Quantitative methylation-specific PCR (TaqMan-MSP) demonstrated differential promoter methylation of LIFR in primary colorectal cancer tissues as compared to normal colon tissues (5%, 4/80). LIFR methylation was not detectable in 13 normal colon mucosa samples obtained from patients without cancer. The mRNA expression of LIFR was significantly down-regulated in colon cancer tissues as compared to corresponding normal tissues. A strong expression of LIFR protein was observed in all non-malignant normal and adjacent normal colon mucosa tissues whereas down-regulated LIFR protein expression was observed in primary tumors. These results demonstrate that cancer-specific methylation and a specific decrease of LIFR expression are a common inactivation event in colon cancer development.
Keywords: leukemia inhibitory factor receptor, methylation, colorectal carcinoma
Introduction
In addition to genetic mutations of oncogenes and TSGs, carcinogenic progression from benign neoplasm to adeno-carcinoma can occur through epigenetic changes in gene promoters (1). Epigenetic alterations have been widely recognized to play an important role in the development of cancer. Aberrant methylation of gene promoters and corresponding loss of gene expression play a critical role in the initiation and progression of colorectal cancer (CRC), and have been recognized as one of the hallmarks of cancer (2). Knowledge of methylation patterns across the genome can help to identify key genes inactivated during tumor formation. Genes that display cancer-specific methylation may serve as biomarkers for early detection, diagnosis and prognosis of cancer (3-6). Genes commonly hypermethylated in CRC include hMLH1, p16INK4a, RAR-β, APC, MGMT, cyclin A1, COX-2 and WT-1 (7-9).
The IL-6 cytokines, including IL-6, leukemia inhibitory factor (LIF), and oncostatin M (OSM), cooperate with a variety of inflammatory cytokines, and play an important role in the local inflammatory process and in the systemic acute phase reaction. IL-6 cytokines have also been implicated in early embryonic development, differentiation as well as tumor growth (10-16). The ability of a cell type to respond to IL-6 cytokines is determined by the expression pattern of the receptor subunit genes (17,18). The leukemia inhibitory factor receptor (LIFR) subunit is a component of cell-surface receptor complexes for the multifunctional cytokines, LIF, cardiotrophin-1, ciliary neurotrophic factor, and human OSM (19). LIF signaling through a receptor heterodimer of LIFR and gp130 induces tyrosine phosphorylation of STAT3 leading to target gene expression (20), and inhibits proliferation of cancer cell lines (18) and non-malignant human breast epithelial cells which express gp130, LIFR and the oncostatin M-specific receptor (OSMR) (21). Recently, suppression of LIFR expression and modification of cellular responsiveness to LIF by epigenetic alterations was reported. Treatment with FR901228, an HDAC inhibitor, induced LIFR transcription in cells that did not express LIFR, and increased the responsiveness to LIF in various cell types, including non-malignant normal and tumor cell lines (22). Interestingly, reduced expression and altered DNA methylation of LIFR was reported in the H-35 rat hepatoma cell line. These cells displayed greatly reduced LIF responsiveness, but when treated with an inhibitor of DNA methyltransferase, 5-Aza-2′-deoxycytidine (5-Aza-dC), displayed increased cellular sensitivity to LIF (23). Epigenetic control of LIFR expression was also observed in human lung cancer and hepatocellular carcinoma cells (24,25). However, little is known about the epigenetic regulation of LIFR in human colon cancer.
We previously reported that LIFR was expressed in a non-tumorigenic human embryonic kidney cell line, HEK293, and HCT116 colon cancer cells, but silenced in most CRC cell lines tested, whereas gp130, the heterodimer of both OSMR and LIFR, was ubiquitously expressed in all the cell lines tested (26). In this report, we examined the methylation status of LIFR gene promoter in human colon cancer. LIFR was frequently methylated in primary CRC tissues but at minimal levels in corresponding normal tissues. A specific decrease of LIFR mRNA and protein in colon cancer was also observed.
Materials and methods
Cell lines and tissues
CRC cell lines were purchased from ATCC (Manassas, VA). CRC cell lines were grown in 5X McCoy medium supplemented with 10% FBS. HEK293 cells were grown in DMEM with 10% FBS. Eighty pairs of DNA from primary colorectal cancers (PT) and matched normal colon mucosa (PN) were described previously (27). Thirteen normal colon epithelial tissues (NN) were obtained from patients without cancer from the Department of Pathology, The Johns Hopkins University. Nine pairs of gDNA used for MSP analysis in this study were from Istanbul University, Turkey.
Bisulfite-sequencing
Bisulfite-modified genomic DNA was amplified by PCR as described previously (28). Primers for the 1st CpG island were 5′-TTATGATTTTTTGAGAGCG-3′ (LIFR Bi-F), 5′-CGCCAAAACCTCCCAAAAA-3′ (LIFR Bi-R). Primers for the 2nd CpG island were 5′-TTTGGTGATTATTTTGTTAGAAAAT-3′ (LIFR 2nd Bi-F) 5′-TACCTAAACAACCCAAAACC-3′ (LIFR 2nd Bi-R). The criteria to determine methylation by bisulfite-sequencing was followed as described previously (26).
Methylation-specific PCR (MSP)
Bisulfite-treated DNA was amplified with either methylation-specific or un-methylation specific primers described previously (26). Primer sequences are 5′-TTTTATGATTTTTTGAGAGCGG-3′ (LIFR M-SP-F), 5′-CCCAAAAACAACGATAACGAC-3′ (LIFR M-SP-R), 5′-TTTTATGATTTTTTGAGAGTGG-3′ (LIFR UnM-SP-F), and 5′-CCCAAAAACAACAATAACAAC-3′ (LIFR UnM-SP-R). Lymphocyte DNA from a healthy individual was methylated in vitro with excess SssI methyltransferase (New England Biolabs Inc., Beverly, MA) to generate completely methylated DNA, and 9 ng of the DNA after bisulfite treatment was used as a positive control for MSP.
Quantitative methylation-specific PCR (TaqMan-MSP)
Quantitative methylation analysis was performed as described (26). LIFR M-SP-F and LIFR M-SP-R primers were used for TaqMan-MSP and the probe sequence was 5′-FAM-GACCGAA TCGCTCCAACGAAACT-TAMRA-3′. The methylation ratio (TaqMan methylation value, TaqMeth V) was defined as the quantity of fluorescence intensity derived from promoter amplification of LIFR divided by fluorescence intensity from β-actin amplification, and multiplied by 100.
5-Aza-dC treatment and RT-PCR
Treatment with 5-aza-2′-deoxycytidine (5-Aza-dC) and/or trichostatin A (TSA), and RNA extraction and cDNA synthesis were followed as described (28). Primer sequences for RT-PCR were as previously reported (29). To determine quantitation of gene expression, the real-time RT-PCR was performed as described (26). Primer sequences for real-time RT-PCR are 5′-TGTATGTGG TGACAAAGGAAAA-3′ (LIFR realT-F), 5′-TGGATT TGGAATATCAG GGTAGA-3′ (LIFR realT-R). Five matched normal and tumor cDNA (a-e) were purchased from Clontech Laboratories, Inc. (Mountain View, CA), and cDNA from a patient without cancer and from an CRC patient were purchased from BioChain Institute, Inc. (Hayward, CA).
Immunohistochemistry
Tissue microarrays were performed with sections (5 μm) of colon cancer tissues, adjacent tissues 1.5 cm away from tumor, and non-malignant normal colon tissues which were purchased from US Biomax, Inc. (Rockville, MD), and IHC was performed as described (26) with anti-LIFR rabbit polyclonal antibody (1:100 dilution) (Santa Cruz Biotechnology, Santa Cruz, CA).
Statistical analysis
We used gene methylation levels (TaqMeth V) to construct a receiver operating characteristic (ROC) curve for the detection of colon cancer as described (26).
Results
LIFR is methylated in CRC cell lines and primary CRC tissues
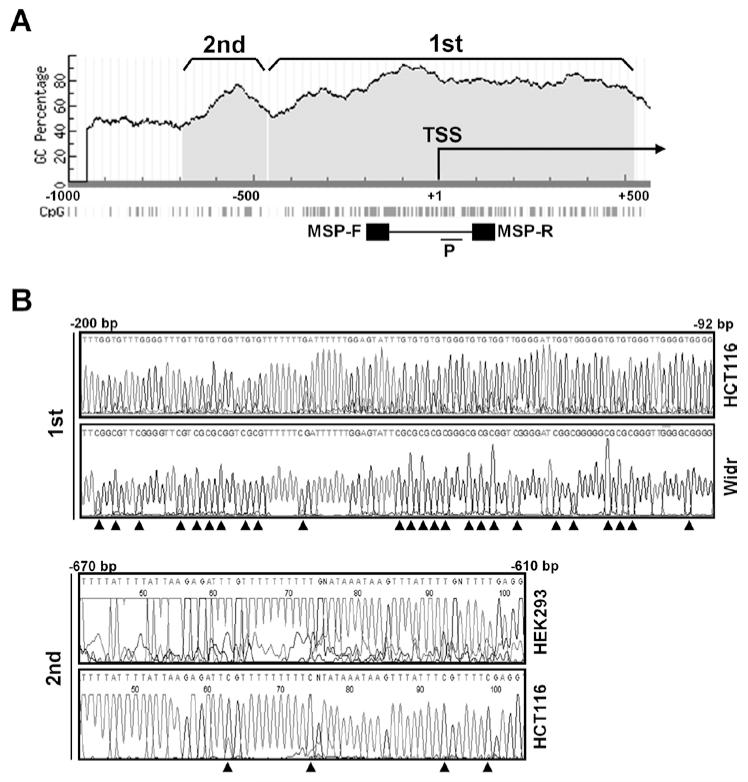
To investigate promoter methylation of LIFR, we searched CpG islands in the LIFR promoter within 5 kb upstream and 5 kb downstream of the transcription start site (TSS) by Methprimer. We found two CpG islands in the LIFR promoter that closely reside at −458 ~ +526 bp (1st CpG island) and −691 ~ −465 (2nd CpG island) with high GC density (50-90%) (Fig. 1A). Two pairs of sequencing primers were designed in the 1st region from −253 bp to +65 bp that harbors a total of 50 CpGs, and in the 2nd region from −843 to −546 bp where 19 CpGs reside. MSP primers were designed at the region from −255 to +54 bp in the 1st CpG island, and a probe (P) for TaqMan-MSP was designed within this region.
Figure 1.
CpG islands upstream and downstream of the TSS in the LIFR promoters. (A) A graphic of CpG islands was taken from Methprimer. bar, PCR-amplified region for MSP. TSS, transcription start site. F, forward; R, reverse. P, a probe used for TaqMan-MSP. (B) Representative bisulfite-sequence results of HCT116 and Widr cells in the 1st region (upper), and HEK293 and HCT116 in the 2nd region. In the 2nd regions, bisulfite-sequencing was performed in 7 CRC and HEK293 cells, whereas in the 1st region, it was performed in HCT116, Widr and SW480 cells. Arrowhead, methyl cytosines that were not converted to thymine after bisulfite treatment.
LIFR methylation status was examined in CRC cell lines and HEK293 cells by bisulfite-sequencing as a pilot study. In the 1st region, HCT116 cells harbored methylation only in one of 50 CpGs, whereas Widr and SW480 cells harbored methylation in 47 of 50 CpGs. In the 2nd region, HEK293 cells harbored no CpG methylation of the LIFR promoter. In contrast, all CRC cell lines examined harbored complete methylation in all 19 CpGs in the 2nd region. Representative sequencing results are shown in Fig. 1B.
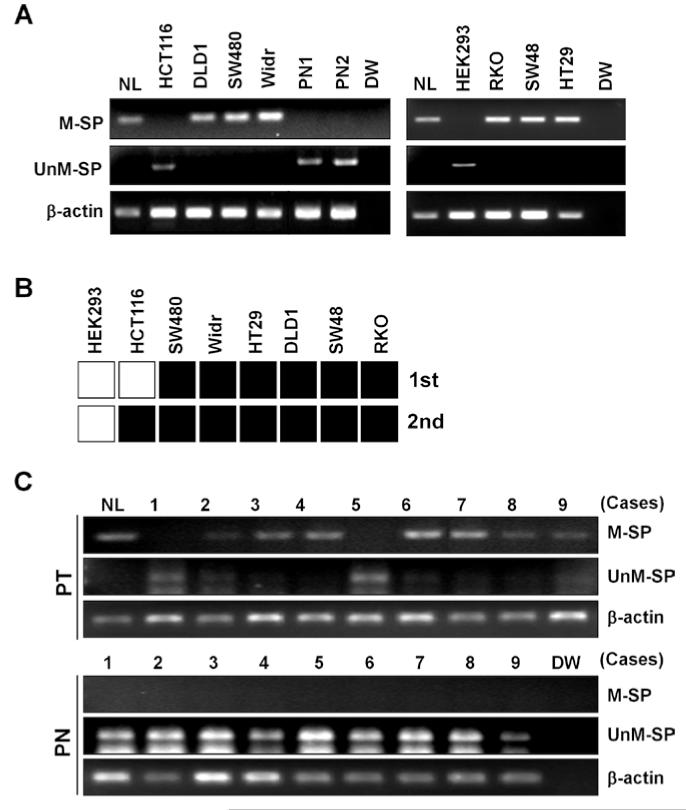
We then performed MSP in the cell lines and two normal colon mucosa tissues collected from CRC patients (PN1 and PN2). PCR-amplification with methylation-specific primers (M-SP) was clearly seen in SW480, DLD1, HT29, Widr, SW48, and RKO cells, whereas no methylated DNA was detected in HCT116, HEK293 cells and two normal colon mucosa tissues (Fig. 2A). Consistent with the results of bisulfite-sequencing, un-methylated DNA was seen in HCT116, HEK293 cells and in the two normal colon mucosa tissues. However, un-methylated DNA was not observed in the remaining 6 CRC cell line, indicating that HEK293 cells harbor only un-methylated LIFR at least at the two regions examined, and that CRC cell lines except for HCT116 cells harbor dense CpG methylation in the promoter (Fig. 2B). Methylation of the LIFR promoter was further investigated in 9 pairs of colon cancer (PT) and matched normal colon mucosa tissues (PN). Methylated DNA was not detected in the normal colon tissues, but clearly observed in 7 of 9 PT cases (77%) (Fig. 2C). Conversely, non-methylated DNA was observed in all 9 PN cases, but only in the two PT cases in which methylated DNA was not observed.
Figure 2.
Methylation of LIFR in CRC cell lines and primary colon tissues. (A) LIFR methylation was examined by MSP after PCR with methylation-(M-SP) and un-methylation-specific (UnM-SP) primers. In vitro methylated, bisulfite-treated human normal lymphocyte DNA (NL) was used as a positive control, and distilled water (DW) was used as a negative control for PCR. β-actin was used to confirm integrity of bisulfite-treated DNA. (B) Results of LIFR methylation in cell lines determined by bisulfite-sequencing or MSP were combined. bisulfite-sequencing was performed in HCT116, Widr, and SW480 for the 1st region, and in HEK293 and CRC cell lines for the 2nd region. MSP was performed in HEK293 and CRC cell lines for the 1st region. Black square, the presence of methylation; white square, the absence of methylation. (C) Methylation results of LIFR in 9 pairs of colon cancer (PT) and matched normal colon mucosa tissues (PN).
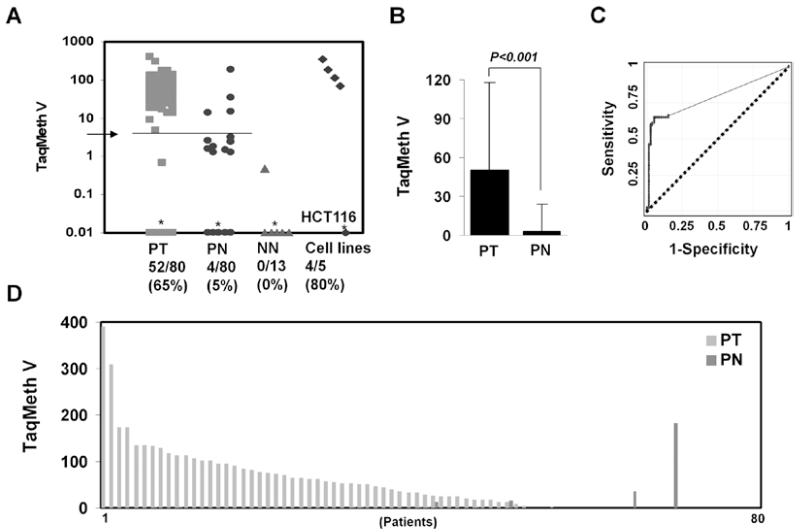
To quantify LIFR promoter methylation, real-time MSP analysis was performed in 80 pairs of colon normal tissues (PN) and primary CRC (PT) as well as 5 CRC cell lines (hCT116, DLD1, SW480, Widr and RKO). Thirteen colon normal mucosa tissues of the non-cancer patients (NN) were also included to compare methylation specificity between cancer and non-cancer patients. In 80 pairs of colon samples, methylation values (TaqMeth V) in tumor ranged from 0 to 390.03 (median value 56.25), and in normal colon from 0 to 181.94 (median value 0.00) (Fig. 3A). The overall TaqMeth V levels detected in primary CRC (50.40±67.21, mean ± SD) were also significantly higher than that in corresponding normal tissues (3.26±20.71, mean ± SD) (P<0.001) (Fig. 3B).
Figure 3.
Quantitative level of LIFR methylation. (A) A Scatter plot of LIFR methylation levels. *Samples with a ratio equal to zero could not be plotted correctly on a log scale, so are presented here as 0.01. TaqMeth v is described in Materials and methods. Arrow, an optimal cut-off value for LIFR calculated from ROC analysis (3.198). (B) The overall TaqMeth V detected in primary CRC were significantly higher than that in corresponding normal tissues (P<0.001, Wilcoxon matched-pairs signed-ranks test). (C) ROC curve analysis of TaqMeth V of LIFR. The area under ROC (AUROC, 0.7928±0.032) conveys the accuracy in distinguishing matched normal colon (PN) from CRC (PT) in terms of its sensitivity and specificity (P<0.001). Solid line, LIFR; dash line, no discrimination. (D) Methylation levels of normal and tumor tissues in individual patients.
Methylation of LIFR yielded a highly discriminative receiver-operator characteristic (ROC) curve profile, clearly distinguishing CRC from corresponding normal mucosa (Fig. 3C). The optimal cut-off (value, 3.198) was calculated from the ROC analysis in order to maximize sensitivity and specificity. At this cut-off, the specificity was 95% (76/80) and sensitivity was 65% (52/80) (P<0.001, Fisher’s exact test). Only 4 of 80 of matched normal colon mucosa displayed TaqMeth V over the cut-off. Only 1 (TaqMeth V = 0.4870) of 13 NN samples and 12 of 80 of PN displayed a TaqMeth V over 0.00. The methylation level in all remaining 12 cases of NN, 27 cases of PT, and 68 cases of PN was undetectable. A high level of LIFR promoter methylation was also found in DLD1, SW480, Widr and RKO cells, but LIFR methylation in HCT116 cells was not detected by TaqMan-MSP. In addition to a simple frequency, the comparison of methylation level of normal and tumor tissues from the same individual patients revealed that the majority of the tumor tissues harbored much higher values than matched normal colon mucosa (Fig. 3D). Taken together, LIFR was frequently methylated in primary CRC tissues but at minimal levels in corresponding normal tissue.
LIFR is down-regulated in colon cancer
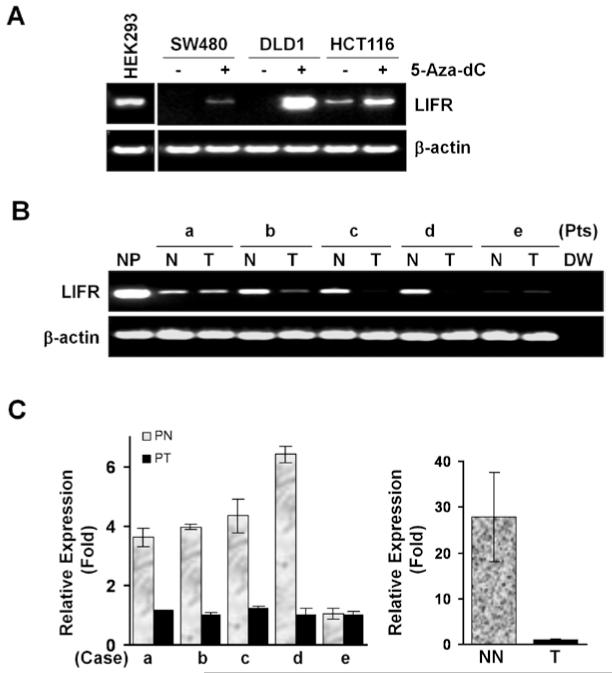
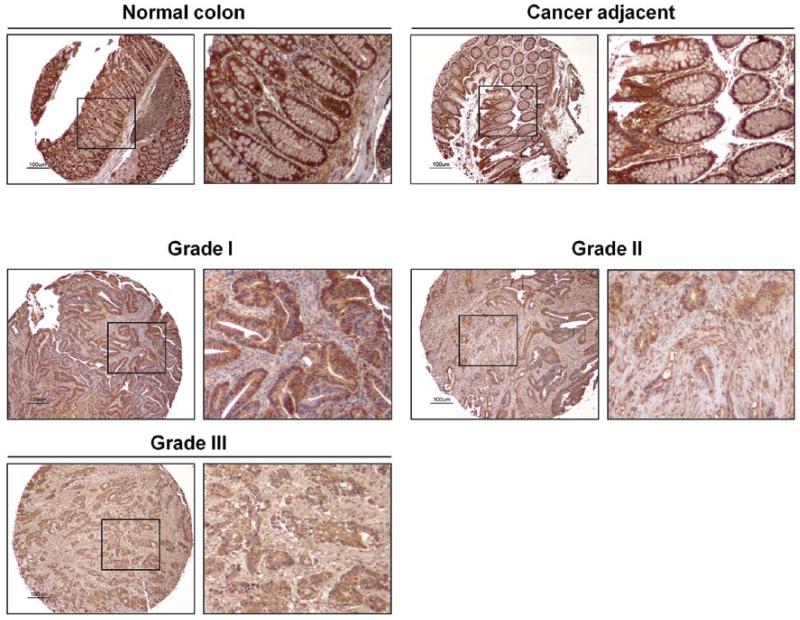
To investigate the mRNA expression of LIFR, RT-PCR analysis was performed. LIFR expression was observed only in the cells where methylation in the 1st region was not found (HCT116 and HEK293) (Fig. 4A) (26), indicating that methylation in the 1st CpG island is enough to silence LIFR gene expression. The expression of LIFR was much higher in HEK293 cells harboring no methylation in the 2nd region than HCT116 cells harboring a dense methylation at the region, implicating that the 2nd CpG island regulates a basal level of LIFR. The other 6 CRC cell lines that harbored LIFR methylation in both regions did not express the LIFR transcript (data not shown) (26). The silenced LIFR in SW480 and DLD1 was re-activated by the treatment of the de-methylating agent, 5-Aza-dC (5 μM), suggesting a tight correlation between promoter methylation and expression of the LIFR. The expression in HCT116 was also further increased by 5-Aza-dC treatment, indicating that the 2nd region of the LIFR promoter might be de-methylated by the treatment. To examine the expression of LIFR in human tissue, we performed RT-PCR in cDNA prepared from tumor (PT) and corresponding normal tissues (PN) of five individual colon cancer patients (matched cDNA). In 3 of 5 tumor cases, LIFR was down-regulated or not detectable (Fig. 4B). To quantify the expression level, we performed real-time RT-PCR in the same panel of cDNA samples. In 4 of 5 tumor cases, LIFR was significantly down-regulated (P=0.043, Wilcoxon matched-pairs signed-ranks test) (Fig. 4C, left). The expression of LIFR in tumor from a CRC patient was 28 times lower than in normal tissue (NN) from a patient without cancer (P<0.001, t-test) (Fig. 4C, right). By immunohistochemical staining of a colon normal and cancer tissue microarray with an anti-LIFR antibody, strong expression of LIFR was detected in all non-malignant normal tissues and adjacent normal colon mucosa from colon cancer patients (Fig. 5). However, faint or only mild expression of LIFR was detected in 9 of 10 primary tumors (Table I). These results suggest a specific decrease of LIFR mRNA and protein in colon cancer development.
Figure 4.
mRNA expression of LIFR in CRC. RT-PCR analysis in cell lines (A), and five pairs (a-e) of normal (N) and tumor cDNA (T) prepared from CRC patients (Pts) (B). NP, cDNA from normal placenta. (C) Real-time RT-PCR in the five pairs of cDNA (left). The LIFR expression between patients with colon cancer (T) or without cancer (NN) (right). Relative expression (fold) was calculated by comparing the ratios of mRNA expression of LIFR to an internal control gene, GAPDh. Experiments were done in duplicate, and values indicate means ± SD.
Figure 5.
Protein expression of LIFR. Immunohistochemical analysis was performed in colon cancer tissue microarray with normal colon, cancer adjacent colon, and colon adenocarcinoma. Small portions of each tissue are shown with a high magnification. Tumor grades are indicated. Scale bar, 100 μm.
Table I.
Immunohistochemical analysis of LIFR in colon cancer tissue microarray with normal tissue controls.
| Tissues | Expression | Tumor grade |
|---|---|---|
| Colon cancer | ||
| 1 | ++ | III |
| 2 | + | III |
| 3 | ++ | III |
| 4 | + + | III |
| 5 | + | II |
| 6 | ++ | II |
| 7 | + | II |
| 8 | + | I |
| 9 | + | I |
| 10 | +++ | I |
| Non-malignant normal colon |
||
| 1 | ++++ | |
| 2 | ++++ | |
| 3 | ++++ | |
| 4 | ++++ | |
| 5 | ++++ | |
| 6 | ++++ | |
| Cancer adjacent normal colon |
||
| 1 | ++++ | |
| 2 | ++++ | |
| 3 | +++ | |
| 4 | +++ | |
| 5 | +++ | |
| 6 | ++++ | |
| 7 | ++++ | |
| 8 | ++++ |
Expression level is indicated as: +, faint; ++, mild; +++, moderate; ++++, strong.
Discussion
Colorectal cancer (CRC) is the second leading cause of cancer in both men and women worldwide (30). At diagnosis, 19% of CRC cases are metastatic, and the overall 5-year survival rate for patients with metastatic CRC is less than 10% (30). Mortality rates for colon cancer have fallen during the past 20 years because of early detection from increased screening. However, more advanced knowledge of the molecular pathogenesis of CRC or key factors in CRC development is still needed. Finding potential diagnostic molecular markers for CRC would thus be invaluable for early detection and potentially treatment of CRC.
Epigenetic events play key roles in the development and progression of CRC, and aberrant DNA methylation of cancer-related genes is one of the major mechanisms in carcinogenesis leading to transcriptional silencing of tumor suppressor genes (31). DNA methylation in gene promoter is among the earliest and most frequent alterations, and occurs at various stages in colorectal neoplasia. A number of genes are commonly hypermethylated in CRC (7-9). However, genes methylated only in neoplastic tissues with high frequency are rare. Previously, we have reported aberrant methylation of OSMR (26), NMDAR2A (27) and DFNA5 (32) in CRC. Methylation of these genes was detected in more than 80% of primary CRC tissues.
In this study, we identified LIFR as a gene harboring cancer-specific promoter methylation in human colorectal cancer. The high frequency of LIFR methylation in CRC (65%) ranks with only a few other genes methylated at high frequency in CRC (Cyclin A1, CDX1, RAR-β, MYOD1, p15INK4b and COX-2) in a cancer-specific manner (8). A clear discrimination of tumor from normal tissues by TaqMan-MSP of the LIFR, indicating that detection of LIFR promoter methylation in blood or stool DNA may have potential for identifying individuals with cancer. Moreover, a strict correlation between promoter methylation and silencing of LIFR in CRC cell lines implies that promoter methylation of LIFR is one of the mechanisms of gene sliencing in CRC.
Loss of LIFR expression has been reported in high Gleason grade carcinomas of the prostate (33), suggesting that LIFR silencing is important to predict increased malignancy in the early stages of cancer. In our results, the mRNA level of LIFR was down-regulated in 4 of 5 colon cancers, and silenced in CRC cell lines that harbored LIFR methylation. Down-regulation of LIFR protein in primary CRC tissues was observed by IHC analysis, but a correlation between decreased LIFR expression and tumor grades was not observed. However, it is still unknown if absence of LIFR may distinguish clinically significant cancers from simple tumors that are unlikely to progress to aggressive cancers. Future work will be focused on LIFR as a diagnostic biomarker for aggressive, metastatic colon cancer.
The interleukin-6 (IL-6)-type cytokines, OSM and LIF, have two specific heterodimeric signaling complexes; gp130/LIFR and gp130/OSMR (34). gp130/OSMR is activated only by OSM, but gp130/LIFR can be activated by either LIF, OSM or both (17). The gp130/OSMR complex activates OSM-specific signaling pathways via the JNK/SAPK and Stat1/Stat5 pathways, whereas both gp130/LIFR and gp130/OSMR complexes activate Stat3 and Erk as common signaling pathways (35). Previously, we reported decreased transcription of OSMR in CRC as well as a significant growth inhibition by OSM in OSMR-expressing HCT116 cells, which was reversed by gene knock-down of OSMR (26). However, the suppression of cell growth by OSM was not observed in SW480 and DLD-1 cells that did not express OSMR due to the promoter methylation of OSMR, whereas it was observed when 5-Aza-dC was treated in those cells. Interestingly, a correlation of LIFR down-regulation with reduced cell responsiveness to LIF has been reported. The H-35 cells that harbored DNA methylation and reduced expression of LIFR had a reduced LIF responsiveness, but exhibited an increased cellular sensitivity to LIF by 5-Aza-dC treatment (23). Treatment with FR901228 (FR), an HDAC inhibitor, induced LIFR transcription in various cell types that did not express LIFR, and increased the responsiveness to LIF (22). The FR-stimulated expression of IL-6-type cytokine receptors in tumor cells supports suppression of cell growth by utilizing the growth inhibitory effect of these cytokines (22). Thus, methylated OSMR and LIFR in CRC might decrease tumor-inhibiting signals from IL-6-type cytokines and key downstream signaling events.
It has been reported that LIF together with LIFR appear to form a more stable complex with gp130 than OSM with gp130/OSMR, and co-activation of LIFR and OSMR results in a predominant LIF-like response (36). These results indicate that a hierarchical order of cytokine receptor action exists in which LIFR ranks as a dominant member. Therefore, loss of LIFR expression by the promoter methylation might have more profound effects on the signaling pathways activated by the IL-6-type cytokines. Further study will be focused on a dominant role of LIFR on downstream signaling pathways contributing to CRC development.
In conclusion, we have demonstrated cancer-specific methylation of LIFR in human colon cancer. A correlation between promoter methylation and expression of LIFR implicates this gene as a common inactivation event in colon tumorigenesis. Inactivation of OSMR and LIFR by promoter methylation appears to constitute different nodal points for abrogation of IL-6 induced tumor suppression and thus identify a targeted therapeutic pathway. Due to its high frequency in primary tumors and near absence in normal tissues, detection and quantification of LIFR methylation thus deserves further attention as a diagnostic biomarker. Detection of LIFR methylation levels in stool and/or plasma DNA may prove valuable in the diagnosis and monitoring of patients with colon cancer.
Acknowledgements
This study was supported by National Cancer Institute (U01-CA84986) and the Flight Attendant Medical Research Institute (Myoung S. Kim) and Oncomethylome Sciences, SA. Under a licensing agreement between Oncomethylome Sciences, SA and the Johns Hopkins University, D. Sidransky is entitled to a share of royalty received by the University upon sales of any products described in this report. D. Sidransky owns Oncomethylome Sciences, SA stock, which is subject to certain restrictions under university policy. D. Sidransky is a paid consultant to Oncomethylome Sciences, SA and is a paid member of the company’s Scientific Advisory Board. The Johns Hopkins University in accordance with its conflict of interest policies is managing the terms of this agreement.
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 3.Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 4.Mandelker DL, Yamashita k, Tokumaru Y, et al. PGP9.5 promoter methylation is an independent prognostic factor for esophageal squamous cell carcinoma. Cancer Res. 2005;65:4963–4968. doi: 10.1158/0008-5472.CAN-04-3923. [DOI] [PubMed] [Google Scholar]
- 5.Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmisano WA, Divine KK, Saccomanno G, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 7.Esteller M, Fraga MF, Guo M, et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum Mol Genet. 2001;10:3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- 8.Xu XL, Yu J, Zhang HJ, et al. Methylation profile of the promoter CpG islands of 31 genes that may contribute to colorectal carcinogenesis. World J Gastroenterol. 2004;10:3441–3454. doi: 10.3748/wjg.v10.i23.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lind GE, Thorstensen L, Løvig T, et al. A CpG island hyper-methylation profile of primary colorectal carcinomas and colon cancer cell lines. Mol Cancer. 2004;3:28. doi: 10.1186/1476-4598-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auernhammer CJ, Melmed S. Leukemia-inhibitory factor-neuroimmune modulator of endocrine function. Endocr Rev. 2000;21:313–345. doi: 10.1210/edrv.21.3.0400. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida K, Taga T, Saito M, et al. Targeted disruption of gp130, a common signal transducer for the interleukin 6 family of cytokines, leads to myocardial and hematological disorders. Proc Natl Acad Sci USA. 1996;93:407–411. doi: 10.1073/pnas.93.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ware CB, Horowitz MC, Renshaw BR, et al. Targeted disruption of the low-affinity leukemia inhibitory factor receptor gene causes placental, skeletal, neural and metabolic defects and results in perinatal death. Development. 1995;121:1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- 13.Barasch J, Yang J, Ware CB, et al. Mesenchymal to epithelial conversion in rat metanephros is induced by LIF. Cell. 1999;99:377–386. doi: 10.1016/s0092-8674(00)81524-x. [DOI] [PubMed] [Google Scholar]
- 14.Bajaj P, Lawry J, Shenton G, et al. Interleukin-6 and tumour necrosis factor alpha synergistically block S-phase cell cycle and upregulate intercellular adhesion molecule-1 expression on MCF7 breast carcinoma cells. Cancer Lett. 1993;71:143–149. doi: 10.1016/0304-3835(93)90109-m. [DOI] [PubMed] [Google Scholar]
- 15.Klausen P, Pedersen L, Jurlander J, et al. Oncostatin M and interleukin 6 inhibit cell cycle progression by prevention of p27kip1 degradation in HepG2 cells. Oncogene. 2000;19:3675–3683. doi: 10.1038/sj.onc.1203707. [DOI] [PubMed] [Google Scholar]
- 16.Nair BC, De Vico AL, Nakamura S, et al. Identification of a major growth factor for AIDS-kaposi’s sarcoma cells as oncostatin M. Science. 1992;255:1430–1432. doi: 10.1126/science.1542792. [DOI] [PubMed] [Google Scholar]
- 17.Mosley B, De Imus C, Friend D, et al. Dual oncostatin M (OSM) receptors. Cloning and characterization of an alternative signaling subunit conferring OSM-specific receptor activation. J biol Chem. 1996;271:32635–32643. doi: 10.1074/jbc.271.51.32635. [DOI] [PubMed] [Google Scholar]
- 18.Douglas AM, Goss GA, Sutherland RL, et al. Expression and function of members of the cytokine receptor superfamily on breast cancer cells. Oncogene. 1997;14:661–669. doi: 10.1038/sj.onc.1200882. [DOI] [PubMed] [Google Scholar]
- 19.Tomida M. Structural and functional studies on the leukemia inhibitory factor receptor (LIF-R): gene and soluble form of LIF-R, and cytoplasmic domain of LIF-R required for differentiation and growth arrest of myeloid leukemic cells. Leuk Lymphoma. 2000;37:517–525. doi: 10.3109/10428190009058503. [DOI] [PubMed] [Google Scholar]
- 20.Boccardo F, Rubagotti A, Conti G, et al. Prednisone plus gefitinib versus prednisone plus placebo in the treatment of hormone-refractory prostate cancer: a randomized phase II trial. Oncology. 2008;74:223–228. doi: 10.1159/000151391. [DOI] [PubMed] [Google Scholar]
- 21.Grant SL, Douglas AM, Goss GA, et al. Oncostatin M and leukemia inhibitory factor regulate the growth of normal human breast epithelial cell. Growth Factors. 2001;19:153–162. doi: 10.3109/08977190109001083. [DOI] [PubMed] [Google Scholar]
- 22.Blanchard F, Kinzie E, Wang Y, et al. FR901228, an inhibitor of histone deacetylases, increases the cellular responsiveness to IL-6 type cytokines by enhancing the expression of receptor proteins. Oncogene. 2002;21:6264–6277. doi: 10.1038/sj.onc.1205777. [DOI] [PubMed] [Google Scholar]
- 23.Blanchard F, Tracy E, Smith J, et al. DNA methylation controls the responsiveness of hepatoma cells to leukemia inhibitory factor. Hepatology. 2003;38:1516–1528. doi: 10.1016/j.hep.2003.09.029. [DOI] [PubMed] [Google Scholar]
- 24.Loewen GM, Tracy E, Blanchard F, et al. Transformation of human bronchial epithelial cells alters responsiveness to inflammatory cytokines. BMC Cancer. 2005;5:145. doi: 10.1186/1471-2407-5-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamura Y, Nomoto S, Kanda M, Li Q, et al. Leukemia inhibitory factor receptor (LIFR) is detected as a novel suppressor gene of hepatocellular carcinoma using double-combination array. Cancer Lett. 2010;289:170–177. doi: 10.1016/j.canlet.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Kim MS, Louwagie J, Carvalho B, et al. Promoter DNA methylation of Oncostatin M receptor-β as a novel diagnostic and therapeutic marker in colon cancer. Plos One. 2009;4:e6555. doi: 10.1371/journal.pone.0006555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MS, Chang X, Nagpal Jk, et al. The N-methyl-D-aspartate receptor type 2A is frequently methylated in human colorectal carcinoma and suppresses cell growth. Oncogene. 2008;27:2045–2054. doi: 10.1038/sj.onc.1210842. [DOI] [PubMed] [Google Scholar]
- 28.Kim MS, Yamashita K, Baek JH, et al. N-methyl-D-aspartate receptor type 2B is epigenetically inactivated and exhibits tumor-suppressive activity in human esophageal cancer. Cancer Res. 2006;66:3409–3418. doi: 10.1158/0008-5472.CAN-05-1608. [DOI] [PubMed] [Google Scholar]
- 29.Rockman SP, Demmler k, Roczo N, et al. Expression of interleukin-6, leukemia inhibitory factor and their receptors by colonic epithelium and pericryptal fibroblasts. J Gastroenterol Hepatol. 2001;16:991–1000. doi: 10.1046/j.1440-1746.2001.02588.x. [DOI] [PubMed] [Google Scholar]
- 30.Jemal A, Murray T, Ward E, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55:10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 31.Toyota M, Ohe-Toyota M, Ahuja N, et al. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci uSA. 2000;97:710–715. doi: 10.1073/pnas.97.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim MS, Chang X, Yamashita K, et al. Aberrant promoter methylation and tumor suppressive activity of the DFNA5 gene in colorectal carcinoma. Oncogene. 2008;27:3624–3634. doi: 10.1038/sj.onc.1211021. [DOI] [PubMed] [Google Scholar]
- 33.Royuela M, Ricote M, Parsons MS, et al. Immunohistochemical analysis of the IL-6 family of cytokines and their receptors in benign, hyperplasic, and malignant human prostate. J Pathol. 2004;202:41–49. doi: 10.1002/path.1476. [DOI] [PubMed] [Google Scholar]
- 34.Gearing DP, Comeau MR, Friend DJ, et al. The IL-6 signal transducer, gp130: an oncostatin M receptor and affinity converter for the LIF receptor. Science. 1992;255:1434–1437. doi: 10.1126/science.1542794. [DOI] [PubMed] [Google Scholar]
- 35.Underhill-Day N, heath JK. Oncostatin M (OSM) cytostasis of breast tumor cells: characterization of an OSM receptor beta-specific kernel. Cancer Res. 2006;66:10891–10901. doi: 10.1158/0008-5472.CAN-06-1766. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Robledo O, Kinzie E, et al. Receptor subunit-specific action of oncostatin M in hepatic cells and its modulation by leukemia inhibitory factor. J Biol Chem. 2000;275:25273–25285. doi: 10.1074/jbc.M002296200. [DOI] [PubMed] [Google Scholar]