Abstract
Heat shock proteins (Hsps) play important roles in the environmental adaptation of various organisms. To explore the functions of Hsps in relation to heat stress and development in Cotesia vestalis, a solitary larval endoparasitoid of Plutella xylostella, four heat shock protein genes, CvHsp40, CvHsc70, CvHsp70 and CvHsp90, were cloned and sequenced from C. vestalis by real-time quantitative PCR and RACE. The cDNA sequence of CvHsp40, CvHsc70, CvHsp70 and CvHsp90 were 1473 bp, 2316 bp, 2279 bp and 2663 bp long, which encode proteins with calculated molecular weights (MW) of 39.1 kDa, 71.2 kDa, 70.1 kDa and 83.3 kDa, respectively. Furthermore, the analysis of genomic DNA confirmed that no introns existed in CvHsp40, CvHsp70 and CvHsp90 while two introns were present in CvHsc70. The amino acid sequence analysis of CvHsps indicated that CvHsp40 is a Type II Hsp40 homolog, CvHsp70 and CvHsc70 are the eukaryotic cytoplasmic Hsp70s, and CvHsp90 is the β-isoform of Hsp90. The divergent transcriptional patterns of CvHsp40, CvHsp70 and CvHsp90 in the different developmental stages suggested that CvHsp transcripts were under different mechanisms of regulation during the development of parasitoid larvae. The dramatic increase of transcripts of CvHsp70 at the third-instar larva coincided with its developmental change in this stage, that is, from inside host to outside host. CvHsp40, CvHsc70 and CvHsp70 showed a trend of sex-specific differences of transcript abundance in the adult stage. All CvHsp transcripts in different developmental stages were significantly induced by heat stress, and the lowest transcript abundances appeared around the temperature 27°C, which probably suggest that this is the most favorable temperature for the development of C. vestalis. Our results suggest that the expression of heat shock proteins reflects to some extent the developmental changes and environmental requirements of insects.
Introduction
The oligophagous solitary larval endoparasitoid, Cotesia vestalis (Haliday) ( = Cotesia plutellae (Kurdjomov))(Hymenoptera: Braconidae) [1], is one of the major natural enemies of the diamondback moth (DBM), Plutella xylostella (L.) (Lepidoptera: Plutellidae), one of the very destructive pests of brassica crops in both small-scale and large-scale farming systems worldwide [2], [3]. C. vestalis is distributed in Europe, China, South Africa, Japan, Pakistan, India and Indonesia, and has been introduced from Europe to several countries, including Australia, Commonwealth of Dominica, Fiji, Thailand and the United States and from South Africa to St. Helena [4]. In Hangzhou (China), it is a major parasitoid of P. xylostella, and the highest parasitism (57.2%) of DBM recorded [5]. Shi and Liu reported that the optimal survival temperature for C. vestalis was 25°C [6]. Above 25°C, the developmental rate increased and the longevity decreased, and no female progeny was produced when the temperature was higher than 35°C.
The heat shock proteins (Hsps) represent a super gene family. On the basis of molecular weight (MW) and homology, Hsps are divided into several families, including Hsp100, Hsp90, Hsp70, Hsp60, Hsp40 and small Hsps (sHsps, the molecular weights ranging from 12 to 43 kDa) [7]–[9]. Hsp40s (also called DnaJs) have been conserved throughout evolution and are important for protein homeostasis, where they stimulate the ATPase activity of the Hsp70s that are involved in protein translation, folding, unfolding, translocation, and degradation [10]. Hsp90s participate in the folding, maintenance of structural integrity, and the proper regulation of a subset of cytosolic proteins, and account for 1% of the soluble protein in most tissues, even in the absence of stress [11].
The Hsps have been widely studied in many fields of biology and a large number of publications describe their molecular and physiological functions, including acting as molecular chaperones that participate in diverse physiological processes including physiological interactions between parasitoid wasps and their host insects [7], [12]–[20]. However, the ecological importance of inducible Hsps has been questioned only recently and was rarely addressed. In the laboratory, it has been shown that very small amounts of induced Hsps from model organisms like Drosophila melanogaster can have effects on life history traits such as development, stress resistance, life span and fecundity [21]–[22]. The research and experiments, especially on Hsp70, are processed from laboratory or natural geographic populations of marine organisms, which were exposed to variable environments including occasional stress exposure and environmental conditions. So far, beyond variations in morphology and DNA sequences, variation of temperature tolerance has been accepted as a new bio-indicator of geographic population variations. Meanwhile, the transcript abundances of Hsp genes provide a link between variation of temperature tolerance and geographic populations.
In the present study, we explore three issues. First, we identify the sequences of four C. vestalis Hsp genes. Second, we describe how the transcript abundances of C. vestalis Hsp genes vary during development. Third, we show that the transcript abundances of C. vestalis Hsp genes reflect temperature adaptations of local populations.
Materials and Methods
Insects and thermal treatments
Pupae and parasitized larvae of P. xylostella by the endoparasitoid C. vestalis were initially collected from cabbage fields in the suburbs of Hangzhou, Zhejiang province, China. Once emerged, both P. xylostella and C. vestalis were raised on cabbage grown at 24°C, 65% relative humidity, and 14 h light: 10 h dark. Adult wasps were fed with 20% (v/v) honey solution and propagated using P. xylostella larvae as hosts.
C. vestalis larvae undergo 3 instars before pupation, and are physiologically staged using previously established morphological criteria [23]. Briefly, the first and second larval instars molted inside the host, and the third instar emerged from the host to spin a cocoon; each instar lasted 2, 5, and 1 day, respectively; the adults emerged at 5 days after pupation.
For thermal treatments (24°C -control, 27°C, 32°C, 37°C and 42°C), groups of 15 first-instar larvae, early second-instar larvae and later second-instar larvae, all developing in host larvae, third-instar larvae, pupae and new emerged (one-day-old) adults were collected into 10 ml cotton-plugged tubes in a glycerol bath (Programmable Temperature Controller DFY-5/10, Nanjing Keer Biotechnology Ltd, Nanjing, China) and set at a selected temperature for 1 h. After thermal treatment, all the treated larvae, pupae and adults were flash-frozen in liquid nitrogen and stored at −70°C until RNA exaction. Each treatment with 15 individuals was replicated 3 times.
Total RNA and Genomic DNA isolation, cDNA Synthesis and Cloning of CvHsps
One-day-old female adults were processed for cDNA cloning. Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA). Residual genomic DNA was removed using RNase-free DNase I (Promega, Germany), and 2 μg RNA was used to generate the cDNAs with a RevertAidTM First Strand cDNA Synthesis Kit (Fermentas, Lithuania). The cDNA fragments of CvHsp40, CvHsp70, CvHsc70 and CvHsp90 were obtained by degenerate primers (Table 1) based on the conserved nucleotide sequences of insects which were deposited in GenBank. The gene specific primers of CvHsp40, CvHsp70, CvHsc70 and CvHsp90 (Table 1) were designed for amplifying the full cDNA sequences using a 5′-Full Race Kit and 3′- Full Race Kit (TaKaRa, Dalian, China) and the full open reading frame (ORF) sequences of CvHsp40, CvHsp70, CvHsc70 and CvHsp90 were verified by PCR. Adult wasp genomic DNA was isolated using the DNeasy Tissue Kit (Qiagen, Germany), and the introns of CvHsc70 were amplified using ORF-verified primers (Table 1).
Table 1. Sequences of Primers.
| Gene | Direction 5′→3′ | Sequence | Used for |
| Cvhsp40 | forward | GCNGARGCNTAYGANGTGCT | degenerate PCR |
| Reward | TTBGTDCCNKCCTTCCAKCC | degenerate PCR | |
| outer primer | AGGCGATCAAGGTCGTGGTA | 3′RACE | |
| inner primer | ACCATTCCCGAAAGAACCATCA | 3′RACE | |
| outer primer | ATGGTTCTTTCGGGAATGGT | 5′RACE | |
| inner primer | CCACGACCTTGATCGCCTTCTT | 5′RACE | |
| forward | ATGGGTAAAGACTACTATAAAACTCTTGGG | verified ORF | |
| Reward | TCAATTAGGTAGAGTGTCATACAGTATGTCTT | verified ORF | |
| forward | CGGTGGTGCTGAAACATA | real-time qPCR | |
| Reward | GGTGGGTCTTGAGCGTGA | real-time qPCR | |
| Cvhsp70 | forward | ACWGTWCCXGCTTAYTTCAA | degenerate PCR |
| Reward | ACATCRAAGGTDCCRCCGCC | degenerate PCR | |
| outer primer | TGCCAGCATACTTCAACGATTC | 3′RACE | |
| inner primer | TGCGATTGCTGGGCTGAACG | 3′RACE | |
| outer primer | CGATACCCAGAGATAGGGGAGCAAC | 5′RACE | |
| inner primer | GGATGCGAGTAGAACCTCCCACGA | 5′RACE | |
| forward | ATGCCTGCCATTGGTATT | verified ORF | |
| Reward | TTAGTCAACTTCTTCAACCGT | verified ORF | |
| forward | GTGGGAGTGTGGCAACAAGGG | real-time qPCR | |
| Reward | GTGTCCGTGAAGGCAACATAGC | real-time qPCR | |
| Cvhsc70 | outer primer | GTCCCTTGTCGTTGGTGATGGTG | 5′RACE |
| inner primer | CGTAGGTGGTGAAGGTCTGGGTT | 5′RACE | |
| forward | AAATGACGAAAGCACCCGC | verified ORF | |
| Reward | ACCTGAATAGGCAGTGGAGTGAC | verified ORF | |
| forward | TTGATTTGGGAACTACATAC | real-time qPCR | |
| Reward | AGTCGCTCAGTGTCTGTAAA | real-time qPCR | |
| Cvhsp90 | forward | GCKGAGATCGCYCAGCTKATGTC | degenerate PCR |
| Reward | GCCTTCATGATRCGYTCCATGTTGGC | degenerate PCR | |
| outer primer | CGTGAGGAAGACAAAGCCAAAT | 3′RACE | |
| inner primer | CCCTGCTGTATCGTTACTTCTC | 3′RACE | |
| outer primer | TTGGCTTTGTCTTCCTCACG | 5′RACE | |
| inner primer | CGATACAGCAGGGCGAGT | 5′RACE | |
| forward | ATGCCGGAAGGAATGGATACCT | verified ORF | |
| Reward | TTAATCGACTTCTTCCATACGAGACG | verified ORF | |
| forward | CTCGCCCTGCTGTATCGT | real-time qPCR | |
| Reward | ATCGTCAAGTGAGAACCC | real-time qPCR | |
| Cv18SrRNA | forward | CGCCTTTCAAGATACCAAAATACGCC | real-time qPCR |
| Reward | TAGCTCTTTCTTGATTCGGTGGGTG | real-time qPCR |
Amplified fragments were purified using the QIAquick Gel Extraction Kit (Qiagen, Germany) and ligated directly into the pGEM-T Cloning Vector (Promega, Madison, WI). Each fragment-containing plasmid was isolated from cultured E. coli cells by an alkaline miniprep method. Insert fragments were verified by PCR using M13 forward and reverse primers. Sequencing was conducted on an automated fluorescence sequencing system ABI3730 (Applied BioSystems, Foster, CA).
Sequence analysis
Nucleotides and deduced amino acid sequences were analyzed using DNASTAR programs (Version 5.02) (DNASTAR, Inc., Madison, WI, USA). The functional domains and motifs of CvHsps were identified using the programs ScanProsite, Motifscan and SignalP4.0 online (http://www.ca.expasy.org). The obtained amino acid sequences of CvHsps were used to search for homologs in GenBank by BLAST (Position-Specific Iterated-BLAST) software available at the NCBI website (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). The sequence alignment was performed with Clustal X version 1.81 using default parameters [24] and edited by GeneDoc (Version 2.04) (Free Software Foundation, Inc., MA, USA). The Maximum parsimony (MP) method was used for phylogenetic analysis with MEGA 5.1 [25], and bootstrap analysis provided support values for the branches [26].
Real-time qPCRs
Real-time qPCR was performed to further compare expression levels of CvHsp40, CvHsp70, CvHsc70 and CvHsp90 in C. vestalis. Total RNA was extracted from whole insect bodies by using the TRIzol reagent (Invitrogen, Carlsbad, CA) and was further cleaned by using an RNeasy MiniElute Cleanup kit (Qiagen). The quality and concentration of the RNA was determined using a NanoDrop ND1000 spectrophotometer (NanoDrop Technologies, Roackland, DE, USA). Total RNA from each developmental stage and thermal treatment was checked for genomic DNA contamination by PCR amplification of each RNA sample using ORF verified primers for CvHsc70. The amplified products and the DNA ladder were analyzed on a 2% agarose gel containing Ethidium Bromide (EB).
Real-time qPCR reactions were run on an EcoTM Thermal Cycler (Illumina) in 10-μl reactions. Each 10 μl reaction contained 1 μl template cDNA, 5 μl Thunderbird Sybr qPCR Mix (TOYOBO, Osaka, Japan), 1 μl each of the corresponding forward and reverse primers (4 μM) and 2 μl ddH2O. Primer pairs used for real-time qPCR experiments were designed from ORF sequences of CvHsps (Table 1). To normalize differences in total RNA amounts that were reverse-transcribed and added to each reaction, 18S rRNA from C. vestalis (Cv18SrRNA) (GenBank accession No. JX399880) was used as an active endogenous control. Based on Tm value of primer pairs, cycling conditions were designed as: 1 min initial denaturation step at 95°C, followed by 40 cycles of 15 s denaturation at 95°C, 35 s annealing at 60°C, then one cycle of 15 s at 95°C, 15 s at 60°C, and 15 s at 95°C in order to produce the melting curves data. Data were acquired during the extension step and analyzed with the EcoTM Real-Time PCR Detection System. Each amplification reaction was carried out in three biological replicates, from which mean threshold cycle (CT) values plus standard deviations were calculated. The plasmid pGEM-T, which contained full ORF sequences of CvHsp genes or a 450 bp fragment of Cv18SrRNA, was diluted 10-fold in PBS buffer with 105 to 101 copies per reaction. Amplification efficiencies (E) of semi-quantitative real-time qPCRs were determined based on slope values obtained from linear regressions, where Ct values were plotted versus the logarithmic values of serially diluted input plasmid DNA templates by employing the equation E = 10(−1/Slope)-1 [27]. Here, amplification efficiencies (E) of CvHsp40, CvHsp70, CvHsc70, CvHsp90 and Cv18SrRNA were 104.2%, 103.2%, 94.7%, 97.1% and 98.7%, respectively.
Relative transcript amounts of CvHsps for each developmental stage and different temperature stresses were determined using the comparative Ct method [28]. First, we normalized the Ct values for differences in the quantity of cDNA in each reaction by subtracting the observed Ct values from our internal control, Cv18SrRNA, to generate ?Ct values. Then, we confirmed that the Ct values of the internal control did not differ between developmental stages (ANOVA, df = 6, F = 0.655, p = 0.687) or different thermal stress temperatures (one day old female adults, ANOVA, df = 4, F = 0.311, p = 0.864).
Statistical analysis
The relative transcript amounts of CvHsps were analyzed using one-way analysis of variance (ANOVA). The differences in relative transcript amounts of CvHsps were compared using Dunnett's multiple comparison and LSD comparison post hoc tests. All statistics were performed using the SPSS software (SPSS 16.0, SPSS Inc., Chicago, IL).
Results
Sequence analysis of the CvHsps
CvHsp40
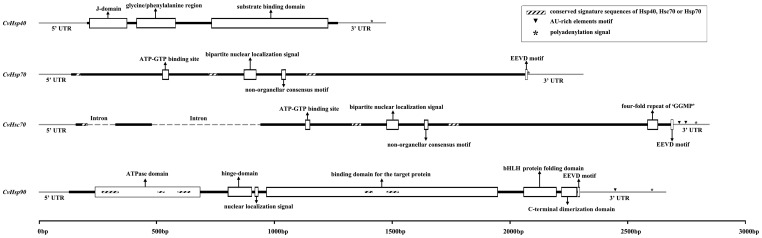
The full length CvHsp40 cDNA (GenBank accession no. JX088376) contains an ORF of 1068 bp encoding a 355 amino acid protein with a predicted molecular weight of 39.1 kDa and theoretical isoelectric point (pI) of 9.12 (Fig. 1 and Fig. S1). Three conserved regions are found in the deduced amino acid sequence of CvHsp40. The first one is a N-terminal J-domain, located at aa 3-57. The second is a glycine/phenylalanine region (G/F domain, aa 70–125). The last region is a C-terminal substrate binding domain (C domain, aa 176–341). Comparing the cDNA and genomic sequences revealed no intron in CvHsp40.
Figure 1. Schematic representation of the full cDNAs for CvHsp40, CvHsc70, CvHsp70 and CvHsp90 of Cotesia vestalis.
Cvhsp70
The full length CvHsp70 cDNA (GenBank accession no. JX088377) contains an ORF of 1938 bp encoding a 645 amino acid protein with a molecular weight of 70.1 kDa and theoretical pI of 5.35 (Fig. 1 and Fig. S2). By Motifscan analysis, we found three conserved characteristic signatures, including IDLGTTYS (aa 6–13), IFDLGGGTFDVSIL (aa 194–207) and VVLVGGSTRIPKIQS (aa 332–346), and four motifs, including an ATP-GTP binding site AEAYLGQK (aa 130-137), a bipartite nuclear localization signal sequence (NLS) ERKYRKNLKTNPRALRRL (aa 244-261), a non-organellar consensus motif RARFEEL (aa 297–303) and a cytoplasmic characteristic motif EEVD (aa 642–645). Comparing the cDNA and genomic sequences revealed no intron in CvHsp70.
CvHsc70
The full length CvHsc70 cDNA (GenBank accession no. JX088378) contains an ORF of 1956 bp encoding a 651 amino acid protein with a predicted molecular weight of 71.2 kDa and a theoretical pI of 5.26 (Fig. 1 and Fig. S3). Additionally, there are two AU-rich elements (ARE; AUUUA motif) located at 25–29 nt and 72–76 nt downstream of the termination codon in the 3′ UTR. By Motifscan analysis, we found three conserved characteristic signatures, including IDLGTTYS (aa 9–16), IFDLGGGTFDVSIL (aa 197–210) and VVLVGGSTRIPKIQS (a 334–348), and five motifs, including an ATP-GTP binding site AEAYLGQK (aa 131–138), a bipartite nuclear localization signal sequence (NLS) KRKYKKDLTSNKRAERRL (aa 246–263), a non-organellar consensus motif RARFEEL (aa 299–305), a four-fold repeat of the tetrapeptide “GGMP” (aa 615–630) and a cytoplasmic characteristic motif EEVD (aa 648–651). The sequence of the Cvhsc70 gene contains 2 introns of 119 and 460 bp length.
CvHsp90
The full length CvHsp90 cDNA (GenBank accession no. JX088379) contains an ORF of 2172 bp encoding a 723 amino acid protein with a predicted molecular weight of 83.3 kDa and a theoretical pI of 4.996 (Fig. 1 and Fig. S4). By Motifscan analysis, we found all five highly conserved signature sequences defining the Hsp90 family of known eukaryotes, NKEIFLRELISNSSDALDKIR (aa 35–55), LGTIAKSGT (aa 102–110), IGQFGVGFYSAYLVAD (aa 126–141), IKLYVRRVFI (aa 351–360) and GVVDSEDLPLNISRE (aa 377–391), as well as a consensus sequence MEEVD at the C-terminus. We also found: (a) a typical histidine kinase-like ATPase domain (aa 37–186) which is ubiquitous in all Hsp90 family members; (b) two highly charged domains, one a hinge-domain (aa 225–259) and the other a C-terminal domain (aa 691–716); (c) a nuclear localization signal (KKKKKK) (aa 263–268); (d) the binding domain for the target protein(s) (aa 279–607) and a basic Helix-Loop-Helix (bHLH) protein folding domain EADKNDKSVKDLVVLLFETALLSSGFSLDDPQVHAARIYRMIKLGLGI (aa 643–690). Comparing the cDNA and genomic sequences revealed no intron in CvHsp90.
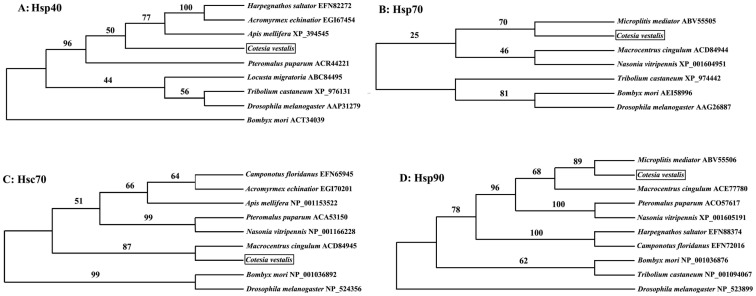
Homologs of CvHsps were found among hymenopteran species by PSI-BLASTP. By using the Parsimony method of tree reconstruction, we revealed that the most phylogenetic closed homolog group of CvHsp40 was (AmHsp40+ (HsHsp40+ AeHsp40)), and they shared identity of 77–79% (Fig. 2A). Meanwhile, the most phylogenetic closed homologs of CvHsp70, CvHsc70 and CvHsp90 were MmHsp70, McHsc70 and MmHsp90, respectively, and they shared identity of 94%, 95% and 86%, respectively (Fig. 2B–D).
Figure 2. Phylogenetic analysis of CvHsps and other correspondence homologs from Hymenoptera.
The Maximum Parsimony (MP) tree is generated from MEGA 5.01, and the numbers on the branch are the bootstrapping values. The positions of Hsps of Cotesia vestalis are boxed.
Transcriptional profiles of CvHsps during different developmental stages
To profile the transcriptional pattern of CvHsps during development at 24°C, mRNA levels of the four CvHsps were analyzed at different developmental stages, including first-instar, early second-instar, later second-instar, and third-instar larvae, pupae, female and male adults. First, the quantity of each CvHsp mRNA was normalized to the abundance of Cv18SrRNA. Then, this normalized value was divided by the amount of the corresponding CvHsp of first-instar larva, and the fold difference was used in the analyses of the relative transcriptional levels of the corresponding CvHsp during the development (Figure 3A).
Figure 3. Relative transcript abundances of CvHsps during developmental stages at 24°C.
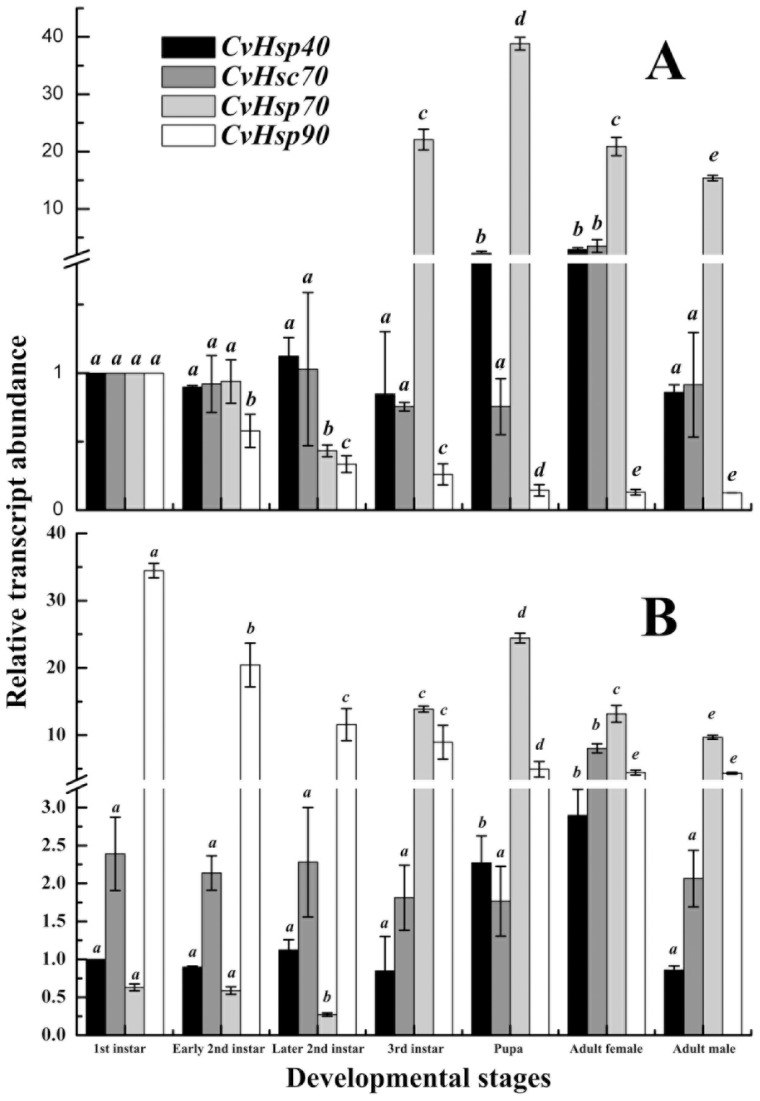
The quantity of each CvHsps mRNA was normalized to the abundance of Cv18SrRNA. Subsequently, the normalized value of each CvHsps was divided by the mount of the corresponding CvHsp of first-instar larva (A) or by the mount of CvHsp40 of first-instar larva (B). Columns topped by different letters indicate significantly different means within the relative transcript abundances of a given CvHsp gene at different developmental stages by ANOVA analysis (p<0.05).
The transcriptional level of CvHsp40 was almost the same throughout the larval stage, but increased significantly at pupal and adult stages (female). The transcriptional level of CvHsc70 was similar during the larval, pupal and male adult stages. The transcriptional level of CvHsp70 was generally low (Fig. 3B) and slightly decreased in early and middle larval stage, including first-instar, early second-instar and later second-instar larval stages, but dramatically increased at the following third-instar larval stage and reached its peak at pupal stage, and then decreased again in adult stages. The transcriptional level of CvHsp90 was highest at the first-instar larval stage, and then dropped approximately 7 folds to a relatively low level at later developmental stages. The transcripts of CvHsp40, CvHsc70 and CvHsp70 in female adult were all significantly more abundant than those in male adult, however the transcript abundance of CvHsp90 in female adult was quite close to that in male adult.
We also tried to compare the transcript abundance within four CvHsps at a given developmental stage. Therefore, the normalized value by the abundance of Cv18SrRNA was then divided by the amount of CvHsp40 of first-instar larva (Figure 3B). We found that CvHsp70 had the lowest transcript abundance in early and middle larval stages while CvHsp90 had its highest transcript abundance. However, in third-instar larval and following developmental stages, CvHsp70 had the highest transcript abundance.
Transcriptional profiles of CvHsps after thermal treatments
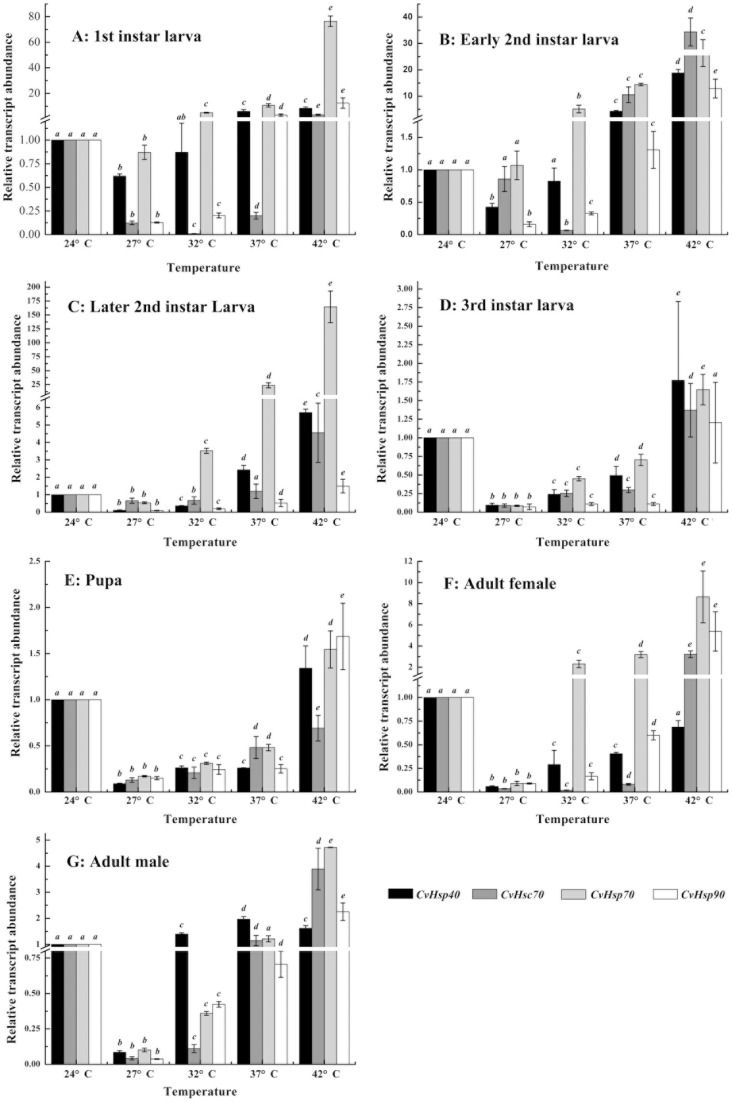
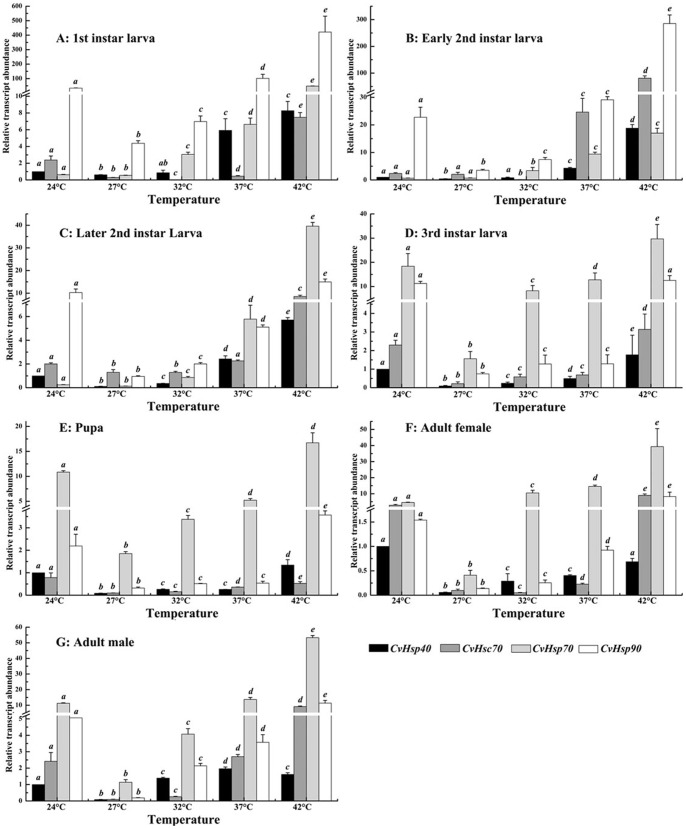
To profile the transcriptional pattern of CvHsps under different temperatures (24°C, 27°C, 32°C, 37°C and 42°C), mRNA levels of the four CvHsps were analyzed at different developmental stages, including all the larval stage, pupae, and female and male adults. First, the quantity of each CvHsps mRNA was normalized to the abundance of Cv18SrRNA. Then, the normalized value of each CvHsps was divided by the amount of the corresponding CvHsp at 24°C of each developmental stage, respectively, and the fold difference was then used in the analyses of the relative transcriptional levels of a given CvHsp at different temperatures (Fig. 4). To further compare the transcript abundance within four CvHsps of a given developmental stage at different heat temperatures, the normalized value of each CvHsps was again divided by the amount of CvHsp40 at 24°C of the corresponding developmental stage (Fig. 5).
Figure 4. Relative transcript abundances of CvHsps of different developmental stage under thermal stress.
The quantity of CvHsp mRNA is normalized to the abundance of Cv18SrRNA. Subsequently, the normalized value of each CvHsp is divided by the amount of the corresponding CvHsp at 24°C of each developmental stage, respectively. Columns topped by different letters indicate significantly different means within the relative transcript abundances of a given CvHsp gene under different temperatures by ANOVA analysis (p<0.05). A-G represents first-instar larva, early second-instar larva, later second-instar larva, third-instar larva, pupa, female adult and male adult, respectively.
Figure 5. Relative transcript abundances of CvHsps of each developmental stage under thermal stress.
The quantity of CvHsp mRNA is normalized to the abundance of Cv18SrRNA. Subsequently, the normalized value of each CvHsp is divided by the mount of Cvhsp40 of the corresponding developmental stage at 24°C. Columns topped by different letters indicate significantly different means within the relative transcript abundances of a given CvHsp gene under different temperatures by ANOVA analysis (p<0.05). A-G represents first-instar larva, early second-instar larva, later second-instar larva, third-instar larva, pupa, female adult and male adult, respectively.
The transcriptional pattern of four CvHsps indicated that 27°C was a “turn-over” temperature, where the transcriptional levels of CvHsp40, CvHsc70, CvHsp70 and CvHsp90 at different developmental stages were significantly lower than those at 24°C, 32°C, 37°C and 42°C, except for the lowest transcript abundance of CvHsc70 of first-instar larva, early-instar larva and female adult were appear at 32°C. When the temperature was higher than 27°C, and 32°C for CvHsc70 of above developmental stages, the transcriptional levels of CvHsp40, CvHsc70, CvHsp70 and CvHsp90 of each developmental stage were increased obviously in response to thermal stress (Fig. 4), suggesting that the transcripts of CvHsp40, CvHsc70, CvHsp70 and CvHsp90 were significantly induced by heat stress. Here, we also noticed that the transcriptional peak of CvHsp40 in male in response to different heat stress showed at 37°C not 42°C (Fig. 4G). CvHsp90 had the highest transcriptional level at first- and early second-instar larvae (Figure 5A–B) while CvHsp70 had its highest transcriptional level at third-instar larval, pupal and adult stages (Figure 5D–G) among four tested CvHsps under all test temperatures. However, there was no such clear transcriptional pattern of CvHsp70 or CvHsp90 in later second-instar larva (Figure 5C). Additionally, the sensitivities of CvHsp40, CvHsc70, CvHsp70 and CvHsp90 to the heat treatment during different developmental stages were different from each other. When the temperatures increased from 27°C to 42°C, the transcriptional levels of four CvHsps were all up-regulated at least 5 folds during larval and pupal stages (Fig. 4A–E), but displaying irregular patterns of heat sensitivity. When comparing the up-regulated ratios of the transcript abundances of CvHsp40, CvHsc70, CvHsp70 or CvHsp90 between female and male adults (Fig. 4 F–G), CvHsp70 and CvHsc70 were greater in females (91.9 folds and 93.9 folds) than in males (47 folds and 69.4 folds) while CvHsp40 was smaller in females (11.7 folds) than in males (18.9 folds), but CvHsp90 exhibited no differences between males and females.
Discussion
Heat shock proteins are key elements of the stress response system at the cellular level in all organisms. They are up-regulated in cells exposed to a wide variety of abiotic stressors, such as heat shock, osmotic stress, and environmental contaminants (heavy metals, pesticides and polycyclic aromatic hydrocarbons), and biotic (bacteria and virus) factors [9]. In the present study, using RACE or direct PCR with primers designed on the basis of conserved Hsp genes sequences, we identified four genes encoding Hsps, including CvHsp90, CvHsp70, CvHsc70 and CvHsp40, in C. vestalis. The predicted amino acid sequences of these proteins showed high similarity to Hsp sequences known from other Hymenoptera, with identity in the range of 76–86% for CvHsp90, 89–94% for CvHsp70, 92–95% for CvHsc70 and 77–79% for CvHsp40. These similarities add confidence to our identifications of genes encoding HSPs in a parasitoid wasp.
Amino acid sequence comparisons revealed that all core signatures or motifs were characterized in these Hsps. We identified five signatures for CvHsp90, three for CvHsp70 and CvHsc70, and two for CvHsp40, plus other motifs. None of the four conserved repeats with the consensus sequence CxxCxGxG (cysteine-rich region or zinc finger motif) was found in the amino acid sequence of CvHsp40, which indicated that it was the Type II Hsp40s [29]. Compared with Type I Hsp40, Type II Hsp40s also can form chaperone pairs with cytosolic Hsp70 and help folding proteins but with much lower efficiency [30]. The well conserved C-terminal motif MEEVD or EEVD argue that these motifs enable CvHsp90, CvHsp70 or CvHsc70 to bind other co-chaperones [31], which also indicated that CvHsp90, CvHsp70 and CvHsc70 are cytosolic Hsps [32]. The non-organellar stress protein motif “RARFEEL” and bipartite nuclear localization signal “(K/R)2(X)nRRLRT” motif suggest that CvHsp70 and CvHsc70 not only belong to the eukaryotic cytosolic-cytoplasmic Hsp70 family but also can selectively translocate into the nucleus of cells [33]. Comparing CvHsp70 and CvHsc70, no “GGXP” motif occurs near the 3′- terminal of CvHsp70, whereas CvHsc70 contains four “GGXP” repeats, which suggests CvHsc70 has a stronger binding affinity in co-chaperone binding activities [34]. There was no glutamine-rich sequence (QTQDQ) be found located at the N-terminus of Cvhsp90, which indicated it was the β-isoform of Hsp90s [35]. Two highly charged domains of CvHsp90 indicate that it more likely to bind to positively charged or hydrophobic protein and the bHLH protein folding domain suggests that CvHsp90 can rapidly convert a basic Helix-Loop-Helix protein from an inactive to an active conformation [36]–[37]. The AU-rich elements (ARE) is found located at 3′-UTR region of CvHsc70 and CvHsp90 suggested that the possible posttranscriptional regulation of them is the mRNA degradation, which is influenced by many exogenous factors, including phorbol esters, calcium ionophores, cytokines, and transcription inhibitors [38].
The role of heat shock proteins in development is less well understood, and earlier studies were only proceeding in model insects and few other insects. For examples, sHsps were continually expressed during development of D. melanogaster [39], expression level of Hsp70 varied among life stages of T. castaneum [40], and three Hsps increased their mRNA expression during the developmental course of P. xylostella [41]. In the current study, transcript abundances of four CvHsps were checked through each developmental stage of C. vestalis. We found that the transcript abundance of CvHsp40 remained a low level during the larval stage, but increased significantly at the pupal and adult stages; the transcript abundance of CvHsc70 remained around the same level during the larval, pupal and male adult stages, but females showed a much higher transcript abundance; the transcript abundance of CvHsp70 is low in early and middle larval stages, and then followed by a sharp increase at later larval stage, third-instar larva; the transcript abundance of CvHsp90 dropped at each consecutive developmental stage. The different transcriptional patterns of CvHsps suggested that they are under differential mechanisms. The life history of C. vestalis showed that the third-instar larva is a special stage [23]. At that time, C. vestalis larva exits the host larva and spins a cocoon outside the host, thus facing very different environment stresses. The transcriptional pattern of CvHsp70, which exhibited a dramatic increase at the third-instar, reveals that CvHsp70 might be a useful biomarker to assess life history traits in future research. The gender-specific transcript increase of CvHsp40, CvHsc70 and CvHsp70 might indicate that they were required in the female reproduction of C. vestalis or female adult of C. vestalis was better at heat tolerance than male. However, it should be noted that the RNA used for the present study was extracted from the whole organism and the data obtained may reveal an average expression of CvHsp40, CvHsc70 or CvHsp70, therefore the examination of expression of CvHsp40, CvHsc70 or CvHsp70 in different tissues and organs is apparently needed to better understand its functions.
Tolerances to extreme environmental factors, particularly temperature, can provide insight into insect biology. In insects and possibly most organisms, Hsps show altered expression profiles during temperature stress, particularly the maximal induction of Hsp transcripts. In this study, our finding that four CvHsp transcripts can be significantly induced by heat stress is similar to previous results [31], [35], [40]–[42]. However, the transcript abundance of CvHsps around 27°C is mostly significantly lower than those of other stress temperature, including 24°C, at every developmental stage, which might indicate that the temperature of 27°C is a suitable condition for development of C. vestalis. The tested population of C. vestalis was originally collected from the Hangzhou area, where this species is an abundant one in the later spring, early summer and autumn in the cruciferous vegetable area, and the average temperature in spring and autumn in this area is approximately 27°C. This might suggest that there is a possible biological relationship between the temperature at which the abundance of the CvHsp transcripts begins to increase and the average temperature of the distribution area of C. vestalis. In conclusion, (1) Four CvHsp genes were characterized from the endoparasitoid wasp, C. vestalis. (2) The divergent transcriptional patterns of CvHsp40, CvHsp70 and CvHsp90 in different developmental stages suggest that CvHsps transcripts are under differential regulation during development. The dramatic increase of transcripts of CvHsp70 at the third-instar larva coincided with its developmental change in this stage. (3) CvHsp40, CvHsc70 and CvHsp70 showed sex-specific differences of transcript abundance in the adult stage; (4) the transcripts of CvHsps at all developmental stages were significantly induced by heat stress; the lowest transcript abundances appeared at 27°C, which probably suggest that this is the most favorable temperature for the development of C. vestalis.
Supporting Information
Full length cDNA and deduced amino acid sequence of CvHsp40 . Asterisk indicates the translational termination codon. The putative polyadenylation signal is grey covered and dash underlined. J-domain is dash underlined. G/F domain is grey covered. C-terminal substrate binding domain is solid underlined.
(TIF)
Full length cDNA and deduced amino acid sequence of CvHsp70 . Asterisk indicates the translational termination codon. The putative polyadenylation signal is grey covered and dash underlined. ATP-GTP binding site is dash underlined. Bipartite nuclear localization signal is solid underlined. Non-organellar consensus motif is grey covered. EEVD motif is double solid underlined.
(TIF)
Full length cDNA and deduced amino acid sequence of CvHsc70 . Asterisk indicates the translational termination codon. The putative polyadenylation signal is grey covered and dash underlined. Two AU-rich elements (ARE) motifs are grey covered and solid underlined. ATP-GTP binding site is dash underlined. Bipartite nuclear localization signal is solid underlined. Non-organellar consensus motif is grey covered. EEVD motif is double solid underlined. Four GGMP motifs are open boxed.
(TIF)
Full length cDNA and deduced amino acid sequence of CvHsp90 . Asterisk indicates the translational termination codon. The putative polyadenylation signal is grey covered and dash underlined. One AU-rich elements (ARE) motifs are grey covered and solid underlined. ATP-GTP binding domain is dash underlined. Charged hinge domain is grey covered. Nuclear localization signal is solid underlined. Target proteins binding domain is light grey covered. Basic Helix-Loop-Helix (bHLH) protein folding domain is open boxed. ATP-GTP binding domain is double dash underlined. EEVD motif is double solid underlined.
(TIF)
Acknowledgments
We thank Dr. Kevin Clark (University of Georgia, USA) for his helping in manuscript writing.
Funding Statement
Funding for this study was provided jointly by the 973 Program (2013CB127603) and Zhejiang Key Program of Agriculture (2009C12048) to XXC, the National Science Foundation of China (30971907) and the Zhejiang Science Fund for Distinguished Young Scholars (R3110049) to MS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Shaw MR (2003) Revised synonymy in the genus Cotesia (Hymenoptera: Braconidae: Microgastrinae): the identity of Microgaster vestalis Haliday, 1834, as a senior synonym of Apanteles plutellae Kurdjumov, 1912. Entomol Gazette 54: 187–189. [Google Scholar]
- 2. Bai SF, Chen XX, Cheng JA, Fu WJ, He JH (2003) Characterization of Cotesia plutellae polydnavirus and its physiological effects on the diamondback moth, Plutella xylostella larvae. Acta Entomol Sinica 46: 401–408. [Google Scholar]
- 3. Bai SF, Chen XX, Cheng JA, Fu WJ, He JH (2005) Effects of wasp-associated factors of Cotesia plutellae on growth and development of Plutella xylostella larvae. J Plant Protect 32: 235–240. [Google Scholar]
- 4. Sarfraz M, Keddie AB, Dosdall LM (2005) Biological control of the diamondback moth, Plutella xylostella: A review. Biocontrol Sci Technol 15: 763–789. [Google Scholar]
- 5. Liu S, Wang X, Guo S, He J, Shi Z (2000) Seasonal abundance of the parasitoid complex associated with the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae) in Hangzhou, China. Bull Entomol Res 90: 221–231. [DOI] [PubMed] [Google Scholar]
- 6. Shi ZH, Liu SS (1999) Influence of temperature on the development, survival and reproduction of Cotesia plutellae, a larval parasite of Plutella xylostella . Acta Phytophylacica Sinica 26: 142–146. [Google Scholar]
- 7. Feder ME, Hofmann GE (1999) Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 61: 243–282. [DOI] [PubMed] [Google Scholar]
- 8. Kim KK, Kim R, Kim S (1998) Crystal structure of small heat-shock protein. Nature 394: 595–599. [DOI] [PubMed] [Google Scholar]
- 9. Sørensen JG, Kristensen GTN, Loeschcke V (2003) The evolutionary and ecological role of heat shock proteins. Ecol Lett 6: 1025–1037. [Google Scholar]
- 10. Qiu XB, Shao YM, Miao S, Wang L (2006) The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci 63: 2560–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Picard D (2002) Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci 59: 1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Asgari S, Zhang GM, Schmidt O (2003) Polydnavirus particle proteins with similarities to molecular chaperones, heat-shock protein 70 and calreticulin. J Gen Virol 84: 1165–1171. [DOI] [PubMed] [Google Scholar]
- 13. Haass C, Klein U, Kloetzel PM (1990) Developmental expression of Drosophila melanogaster small heat-shock proteins. J Cell Sci 96: 413–418. [DOI] [PubMed] [Google Scholar]
- 14. Joanisse DR, Michaud S, Inaguma Y, Tanguay RM (1998) Small heat shock proteins of Drosophila: developmental expression and functions. J Biosci 23: 369–376. [DOI] [PubMed] [Google Scholar]
- 15. Johnston JA, Ward CL, Kopito RR (1998) Aggresomes: a cellular response to misfolded proteins. J Cell Biol 143: 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pockley AG (2003) Heat shock proteins as regulators of the immune response. Lancet 362: 469–476. [DOI] [PubMed] [Google Scholar]
- 17. Rinehart JP, Denlinger DL, Rivers DB (2002) Upregulation of transcripts encoding select heat shock proteins in the flesh fly Sarcophaga crassipalpis in response to venom from the ectoparasitoid wasp Nasonia vitripennis . J Invertebr Pathol 79: 62–63. [DOI] [PubMed] [Google Scholar]
- 18. Rinehart JP, Li A, Yocum GD, Robich RM, Hayward SA, Denlinger DL (2007) Up-regulation of heat shock proteins is essential for cold survival during insect diapauses. Proc Natl Acad Sci USA 104(27): 11130–11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shim JK, Ha DM, Nho SK, Song KS, Lee KY (2008) Upregulation of heat shock protein genes by envenomation of ectoparasitoid Bracon hebetor in larval host of Indian meal moth Plodia interpunctella . J Invertebr Pathol 97: 306–309. [DOI] [PubMed] [Google Scholar]
- 20. Zhu JY, Fang Q, Wang L, Hu C, Ye GY (2010) Proteomic analysis of the venom from the endoparasitoid wasp Pteromalus Puparum (Hymenoptera: Pteromalidae). Arch Insect Biochem Physiol 75: 28–44. [DOI] [PubMed] [Google Scholar]
- 21. Rutherford SL (2003) Between genotype and phenotype: Protein chaperones and evolvability. Nat Rev Genet 4: 263–274. [DOI] [PubMed] [Google Scholar]
- 22. Sørensen JG, Loeschcke V (2001) Larval crowding in Drosophila melanogaster induces Hsp70 expression, and leads to increased adult longevity and adult thermal stress resistance. J Insect Physiol 47: 1301–1307. [DOI] [PubMed] [Google Scholar]
- 23. Yu RX, Shi M, Huang F, Chen XX (2008) Immature Development of Cotesia vestalis (Hymenoptera: Braconidae), an Endoparasitoid of Plutella xylostella (Lepidoptera: Plutellidae). Ann Entomol Soc Am 101(1): 189–196. [Google Scholar]
- 24. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24): 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9(4): 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- 27. Peirson SN, Butler JN, Foster RG (2003) Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res 31: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitayive PCR and the 2T-Delta Delta C method. Methids 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 29. Caplan AJ, Cyr DM, Douglas MG (1993) Eukaryotic homologs of Escherichia coli DnaJ: a diverse protein family that functions with Hsp70 stress proteins. Mol Biol Celt 4: 555–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fan CY, Lee S, Ren HY, Cyr DM (2004) Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell 15: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang QR, Denlinger DL (2010) Molecular characterization of heat shock protein 90, 70 and 70cognate cDNAs and their expression patterns during thermal stress and pupal diapause in the corn earworm. J Insect Physiol 56: 138–150. [DOI] [PubMed] [Google Scholar]
- 32. Gupta RS (1995) Phylogenetic analysis of the 90 kD heat-shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol Biol Evol 12: 1063–1073. [DOI] [PubMed] [Google Scholar]
- 33. Vayssier M, Le Guerhier F, Fabien JF, Philippe H, Vallet C, Ortega-Pierres G, Soule C, Perret C, Liu M, Vega-Lopez M, Boireau P (1999) Cloning and analysis of a Trichinella britovi gene encoding a cytoplasmic heat shock protein of 72 kDa. Parasitology 119: 81–93. [DOI] [PubMed] [Google Scholar]
- 34. Demand J, Lüders J, Höhfeld J (1998) The carboxy-terminal domain of HSC70 provides binding sites for a distinct set of chaperone cofactors. Mol Cell Biol 18: 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gao Q, Zhao J, Song L, Qiu L, Yu Y, Zhang H, Ni D (2008) Molecular cloning, characterization and expression of heat shock protein 90 gene in the haemocytes of bay scallop Argopecten irradians . Fish Shellfish Immunol 24: 379–385. [DOI] [PubMed] [Google Scholar]
- 36. Csermely P, Schnaider T, Soti C, Prohászka Z, Nardai G (1998) The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharmacol Ther 79(2): 129–68. [DOI] [PubMed] [Google Scholar]
- 37. Shaknovich R, Shue G, Kohtz DS (1992) Conformational activation of a basic helix-loop-helix protein (MyoD1) by the C-terminal region of murine HSP90 (HSP84). Mol Cell Biol 12(11): 5059–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen CYA, Shyu AB (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20 (11): 465–470. [DOI] [PubMed] [Google Scholar]
- 39. Kurzik-Dumke U, Lohmann E (1995) Sequence of the new Drosophila melanogaster small heat-shock-related gene, lethal (2) essential for life [l(2)efl], at locus 59F4,5. Gene 154(2): 171–175. [DOI] [PubMed] [Google Scholar]
- 40. Mahroof R, Zhu KY, Subramanyam B (2005) Changes in Expression of Heat Shock Proteins in Tribolium castaneum (Coleoptera: Tenebrionidae) in Relation to Developmental Stage, Exposure Time, and Temperature. Ann Entomol Soc Am 98(1): 100–107. [Google Scholar]
- 41. Sonoda S, Ashfaq M, Tsumuki H (2006) Cloning and nucleotide sequencing of three heat shock protein genes (hsp90, hsc70, and hsp19.5) from the diamondback moth, Plutella xylostella (L.) and their expression in relation to developmental stage and temperature. Arch Insect Biochem Physiol 62: 80–90. [DOI] [PubMed] [Google Scholar]
- 42. Wang H, Li K, Zhu JY, Fang Q, Ye GY (2012) Cloning and expression pattern of heat shock protein genes from the endoparasitoid wasp, Pteromalus puparum in response to environmental stresses. Arch Insect Biochem Physiol 79(4–5): 247–263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full length cDNA and deduced amino acid sequence of CvHsp40 . Asterisk indicates the translational termination codon. The putative polyadenylation signal is grey covered and dash underlined. J-domain is dash underlined. G/F domain is grey covered. C-terminal substrate binding domain is solid underlined.
(TIF)
Full length cDNA and deduced amino acid sequence of CvHsp70 . Asterisk indicates the translational termination codon. The putative polyadenylation signal is grey covered and dash underlined. ATP-GTP binding site is dash underlined. Bipartite nuclear localization signal is solid underlined. Non-organellar consensus motif is grey covered. EEVD motif is double solid underlined.
(TIF)
Full length cDNA and deduced amino acid sequence of CvHsc70 . Asterisk indicates the translational termination codon. The putative polyadenylation signal is grey covered and dash underlined. Two AU-rich elements (ARE) motifs are grey covered and solid underlined. ATP-GTP binding site is dash underlined. Bipartite nuclear localization signal is solid underlined. Non-organellar consensus motif is grey covered. EEVD motif is double solid underlined. Four GGMP motifs are open boxed.
(TIF)
Full length cDNA and deduced amino acid sequence of CvHsp90 . Asterisk indicates the translational termination codon. The putative polyadenylation signal is grey covered and dash underlined. One AU-rich elements (ARE) motifs are grey covered and solid underlined. ATP-GTP binding domain is dash underlined. Charged hinge domain is grey covered. Nuclear localization signal is solid underlined. Target proteins binding domain is light grey covered. Basic Helix-Loop-Helix (bHLH) protein folding domain is open boxed. ATP-GTP binding domain is double dash underlined. EEVD motif is double solid underlined.
(TIF)