Abstract
There is no licensed vaccine or cure for human cytomegalovirus (CMV), a ubiquitous β-herpesvirus infecting 60–95% of adults worldwide. Infection can cause congenital abnormalities, result in severe disease in immunocompromised patients, and is a major impediment during successful organ transplantation. In addition, it has been associated with numerous inflammatory diseases and cancers, as well as being implicated in the development of essential hypertension, a major risk factor for heart disease. To date, limited data regarding the identification of immunogenic viral targets has frustrated CMV vaccine development. Based upon promising clinical data suggesting an important role for T cells in protecting against disease in the transplantation setting, we designed a novel panel of highly-optimized synthetic vaccines encoding major CMV proteins and evaluated their immune potential in murine studies. Vaccination induced robust CD8+ and CD4+ T cells of great epitopic breadth as extensively analyzed using a novel modified T cell assay described herein. Together with improved levels of CMV-specific T cells as driven by a vaccine, further immune evaluation of each target is warranted. The present model provides an important tool for guiding future immunization strategies against CMV.
Keywords: CMV, T cell, vaccine, gene therapy, electroporation, animal model
Human cytomegalovirus (CMV), also known as human herpesvirus 5, is a ubiquitous betaherpesvirus that has been the subject of roughly 50 y of clinical research.1 Infection is lifelong and there is no licensed vaccine or cure.2-5 It is highly seroprevalent in the human population (> 95% in developing countries) and has been estimated to infect 50–80% of adults over the age of 40 in the US alone (www.cdc.gov/CMV/index.html). Overt infection occurring during primary infection or reactivation can lead to a number of clinical diseases.2,6 As a member of the ‘TORCH’ infections, it can lead to numerous congenital abnormalities including death,7 and has been associated with cerebral palsy8 and medulloblastoma.9 It can also result in severe disease and death in immunocompromised and in HIV-infected individuals,10,11 as well as being a major impediment during successful organ transplantation.2 Haematopoietic and solid organ transplant recipients are most at risk for disease ranging from acute and chronic graft rejection, accelerated coronary artery disease after heart transplantation, and new onset diabetes mellitus.12 Recently, a therapeutic CMV DNA vaccine encoding the gB and pp65 proteins, and formulated with poloxamer CRL1005 and benzalkonium chloride was shown to be immunogenic in a Phase I trial,13 then later demonstrated a significant reduction in occurrence and duration of episodes of CMV viremia for patients undergoing hemopoietic stem-cell transplantation in a recent Phase 2, placebo-controlled trial.14 Despite promising clinical data, the lack of an effective vaccine requires in some cases that antiviral treatments during transplantation be used which have demonstrated low clinical efficacy, can be highly toxic, can lead to invasive bacterial and fungal infection, are not suitable for long-term use, and contribute to the development of CMV drug resistance.15-17 Development of an effective CMV vaccine would he highly desirable and may allow the reduction and restriction of antiviral treatments.
While it was widely held that latent or asymptomatic infection was virtually benign in healthy individuals, low-grade “microinfection” with CMV can only be detected by highly sensitive assays3,18 and has been implicated as a causative factor rather than an epiphenomenon in certain cancers, inflammatory, and hypertensive and pulmonary diseases.3,9,18-20 As such, CMV has been directly implicated in essential hypertension,3 which is a prevalent risk factor for a variety of cardiovascular diseases including stroke, coronary heart disease and renal and heart failure, affecting > 1 billion adults worldwide and costing the US an estimated $444 billion in 2010 (www.cdc.gov/chronicdisease/resources/publications/aag/dhdsp.htm).21 This new evidence supports previous studies that have demonstrated pulmonary shedding of virus in asymptomatic patients22 and has implicated CMV infection in atherosclerosis, coronary heart disease and cardiac transplant arteriopathy.20,23-25 Furthermore, CMV genome and proteins are present in certain malignant tumors including colon cancer,19 malignant glioblastoma,26 medulloblastoma,9 EBV-negative Hodgkins lymphoma,27 prostatic carcinoma,28 and colon and breast cancer,29 in which virus-mediated “oncomodulation” may or may not have played a role in the cancer itself, but certainly in altering the progression of disease. CMV microinfection has also been associated with inflammatory bowel disease (90% of patients had active infection in bowel)30 and can be detected in inflamed tissues in patients with rheumatoid arthritis, Sjögren’s syndrome, dermato- and polymyositis, psoriasis, Wegener’s granulomatosis, ulcerative colitis, and Crohn’s disease.18 The US Institute of Medicine and the US National Vaccine Program Office have ranked it with the highest priority in terms of health care dollars saved and improvement in quality adjusted life years.31 An effective CMV vaccine could prove highly valuable, reducing congenital diseases and long-term sequelae, improving longevity of transplant patients and the transplants, as well as saving billions in annual healthcare costs and disability adjusted loss years in the developing world.21
The lack of a licensed vaccine for CMV has been complicated by the task of vaccine target selection due to the relative complexity of the CMV genome and its numerous glycoproteins associated with cell tropism and entry.32 However, promising clinical14,33,34 and preclinical35-37 data suggest that an effective vaccine will induce both cellular and humoral immune responses. Recently, a purified recombinant CMV gB protein plus MF59 adjuvant (a sequalene-in-water emulsion) vaccine was immunogenic in Phase I trials,38-40 and in a Phase II trial, demonstrated a 50% protection against virus transmission to women of child-bearing age.34 While the protection rate achieved by this vaccine was much lower than desired for women at risk for CMV during pregnancy, it is the first to demonstrate protective efficacy of the gB immunogen in the clinic. Futher, CTLs and NAbs have been shown to aid in control and reduction of disease2,14,41-44 and are critically dependent on adequate CD4 T cell help.37,42,45 NAbs are also likely crucial for providing sterilizing immunity against viral transmission.33,34,46,47
To help in the identification and development of target CMV immunogens, we assembled a novel panel of CMV Ags including mainly glycoproteins, as well as matrix and charerones.32,48 Each Ag gene was genetically-optimized and consensus-engineered,49-51 and then administered as enhanced DNA (E-DNA) vaccines for immune evaluation in preclinical ‘proof-of-concept’ rodent studies. Using a novel T cell assay described herein, it was determined that constructs elicited robust and highly-diverse T cell responses. Responses were constituted by both CD8+ and CD4+ T cells, both of which are considered critical in providing protection against post-transplant occurrence of CMV disease.37,42,45 In addition, a majority of murine T cell epitopes identified herein also contained HLA that have previously been reported to contribute to the suppression of viremia and amelioration of overt disease.2,14,41-44 These data suggest studies investigating the potential for generating vaccine-induced humoral immunity as well as protection in preclinical challenge models are warranted.
Results
Construction of CMV immunogen panel
We developed a novel panel of CMV immunogens comprised of mainly surface-associated proteins, as well as matrix and chaperones. Due to the complexity of the glycoproteins that may be involved in viral entry,32,52 we selected a panel of traditional and novel neutralization-sensitive candidates for screening as vaccine targets for cellular immunity.48 Structural glycoproteins typically grouping in high molecular weight complexes (gC) included gB (ORF UL55), gH (ORF UL75), gL (ORF UL115), gM (ORF UL100), gN (ORF UL73), and gO (ORF UL74).46,47 The gB (gC-I) is an integral membrane protein that homodimerizes to form the Type 1 membrane protein, is essential for both in vivo and in vitro replication, is implicated in virus attachment, is required for entry and cell fusion, can be detected on the surface of both infected cells and virions,53 represents a major target for NAbs,46,54-56 and has been a predominant core component of CMV vaccine platforms.35 Glycoproteins gM and gN (gC-II) heterodimerize to form the CMV “viral infectivity complex” required for virion assembly and egress and have been shown to elicit binding Abs during infection.57 As a novel component for an CMV vaccine, glycoproteins gH, gL, and gO (gC-III) can heterotrimerize to form the gCIII fusion complex. During infection, gH is a potent immunogen as the target of NAbs that likely function by blocking a CMV post-attachment event such as membrane fusion or virus penetration.58 However, it is known that co-expression of gL and gH is important for macromolecule expression since as gL serves as a chaperone for gH.48 In addition, the chaperone proteins encoded by ORFs UL128-UL131A (UL128, UL130 and UL131A) were chosen since they facilitate gO in the formation of the gCIII complex, can incorporate into gH/gL or gH/gL/gO which alters viral tropism from fibroblasts to epithelial and endothelial cells,46,56,59 and may serve as T cell targets. Expression of gO is not required for the production of infectious virus in vitro, but deficient mutant viruses are growth impaired.60 While co-expression of gH and gL along with gB constitute the core fusion machinery, both are necessary and sufficient for fusion.32 Lastly, the lower matrix protein pp65 (ORF UL83) was included in this study since it has been reported to elicit strong T cell epitopes and is a major component of current vaccine strategies.35
CMV immunogen consensus-engineering
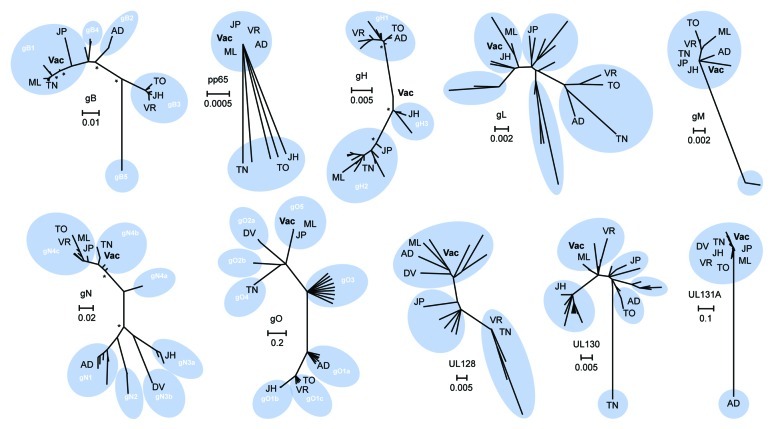
To maximize the potential for broadly-reactive immunity, we generated amino acid consensus sequences from CMV clinical strains, and excluded those from potentially-divergent, highly-passaged lab-adapted strains,49-51,61,62 a strategy shown previously to enhance protection against divergent strains of influenza and HIV.63,64 Phylogenetic diversity for each CMV immunogen was determined using published sequences and results are displayed in Figure 1. Analysis of the gB confirmed the presence of four main variants (gB1-gB4)65 and one nonprototypic variant (gB5).66-69 Since the gB protein is relatively conserved among clinical and low-passage strains (~86% identical), we chose the consensus of these sequences. The vaccine sequence (Vac) was phylogenetically closest to the gB1 genotype which has been found to account for the majority of highly symptomatic individuals in the clinic.70-72
Figure 1. Phylogenetic analysis and consensus engineering of CMV immunogens. Phylogenetic trees of selected CMV immunogens are shown. The significance of the unrooted phylogenetic trees was verified by bootstrap analysis and significant support values (≥ 80%; 1,000 bootstrap replicates) are indicated by asterisks at major nodes. Previously reported genotypes are illustrated (white) and reference strains are indicated; AD - AD169, DV - Davis, JH - JHC, JP - JP, ML - Merlin, TO - Toledo, TN - Towne, VR - VR1814. CMV vaccine immunogens as derived by alignment of clinical strain sequence data are displayed (Vac) and scale bars signify distance of amino acids per site.
Components of the CMV gCIII fusion complex, gH, gL and gO were developed herein as novel candidate immunogens. Phylogenetic analysis of gH confirmed the presence of two main genotypes in addition to a possible third group including the newly reported JHC strain that was isolated from a bone marrow transplant patient.73 Analysis confirmed a low level of variation among the gHs (~93% identical) which may explain why anti-gH MAbs appear broadly reactive.58,74 Due to this high level of conservation, the DNA vaccine consensus immunogen fell between gH1 and gH2, and was closest to the putative third gH group (gH3) including with the JHC clinical isolate. Phylogenetic analysis of the gL protein, while similarly highly conserved (~91%), was less distinctly grouped. Upon removal of amino acid sequences of gLs from strains extensively passaged, the resultant DNA consensus immunogen fell closest to the JHC and Merlin clinical isolates. The gO, which is highly glycosylated, is highly variable at the 5′ end50 contributing to high polymorphism (~55% divergence). Thus, we chose the consensus sequence of the gO5 genotype group for our target immunogen since it has been described to be genetically linked with the gN4c genotype, the largest gN4 variant group and the most seroprevalent.75 Identity within the gO5 subgroup was ~99% and thus, the consensus Ag was phylogenetically grouped within this subgroup that also included the Merlin and JP clinical isolates.
Recently, it was shown that UL128, UL130 and UL131A can form a pentameric complex with gH and gL, instead of the classically defined association of gH/gL/gO constituting the gCIII fusion complex.76-79 Furthermore, that this complex has been described to elicit potent nAbs.58,80 Due to the relatively high level of amino acid conservation upon removal of highly-passaged and lab-adapted strains (~87% for UL128, ~86% for UL130, and ~73% for UL131A), we used consensus sequences for each candidate vaccine immunogens. The UL128 vaccine sequence was phylogenetically grouped with the Merlin and Davis isolates, as well as the AD169 strain. However, both of the UL130 and UL131A sequences were phylogenetically distant from the Towne and AD169 lab strains, respectively, which have lost their ability to infect endothelial cells, epithelial cells, and leukocytes due to deletions or mutations of these genes.79,81
CMV gM and gN heterodimerize in the ER by both covalent disulfide bonding and noncovalent interaction to form the viral infectivity complex. While the gM is highly conserved among the CMV (~95%), the gN is extremely variable (~45%). Due to this relatively high identity among the gM, consensus of all clinical sequences determined our candidate vaccine immunogen. Conversely, due to the highly modified nature of gN, characterized by almost exclusive O-linked sugars, consensus of the gN4 subtype was used as the vaccine immunogen since this subgroup was reported to be the most prevalent of all clinical isolates in North America, Europe, China, and Australia.75
CMV pp65 was studied due to its current use in recent vaccine strategies as a T cell target based upon its elicitation of a dominant cellular immune response.82,83 This protein is highly conserved among the CMV and was ~97% identical when not accounting for the 3′ truncation associated with many published sequences. Thus, a synthetic, optimized, and consensus of the pp65 protein was developed as a target vaccine Ag that was phylogenetically similar to the JP, VR1814, Merlin and AD169 strains.
DNA vaccine construction
To potentially enhance the induction of NAbs upon vaccination, an additional strategy was employed consisting of the co-inclusion of multiple proteins of structural significance within the same vaccine construct (Fig. 2). Candidate viral glycoproteins requiring heterologous interaction for the construction of multimeric functional virion surface complexes were encoded in combination within the same DNA vaccine plasmid. Multiple protein-expressing plasmids gHgL, gMgN and UL encoded ubiquitous endo-proteolytic furin cleavage sites between immunogens to facilitate post-translational cleavage and modification. In this way, co-expression of structurally and functionally relevant proteins may hypothetically facilitate the formation of macromolecular complexes that express clinically- and virologically-relevant B cell epitopic determinants. This may be particularly critical in cases where coexpression is required for productive expression; gH requires coexpression of gL for intracellular transport and terminal carbohydrate modifications, and similarly, gL remains localized in the ER when expressed in the absence of gH.48 Thus, plasmids were constructed for optimizing nascent coexpression of relevant proteins. In total, six amino acid target sequences were constructed and included the consensus-engineered gB, pp65, gHgL, gO, gMgN and UL (including the UL128–131A). Target sequences were genetically optimized at the nucleic acid level which included codon and RNA optimization, among others, for maximal protein expression. Transgenes were then synthesized and subcloned into modified pVAX1 mammalian expression vectors yielding the following plasmid vaccines: pHCMV-gB, pHCMV-pp65, pHCMV-gHgL, pHCMV-gO, pHCMV-gMgN, pHCMV-UL.
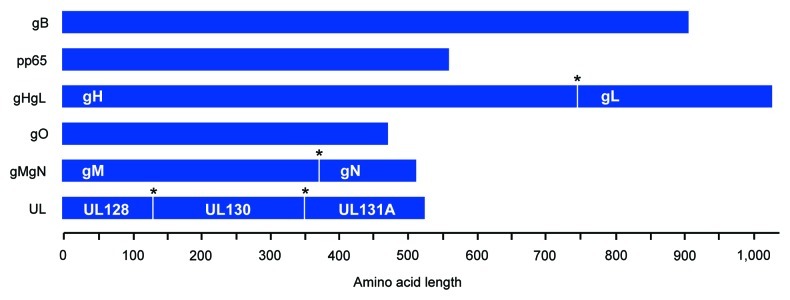
Figure 2. Construction of consensus CMV immunogens and structurally-relevant poly-proteins. Cartoon of consensus CMV immunogens. Poly-proteins are displayed and express multiple structurally-relevant proteins (labeled in white) that are separated by a furin cleavage site (*) for host cell post-translational modification. Amino acid length is defined. All proteins were subsequently genetically-optimized for expression in humans, commercially synthesized, and then subcloned into a modified pVAX mammalian expression vector.
E-DNA vaccination was highly T cell immunogenic
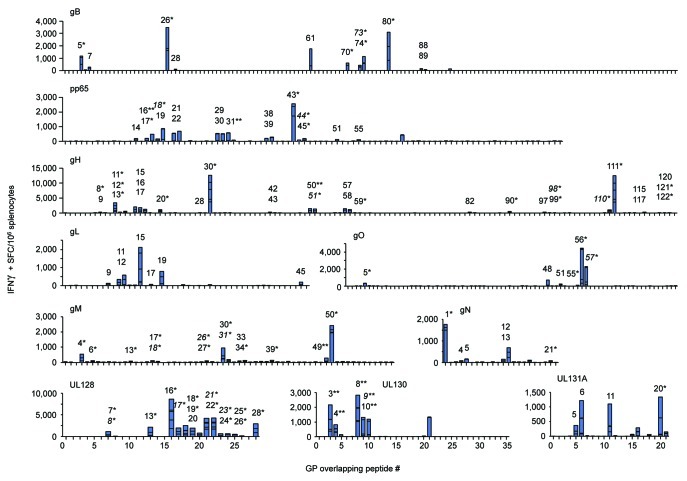
Vaccines were evaluated for T cell immunogenicity in preclinical ‘proof-of-concept’ rodent studies. Since it has been established that the diversity, or breadth, of the T cell response may be important for conferring protection,84-86 we aimed to evaluate the comprehensive epitopic T cell response as induced by each vaccine construct. We developed a modified IFNγ ELISPOT assay that identified and measured subdominant and immunodominant CMV T cell epitopes. This was achieved by incubating samples with individual peptides, as opposed to whole or matrix peptide pools, to increase assay resolution; the traditional practice of pooling peptides for the sake of sample preservation, such as the use of matrix array pools, results in a reduction of assay sensitivity since total functional responses in pools containing multiple epitope-displaying peptides will effectively lower assay resolution, i.e., ‘drown-out’ those of lower magnitude. Mice (n = 5/group) were vaccinated twice with in vivo electroporation (EP) delivery of the indicated CMV vaccines and IFNγ responses were measured 8 d later. E-DNA vaccination with each construct induced robust and broad IFNγ responses that recognized a diversity of T-cell epitopes (Fig. 3 and Table 1). All positive epitope-comprising peptides were subsequently confirmed and further characterized by FACS (data not shown). This modified approach was vaccine-specific and extremely sensitive since little IFNγ production was observed in control wells by stimulation with an irrelevant h-Clip peptide (Control).
Figure 3. E-DNA vaccination was highly T cell immunogenic. Mice (n = 5/group) were immunized twice with EP delivery of the indicated CMV vaccine plasmid and IFNγ responses were measured by modified IFNγ ELISPOT. Splenocytes were incubated in the presence of individual peptides spanning each consensus CMV immunogen and results are shown in stacked bar graphs (summarized in Table 1). Peptides eliciting CD4-restriced IFNγ responses, CD8-restricted (*), and dual CD4/CD8-restricted responses (**), as determined by FACS analysis, are individually numbered and displayed. Putative shared epitopes for contiguous positive peptide responses are italicized.
Table 1. Identification and characterization of CMV vaccine-induced H-2b T cell epitopes.
| |
Best con. % rank (H-2b) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
ELISPOT |
FACS |
CD8+ |
CD4+ |
|
||||||
|
(≤ 0.6) |
(≤ 28) |
||||||||||
| Enhanced Plasmid Vaccine | CMV Ag | Pep # | Position | Sequence | AVE | ± SEM | T cell Restr. | Db | Kb | I-Ab | Previously defined CMV epitopes (Blast - 90%; Allele (Ref)) |
|
pHCMV-gB |
gB |
5 |
25–39 |
SSSTRGTSATHSHHS |
388 |
140 |
8+ |
|
|
14.5 |
|
| |
|
7 |
37–51 |
HHSSHTTSAAHSRSG |
37 |
35 |
4+ |
|
|
18.4 |
|
| |
|
26 |
151–165 |
RRSYAYIHTTYLLGS |
1,105 |
472 |
8+ |
0.1 |
0.2 |
13.5 |
HLA-A*24:0294 |
| |
|
28 |
163–177 |
LGSNTEYVAPPMWEI |
30 |
18 |
4+ |
|
|
4 |
|
| |
|
61 |
361–375 |
AEDSYHFSSAKMTAT |
577 |
430 |
4+ |
|
0.1 |
1.2 |
|
| |
|
70 |
415–429 |
KYGNVSVFETTGGLV |
183 |
89 |
8+ |
|
0.4 |
|
HLA-DR795 |
| |
|
73* |
433–447 |
QGIKQKSLVELERLA |
95 |
73 |
8+ |
|
|
|
|
| |
|
74 |
439–453 |
SLVELERLANRSSLN |
360 |
146 |
8+ |
|
|
|
|
| |
|
80 |
475–489 |
SVHNLVYAQLQFTYD |
1,045 |
169 |
8+ |
|
0.2 |
|
|
| |
|
88 |
523–537 |
INPSAILSAIYNKPI |
53 |
31 |
4+ |
|
|
20.7 |
HLA-A*24:0294 |
| |
|
89 |
529–543 |
LSAIYNKPIAARFMG |
18 |
13 |
4+ |
0.3 |
|
2.5 |
HLA-A*24:0294 |
| |
|
|
|
|
3,798 |
1,544 |
|
|
|
|
|
|
pHCMV-pp65 |
pp65 |
14 |
79–93 |
HTYFTGSEVENVSVN |
130 |
74 |
4+ |
|
|
11.4 |
HLA-A6996 |
| |
|
16 |
91–105 |
SVNVHNPTGRSICPS |
106 |
46 |
8+/4+ |
|
|
|
HLA-A3397 |
| |
|
17 |
97–111 |
PTGRSICPSQEPMSI |
161 |
127 |
8+ |
0.6 |
|
|
TMTL |
| |
|
18* |
103–117 |
CPSQEPMSIYVYALP |
39 |
28 |
8+ |
|
0.3 |
|
TMTL |
| |
|
19 |
109–123 |
MSIYVYALPLKMLNI |
427 |
196 |
4+ |
0.3 |
0.1 |
0.4 |
TMTL |
| |
|
21 |
121–135 |
LNIPSINVHHYPSAA |
192 |
132 |
4+ |
|
|
16.9 |
TMTL |
| |
|
22 |
127–141 |
NVHHYPSAAERKHRH |
277 |
178 |
4+ |
|
|
15.2 |
TMTL |
| |
|
29 |
169–183 |
TRQQNQWKEPDVYYT |
192 |
115 |
4+ |
|
|
|
TMTL |
| |
|
30 |
175–189 |
WKEPDVYYTSAFVFP |
216 |
117 |
4+ |
|
0.6 |
23.1 |
TMTL |
| |
|
31 |
181–195 |
YYTSAFVFPTKDVAL |
338 |
161 |
8+/4+ |
0.6 |
0.5 |
4.8 |
TMTL |
| |
|
38 |
223–237 |
YVKVYLESFCEDVPS |
87 |
50 |
4+ |
|
|
|
TMTL |
| |
|
39 |
229–243 |
ESFCEDVPSGKLFMH |
117 |
62 |
4+ |
|
|
|
TMTL |
| |
|
43 |
253–267 |
DLTMTRNPQPFMRPH |
994 |
468 |
8+ |
0.5 |
|
|
TMTL |
| |
|
44* |
259–273 |
NPQPFMRPHERNGFT |
190 |
177 |
8+ |
|
|
|
TMTL |
| |
|
45 |
265–279 |
RPHERNGFTVLCPKN |
150 |
97 |
8+ |
|
|
|
TMTL |
| |
|
51 |
301–315 |
HFGLLCPKSIPGLSI |
45 |
22 |
4+ |
|
|
27 |
TMTL |
| |
|
55 |
325–339 |
QIFLEVQAIRETVEL |
34 |
8 |
4+ |
|
|
|
TMTL |
| |
|
|
|
|
3,466 |
1,853 |
|
|
|
|
|
|
pHCMV-gHgL |
gH |
8 |
43–57 |
LNTYGRPIRFLRENT |
38 |
33 |
8+ |
|
|
|
HLA-A*24:0294; HLA-II98 |
| |
|
9 |
49–63 |
PIRFLRENTTQCTYN |
26 |
15 |
4+ |
|
|
|
HLA-II98 |
| |
|
11 |
61–75 |
TYNSSLRNSTVVREN |
776 |
141 |
8+ |
0.1 |
|
24.5 |
HLA-A399 |
| |
|
12 |
67–81 |
RNSTVVRENAISFNF |
72 |
46 |
8+ |
|
|
|
HLA-A399 |
| |
|
13 |
73–87 |
RENAISFNFFQSYNQ |
94 |
28 |
8+ |
0.1 |
0.1 |
|
|
| |
|
15 |
85–99 |
YNQYYVFHMPRCLFA |
559 |
231 |
4+ |
|
|
3.5 |
HLA-B799; HLA-A*24:0294 |
| |
|
16 |
91–105 |
FHMPRCLFAGPLAEQ |
419 |
199 |
4+ |
|
|
6.9 |
HLA-B799; HLA-A*24:0294 |
| |
|
17 |
97–111 |
LFAGPLAEQFLNQVD |
281 |
139 |
4+ |
|
|
25.5 |
HLA-B799 |
| |
|
20 |
115–129 |
TLERYQQRLNTYALV |
153 |
36 |
8+ |
0.6 |
|
|
HLA-A399 |
| |
|
28 |
163–177 |
SIPHVWMPPQTTPHG |
20 |
5 |
4+ |
|
|
1.2 |
|
| |
|
30 |
175–189 |
PHGWKESHTTSGLHR |
2,942 |
81 |
8+ |
|
|
25 |
|
| |
|
42 |
247–261 |
MLLIFGHLPRVLFKA |
78 |
58 |
4+ |
0.6 |
0.3 |
27.6 |
HLA-B7/A2/A399 |
| |
|
43 |
253–267 |
HLPRVLFKAPYQRDN |
24 |
9 |
4+ |
|
|
26.8 |
HLA-B799 |
| |
|
50 |
295–309 |
DPDFLDAALDFNYLD |
331 |
187 |
8+/4+ |
|
0.5 |
|
HLA-II98 |
| |
|
51* |
301–315 |
AALDFNYLDLSALLR |
307 |
181 |
8+ |
|
0.5 |
16.1 |
HLA-A*24:02
94
|
| |
|
57 |
337–351 |
RTVEMAFAYALALFA |
340 |
190 |
4+ |
|
0.4 |
1.6 |
HLA-II98 |
| |
|
58 |
343–357 |
FAYALALFAAARQEE |
265 |
157 |
4+ |
0.4 |
|
5.2 |
|
| |
|
59 |
349–363 |
LFAAARQEEAGAEVS |
27 |
17 |
8+ |
|
|
12.9 |
|
| |
|
82 |
487–501 |
EIFIVETGLCSLAEL |
64 |
31 |
4+ |
|
|
|
|
| |
|
90 |
535–549 |
RLTRLFPDATVPATV |
81 |
32 |
8+ |
|
|
6.5 |
|
| |
|
97 |
577–591 |
ESFSALTVSEHVSYV |
51 |
21 |
4+ |
|
|
15.9 |
HLA-A399 |
| |
|
98* |
583–597 |
TVSEHVSYVVTNQYL |
10 |
5 |
8+ |
|
|
|
HLA-A399; HLA-A*24:0294 |
| |
|
99 |
589–603 |
SYVVTNQYLIKGISY |
17 |
4 |
8+ |
0.1 |
|
|
|
| |
|
110* |
655–669 |
LLEYDDTQGVINIMY |
191 |
83 |
8+ |
|
|
|
|
| |
|
111 |
661–675 |
TQGVINIMYMHDSDD |
2,864 |
136 |
8+ |
0.4 |
|
|
HLA-A*24:0294 |
| |
|
115 |
685–699 |
EVVVSSPRTHYLMLL |
22 |
14 |
4+ |
|
|
13.1 |
HLA-B799 |
| |
|
117 |
697–711 |
MLLKNGTVLEVTDVV |
58 |
23 |
4+ |
0.4 |
|
|
HLA-A299 |
| |
|
120 |
715–729 |
TDSRLLMMSVYALSA |
14 |
4 |
4+ |
|
|
|
|
| |
|
121 |
721–735 |
MMSVYALSAIIGIYL |
32 |
16 |
8+ |
|
|
7.4 |
HLA-A399 |
| |
|
122 |
727–741 |
LSAIIGIYLLYRMLK |
13 |
9 |
8+ |
0.5 |
0.2 |
|
HLA-A399 |
| 9,661 | 1,858 | ||||||||||
Epitope-containing peptides were identified by IFNγ ELISPOT (≥ 10 SFC/106 splenocytes AND ≥ 80% animal response rate) and then confirmed/characterized by FACS for function (≥ 3–5 x 104 CD3+ cells were acquired) and phenotype (CD4 and/or CD8 expression by CD3+/CD44+/IFNγ+ cells). Predicted epitopes of peptides confirmed to be CD8-restricted are underlined (as determined by best consensus % rank). All previously-described epitopes (by 90% Blast) are referenced; no H-2b epitopes reported herein, as well as any for UL128, UL130, UL131A, gM, gN and gO, have been previously described (IEDB; www.iedb.org). Contiguous peptides with putative shared and/or partial epitopes as confirmed by ELISPOT (data not shown) are italicized and indicated (*). Immunodominant T cell epitopes are boldface and total responses on average ± SEM are displayed and do not include putative shared and/or partial epitopic responses. TMTL, Too many to list.
Results from the modified ELISPOT assay showed that vaccination induced measurable T cell epitopes in all animals; gB induced 10 total epitopes, pp65 - 17, gH - 28, gL - 7, gO - 5, gM - 12, gN - 6, UL128 - 11, UL130 - 9, and UL131A - 4 (Table 1). When enumerated for each multi-protein expressing construct, pHCMV-gH induced a total of 35 epitopes, pHCMV-gMgN - 18, and pHCMV-UL - 24. These data also account for possible shared/partial T cell epitopes which were investigated for all instances of contiguous peptide responses as identified by modified ELISPOT assay (data not shown). Importantly, diverse epitope hierarchies were consistent and reproducible, and were comprised by both activated CD4+ and CD8+ T cells in all cases except for the gL immunogen which elicited CD4+ epitopes. Indeed, 6 of the 10 immunogens elicited T cell responses that were dominated by CMV-specific CD8+ T cells; approximately 81% of the average total gB-specific response was CD8-restricted, as well as at least 77% of gH, 86% of gO, at least 92% of gM, 60% of gN, and 98% of the UL128 response. Furthermore, vaccination with pHCMV-gHgL, pHCMV-gMgN, or pHCMV-UL generated CD8-dominant responses that constituted 67%, 80%, and 68% of the total average T cell response, respectively. Conversely, the CD4-restricted response was dominant after vaccination with pp65 (at least 50% was CD4+), gL (100%), and UL131A (74%). It was difficult to determine the composition of the T cell response for the UL130 immunogen since all 5 peptides contained dual CD4+/CD8+ epitopes. In most cases, the phenotypic composition of the vaccine-induced T cell response was not directly related to frequency of respective T cell epitopes; while gB vaccination elicited 5 of 10 (50%) epitopes that were CD4-restricted, they accounted for only 19% of the total average response. Similarly, after vaccination with gH or gN, 15 of 18 (54%) and 4 of 6 (67%) of CD4 T cell epitopes induced only 23% and 40% of the total average response, respectively. These data are likely explained by establishment of diverse epitope hierarchies constituted by an array of immunodominant and subdominant epitopes as driven by vaccination with the highly-optimized CMV constructs.
Each immunogen elicited at least 1 immunodominant epitope which was loosely defined as generating an IFNγ response approximately 2-fold over the highest subdominant response within that immunogen; single immunodominant epitopes were detected after vaccination with pp65 [pp65235–267; CD8+ (peptide #43)], gL [gL85–99; CD4+ (#15)], gO [gO331–345; CD8+ (#56)], gM [gM295–309; CD8+ (#50)], gN [gN1–15; CD8+ (#1)], UL128 [UL12891–105; CD8+ (#16)], UL130 [UL13043–57; CD8+/CD4+ (#8)], and UL131A [UL131A61–75; CD4+ (#11)]. Two CD8-restricted immunodominant epitopes were detected following vaccination with either gB or gH that were comprised by peptides gB151–165 (#26), gB475–489 (#80), gH175–189 (#30), and gH661–675 (#111). When considering the multi-protein expression strategy employed by several of these vaccines, immunodominance among the T cell epitopes occurring within multiple immunogens may shift. While both the gH and the gL contain immunodominant epitopes, that from the gL becomes subdominant to the two within the gH since it induces responses that are 2-fold lower, and the immunodominance hierarchy shifts from a total of 3 to 2. This is also true for the pHCMV-UL in which the immunodominant epitope occurring within the UL128 is over 2-fold greater than those within the UL130 and the UL131A, and thus the total immunodominant epitopes elicited by vaccination of all three immunogens is 1 instead of 3. However, immunodominance does not change following vaccination with the pHCMV-gMgN since each of the single immunodominant epitopes remains approximately 2-fold higher than the greatest subdominant response between the two immunogens. Altogether, these data show that each of the plasmids was immune potent and drove CMV-specific T cell responses that were CD8-dominant and recognized a broad array of T cell epitopes.
Discussion
Based upon promising clinical14,33,34 and preclinical35-37 data suggesting that an effective CMV vaccine will induce cellular immune responses, a combination of traditional and surface glycoproteins, matrix and chaperones were selected for evaluation for T cell immunogenicity following delivery as E-DNA vaccines.32,48 While ‘first-generation’ DNA vaccines were poorly immunogenic and elicited mainly CD4-dominant T cell responses, recent technological advancements have dramatically improved their immunogenicity in recent clinical trials.87,88 Nucleic acid synthesis technologies have revolutionized the way genes can be developed via elimination of destabilizing RNA structures, implementation of species-specific codon-usage, modification of GC content and others, rendering these genes capable of strong in vivo expression when inserted into ‘highly-optimized’ plasmid DNA.87 In response to polymorphism, likely attributed to the accumulation of spontaneous mutations and/or by inter- and intra-strain homologous recombination induced by selective pressure (Fig. 1),50,51,89 immunity can be further directed toward multiple circulating strains by “consensus-engineering” of the amino acid sequence of the vaccine immunogen.63,64 Furthermore, structurally-relevant immunogens can be incorporated into the same plasmid vaccines for the co-expression of virologically-relevant macromolecular complexes (Fig. 2). Cellular uptake and subsequent Ag expression are substantially increased when highly-concentrated plasmid vaccine formulations are administered with EP, a technology that uses brief square-wave electric pulses within the vaccination site to drive plasmids into transiently permeabilized cells.90 In theory, a cocktail of DNA plasmids could be assembled for directing a highly-specialized immune response against any number of variable Ags.
E-DNA vaccination with each CMV construct herein was highly T cell immunogenic in preclinical ‘proof-of-concept’ murine studies, generating robust and broad T cell responses as extensively analyzed by a novel modified ELISPOT assay developed herein (Fig. 3 and Table 1). This may be critical since prevention of CMV infection and disease in the transplantation setting may require a greater cellular response than is needed to prevent congenital infection. While little quantitative data regarding the induction of CMV-specific T cell responses in mice as driven by a vaccine exist, vaccination herein with the pHCMV-pp65 generated a T cell response that was on average over 40% greater than a previous pp65 DNA vaccine administered three times with a poloxamer-based delivery system that induced ~2,000 IFNγ+ SFC/106 cells.91 Importantly, responses from both CD8+ and CD4+ helper cells responses were observed which may require for protection against post-transplant occurrence of CMV disease,37,42,45 as well as driving diverse T cell hierarchies of which subdominant responses can make significant contributions to protection.84-86 These data demonstrate that ‘next-generation’ DNA vaccine technologies are effective at inducing CD8+ T cell responses in contrast to prior strategies that induced mainly CD4-dominant responses. Additionally, a majority of epitopes identified for gB, pp65, gH, and gL also contained HLA that have previously been reported to contribute to the suppression of viremia and amelioration of overt disease (Table 1).2,14,41-44 In summary, the elicitation, identification, and characterization of extensive T cell responses as driven by a vaccine herein provide an important tool for guiding CMV vaccine development. These data suggest further study of these constructs as vaccine components in strategies aiming to generate broad T cell responses, and investigation of their potential for generating Abs capable of neutralization and protection in preclinical challenge models is warranted.
Methods
Ethics statement
Animal experimentation was conducted following UPenn IACUC and School of Medicine Animal Facility guidelines for housing and care of laboratory animals and performed in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals of NIH.
E-DNA vaccine construction
Plasmid constructs encoded full-length CMV proteins: pHCMV-gB, pHCMV-pp65, pHCMV-gHgL, pHCMV-gO, pHCMV-gMgN, pHCMV-UL (UL128, UL130 and UL131A). Immunogens were consensus, as determined by alignment using Vector NTI® (Invitrogen), of publically available (GenBank) CMV clinical strains (passaged < 6 times in tissue culture), and resultant sequences are listed below. This strategy excluded sequences from highly-passaged lab-adapted strains that may diverge in sequence from clinical strains, thus maximizing the potential for broadly-reactive immunity. Phylogenetic analysis was performed by multiple-alignment with ClustalW using MEGA version 5 software. Commercial genetic optimization was proprietary and included codon and RNA optimization, among others, for species-specific protein expression in humans, and all genes were synthesized and subcloned into a modified pVAX1 mammalian expression vector (GeneArt or GenScript). Several plasmids (pHCMV-gHgL, pHCMV-gMgN and pHCMV-UL) encoded multiple immunogens separated by a furin cleavage site (RGRKRRS).
Consensus CMV immunogen sequences
gB
MESRIWCLVVCVNLCIVCLGAAVSSSSTRGTSATHSHHSSHTTSAAHSRSGSVSSQRVTSSEAVSHRANETIYNTTLKYGDVVGVNTTKYPYRVCSMAQGTDLIRFERNIVCTSMKPINEDLDEGIMVVYKRNIVAHTFKVRVYQKVLTFRRSYAYIHTTYLLGSNTEYVAPPMWEIHHINSHSQCYSSYSRVIAGTVFVAYHRDSYENKTMQLMPDDYSNTHSTRYVTVKDQWHSRGSTWLYRETCNLNCMVTITTARSKYPYHFFATSTGDVVDISPFYNGTNRNASYFGENADKFFIFPNYTIVSDFGRPNSALETHRLVAFLERADSVISWDIQDEKNVTCQLTFWEASERTIRSEAEDSYHFSSAKMTATFLSKKQEVNMSDSALDCVRDEAINKLQQIFNTSYNQTYEKYGNVSVFETTGGLVVFWQGIKQKSLVELERLANRSSLNLTHRTKRSTDGNNTTHLSNMESVHNLVYAQLQFTYDTLRGYINRALAQIAEAWCVDQRRTLEVFKELSKINPSAILSAIYNKPIAARFMGDVLGLASCVTINQTSVKVLRDMNVKESPGRCYSRPVVIFNFANSSYVQYGQLGEDNEILLGNHRTEECQLPSLKIFIAGNSAYEYVDYLFKRMIDLSSISTVDSMIALDIDPLENTDFRVLELYSQKELRSSNVFDLEEIMREFNSYKQRVKYVEDKVVDPLPPYLKGLDDLMSGLGAAGKAVGVAIGAVGGAVASVVEGVATFLKNPFGAFTIILVAIAVVIITYLIYTRQRRLCTQPLQNLFPYLVSADGTTVTSGSTKDTSLQAPPSYEESVYNSGRKGPGPPSSDASTAAPPYTNEQAYQMLLALARLDAEQRAQQNGTDSLDGQTGTQDKGQKPNLLDRLRHRKNGYRHLKDSDEEENV
pp65
MESRGRRCPEMISVLGPISGHVLKAVFSRGDTPVLPHETRLLQTGIHVRVSQPSLILVSQYTPDSTPCHRGDNQLQVQHTYFTGSEVENVSVNVHNPTGRSICPSQEPMSIYVYALPLKMLNIPSINVHHYPSAAERKHRHLPVADAVIHASGKQMWQARLTVSGLAWTRQQNQWKEPDVYYTSAFVFPTKDVALRHVVCAHELVCSMENTRATKMQVIGDQYVKVYLESFCEDVPSGKLFMHVTLGSDVEEDLTMTRNPQPFMRPHERNGFTVLCPKNMIIKPGKISHIMLDVAFTSHEHFGLLCPKSIPGLSISGNLLMNGQQIFLEVQAIRETVELRQYDPVAALFFFDIDLLLQRGPQYSEHPTFTSQYRIQGKLEYRHTWDRHDEGAAQGDDDVWTSGSDSDEELVTTERKTPRVTGGGAMAGASTSAGRKRKSASSATACTAGVMTRGRLKAESTVAPEEDTDEDSDNEIHNPAVFTWPPWQAGILARNLVPMVATVQGQNLKYQEFFWDANDIYRIFAELEGVWQPAAQPKRRRHRQDALPGPCIASTPKKHRG
gH
MRPGLPSYLTVFAVYLLSHLPSQRYGADAASEALDPHAFHLLLNTYGRPIRFLRENTTQCTYNSSLRNSTVVRENAISFNFFQSYNQYYVFHMPRCLFAGPLAEQFLNQVDLTETLERYQQRLNTYALVSKDLASYRSFSQQLKAQDSLGEQPTTVPPPIDLSIPHVWMPPQTTPHGWKESHTTSGLHRPHFNQTCILFDGHDLLFSTVTPCLHQGFYLIDELRYVKITLTEDFFVVTVSIDDDTPMLLIFGHLPRVLFKAPYQRDNFILRQTEKHELLVLVKKDQLNRHSYLKDPDFLDAALDFNYLDLSALLRNSFHRYAVDVLKSGRCQMLDRRTVEMAFAYALALFAAARQEEAGAEVSVPRALDRQAALLQIQEFMITCLSQTPPRTTLLLYPTAVDLAKRALWTPNQITDITSLVRLVYILSKQNQQHLIPQWALRQIADFALKLHKTHLASFLSAFARQELYLMGSLVHSMLVHTTERREIFIVETGLCSLAELSHFTQLLAHPHHEYLSDLYTPCSSSGRRDHSLERLTRLFPDATVPATVPAALSILSTMQPSTLETFPDLFCLPLGESFSALTVSEHVSYVVTNQYLIKGISYPVSTTVVGQSLIITQTDSQTKCELTRNMHTTHSITAALNISLENCAFCQSALLEYDDTQGVINIMYMHDSDDVLFALDPYNEVVVSSPRTHYLMLLKNGTVLEVTDVVVDATDSRLLMMSVYALSAIIGIYLLYRMLKTC
gL
MCRRPDCGFSFSPGPVILLWCCLLLPIVSSAAVSVAPTAAEKVPAECPELTRRCLLGEVFQGDKYESWLRPLVNVTGRDGPLSQLIRYRPVTPEAANSVLLDEAFLDTLALLYNNPDQLRALLTLLSSDTAPRWMTVMRGYSECGDGSPAVYTCVDDLCRGYDLTRLSYGRSIFTEHVLGFELVPPSLFNVVVAIRNEATRTNRAVRLPVSTAAAPEGITLFYGLYNAVKEFCLRHQLDPPLLRHLDKYYAGLPPELKQTRVNLPAHSRYGPQAVDAR
gO
MGKKEMIMVKGIPKIMLLISITFLLLSLINCNVLVNSKGTRRSWPYTVLSYRGKEILKKQKEDILKRLMSTSSDGYRFLMYPSQQKFHAIVISMDKFPQDYILAGPIRNDSITHMWFDFYSTQLRKPAKYVYSEYNHTAHKITLRPPPCGTVPSMNCLSEMLNVSKRNDTGEKGCGNFTTFNPMFFNVPRWNTKLYIGSNKVNVDSQTIYFLGLTALLLRYAQRNCTRSFYLVNAMSRNLFRVPKYINGTKLKNTMRKLKRKQALVKEQPQKKNKKSQSTTTPYLSYTTSTAFNVTTNVTYSATAAVTRVATSTTGYRPDSNFMKSIMATQLRDLATWVYTTLRYRNEPFCKPDRNRTAVSEFMKNTHVLIRNETPYTIYGTLDMSSLYYNETMSVENETASDNNETTPTSPSTRFQRTFIDPLWDYLDSLLFLDKIRNFSLQLPAYGNLTPPEHRRAANLSTLNSLWWWLQYP
gM
MAPSHVDKVNTRTWSASIVFMVLTFVNVSVHLVLSNFPHLGYPCVYYHVVDFERLNMSAYNVMHLHTPMLFLDSVQLVCYAVFMQLVFLAVTIYYLVCWIKISMRKDKGMSLNQSTRDISYMGDSLTAFLFILSMDTFQLFTLTMSFRLPSMIAFMAAVHFFCLTIFNVSMVTQYRSYKRSLFFFSRLHPKLKGTVQFRTLIVNLVEVALGFNTTVVAMALCYGFGNNFFVRTGHMVLAVFVVYAIISIIYFLLIEAVFFQYVKVQFGYHLGAFFGLCGLIYPIVQYDTFLSNEYRTGISWSFGMLFFIWAMFTTCRAVRYFRGRGSGSVKYQALATASGEEVAALSHHDSLESRRLREEEDDDDDEDFEDA
gN
MEWNTLVLGLLVLSVAASSNNTSTASTPSPSSSTHTSTTVKATTTATTSTTTATSTTSSTTSTKPGSTTHDPNVMRPHAHNDFYKAHCTSHMYELSLSSFAAWWTMLNALILMGAFCIVLRHCCFQNFTATTTKGY
UL128
SPKDLTPFLTALWLLLGHSRVPRVRAEECCEFINVNHPPERCYDFKMCNRFTVALRCPDGEVCYSPEKTAEIRGIVTTMTHSLTRQVVHNKLTSCNYNPLYLEADGRIRCGKVNDKAQYLLGAAGSVPYRWINLEYDKITRIVGLDQYLESVKKHKRLDVCRAKMGYMLQ
UL130:
LRLLLRHHFHCLLLCAVWATPCLASPWSTLTANQNPSPPWSKLTYSKPHDAATFYCPFLYPSPPRSPLQFSGFQRVSTGPECRNETLYLLYNREGQTLVERSSTWVKKVIWYLSGRNQTILQRMPRTASKPSDGNVQISVEDAKIFGAHMVPKQTKLLRFVVNDGTRYQMCVMKLESWAHVFRDYSVSFQVRLTFTEANNQTYTFCTHPNLIV
UL131A:
MRLCRVWLSVCLCAVVLGQCQRETAEKNDYYRVPHYWDACSRALPDQTRYKYVEQLVDLTLNYHYDASHGLDNFDVLKRINVTEVSLLISDFRRQNRRGGTNKRTTFNAAGSLAPHARSLEFSVRLFAN
Immunization, mice, and challenge
Adult female C57BL/6 (H-2b) mice 6–8 weeks of age were purchased from The Jackson Laboratory. Mice were immunized i.m. by needle injection of 45 µg plasmid DNA resuspended in water and immediately followed by EP at the same site, using a three-pronged Minimally Invasive Device inserted approximately 2 mm intramuscularly as previously described92 using the CELLECTRA® adaptive constant current device (Inovio Pharmaceuticals, Inc.). Square-wave pulses were delivered through a triangular 3-electrode array consisting of 26-gauge solid stainless steel electrodes and two constant-current pulses of 0.1 Amps were delivered for 52 msec/pulse separated by a 1 sec delay. For T cell studies, spleens were harvested 8 days post-immunization.93 Briefly, splenocytes were resuspended in RPMI 1640 medium (Mediatech Inc.) supplemented with 10% FBS, 1X Anti-Anti, and 1X β-ME (Invitrogen).
ELISPOT assays
Standard IFNγ ELISPOT assay has been described93 and was modified herein for comprehensive analysis of T cell breadth. Identification and measurement of subdominant and immunodominant CMV T cell epitopes were assessed by stimulating splenocytes with individual peptides (15-mers overlapping by 9 amino acids; 2.5 µg/ml final) spanning each consensus CMV immunogen, as opposed to whole or matrix peptide pools. Peptides containing T cell epitopes were identified (≥ 10 AVE IFNγ+ spots AND ≥ 80% animal response rate; summarized in Table 1) and then later confirmed functionally and phenotypically by FACS. Possible shared/partial T cell epitopes were addressed for all instances of contiguous peptide responses as identified by modified ELISPOT assay. Here, cells were stimulated individually with each of the contiguous peptides, as well as in combination for comparison (data not shown), and were defined as ‘shared/partial’ if the combined response was not greater than either of the two individual responses.
Flow cytometry
For splenocyte ICS, 1 × 106 cells/well were added to 96-well plates and stimulated for 5 h for stimulation with individual peptides (2.5 µg/ml final) for functional confirmation of all peptides identified by modified ELISPOT as well as phenotypic characterization (Table 1). Immunostaining of splenocytes and transfected 293Ts was performed in 96-well plates and cells were pre-stained with LIVE/DEAD® Fixable Violet Dead Cell Stain Kit (Invitrogen). Splenocytes were surface-stained for CD19 (V450; clone 1D3), CD4 (PE-Cy7; clone RM4-5), CD8α (APC-Cy7; clone 53-6.7) and CD44 (PE-Cy5; clone IM7) (BD Biosciences), washed three times in PBS + 1% FBS, permeabilized with BD Cytofix/Cytoperm™ kit, and then stained intracellularly with IFNγ (APC; clone XMG1.2) and CD3 (PE-cy5.5; clone 145-2C11) (eBioscience). All cells were fixed in 1% paraformaldehyde. LSRII flow cytometer (BD Biosciences) was used for acquisition and FlowJo (Tree Star) for data analysis. Activated T cells were gated by FACS: total lymphocytes, live (LD) CD3+ cells that were negative for CD19 and LIVE-DEAD (dump channel), singlets (excludes cell doublets), CD4+ and CD8+ cells, activated cells (CD44+), and peptide-specific IFNγ-producing T cells were gated.
Statistical analyses
Significance for unrooted phylogenetic trees was determined by maximum-likelihood method and verified by bootstrap analysis and significant support values (≥80%; 1,000 bootstrap replicates) were determined by MEGA version 5 software.
Acknowledgements
We would like to acknowledge Steven E. Bosinger, Yvonne Suessmuth, and members of the Weiner laboratory for contributions and/or critical reading of this manuscript. This work was supported by NIH, NIAID, and/or DAIDS grants: HIV Vaccine Research and Design (HIVRAD) (P01-AI071739) and “Development of a Universal Influenza Seasonal Vaccine” (R01-AI092843) to DBW, and by funding to DBW and DJS (T32-AI070099). In addition, funding from Inovio Pharmaceuticals was received as part of a Sponsored Research Agreement entitled “Study and Development of Consensus Immunogens in the area of Viral Vaccines”.
Conflict-of-interest disclosure: DBW notes that he and his laboratory has several commercial relationships with companies in the area of vaccines. These include him receiving consulting fees or stock ownership for Advisory/Review Board service, speaking support, or research support from commercial entities including Inovio Pharmaceuticals, Bristol-Myers Squibb, VGXI, Pfizer, Virxsys Co., Johnson & Johnson, Merck & Co., Sanofi Pasteur, Althea, Novo Nordisk, Statens Serum Institut, Aldevron, Novartis, Incyte, and possibly others. No writing assistance was utilized in the production of this manuscript.
Glossary
Abbreviations:
- CMV
human cytomegalovirus
- E-DNA
enhanced DNA
- EP
in vivo electroporation
- Nabs
neutralizing Abs
- gC
high molecular weight complexes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/vaccines/article/22447
References
- 1.Sung H, Schleiss MR. Update on the current status of cytomegalovirus vaccines. Expert Rev Vaccines. 2010;9:1303–14. doi: 10.1586/erv.10.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson C, Emery VC. Cytomegalovirus quantification: where to next in optimising patient management? J Clin Virol. 2011;51:223–8. doi: 10.1016/j.jcv.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, et al. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124:175–84. doi: 10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- 4.Sandalova E, Laccabue D, Boni C, Tan AT, Fink K, Ooi EE, et al. Contribution of herpesvirus specific CD8 T cells to anti-viral T cell response in humans. PLoS Pathog. 2010;6:e1001051. doi: 10.1371/journal.ppat.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202:673–85. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinclair J, Sissons P. Latency and reactivation of human cytomegalovirus. J Gen Virol. 2006;87:1763–79. doi: 10.1099/vir.0.81891-0. [DOI] [PubMed] [Google Scholar]
- 7.Revello MG, Gerna G. Diagnosis and management of human cytomegalovirus infection in the mother, fetus, and newborn infant. Clin Microbiol Rev. 2002;15:680–715. doi: 10.1128/CMR.15.4.680-715.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McMichael G, Maclennan A, Gibson C, Alvino E, Goldwater P, Haan E, et al. Cytomegalovirus and Epstein-Barr virus may be associated with some cases of cerebral palsy. J Matern Fetal Neonatal Med 2012:Early Online: 1-4. [DOI] [PubMed]
- 9.Baryawno N, Rahbar A, Wolmer-Solberg N, Taher C, Odeberg J, Darabi A, et al. Detection of human cytomegalovirus in medulloblastomas reveals a potential therapeutic target. J Clin Invest. 2011;121:4043–55. doi: 10.1172/JCI57147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komanduri KV, Donahoe SM, Moretto WJ, Schmidt DK, Gillespie G, Ogg GS, et al. Direct measurement of CD4+ and CD8+ T-cell responses to CMV in HIV-1-infected subjects. Virology. 2001;279:459–70. doi: 10.1006/viro.2000.0697. [DOI] [PubMed] [Google Scholar]
- 11.Riddell SR. Pathogenesis of cytomegalovirus pneumonia in immunocompromised hosts. Semin Respir Infect. 1995;10:199–208. [PubMed] [Google Scholar]
- 12.Fishman JA, Rubin RH. Infection in organ-transplant recipients. N Engl J Med. 1998;338:1741–51. doi: 10.1056/NEJM199806113382407. [DOI] [PubMed] [Google Scholar]
- 13.Wloch MK, Smith LR, Boutsaboualoy S, Reyes L, Han C, Kehler J, et al. Safety and immunogenicity of a bivalent cytomegalovirus DNA vaccine in healthy adult subjects. J Infect Dis. 2008;197:1634–42. doi: 10.1086/588385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kharfan-Dabaja MA, Boeckh M, Wilck MB, Langston AA, Chu AH, Wloch MK, et al. A novel therapeutic cytomegalovirus DNA vaccine in allogeneic haemopoietic stem-cell transplantation: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Infect Dis. 2012;12:290–9. doi: 10.1016/S1473-3099(11)70344-9. [DOI] [PubMed] [Google Scholar]
- 15.Boeckh M, Ljungman P. How we treat cytomegalovirus in hematopoietic cell transplant recipients. Blood. 2009;113:5711–9. doi: 10.1182/blood-2008-10-143560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drew WL. Is combination antiviral therapy for CMV superior to monotherapy? J Clin Virol. 2006;35:485–8. doi: 10.1016/j.jcv.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 17.Limaye AP, Corey L, Koelle DM, Davis CL, Boeckh M. Emergence of ganciclovir-resistant cytomegalovirus disease among recipients of solid-organ transplants. Lancet. 2000;356:645–9. doi: 10.1016/S0140-6736(00)02607-6. [DOI] [PubMed] [Google Scholar]
- 18.Söderberg-Nauclér C. HCMV microinfections in inflammatory diseases and cancer. J Clin Virol. 2008;41:218–23. doi: 10.1016/j.jcv.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Harkins L, Volk AL, Samanta M, Mikolaenko I, Britt WJ, Bland KI, et al. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet. 2002;360:1557–63. doi: 10.1016/S0140-6736(02)11524-8. [DOI] [PubMed] [Google Scholar]
- 20.Zhou YF, Leon MB, Waclawiw MA, Popma JJ, Yu ZX, Finkel T, et al. Association between prior cytomegalovirus infection and the risk of restenosis after coronary atherectomy. N Engl J Med. 1996;335:624–30. doi: 10.1056/NEJM199608293350903. [DOI] [PubMed] [Google Scholar]
- 21.Heidenreich PA, Trogdon JG, Khavjou OA, Butler J, Dracup K, Ezekowitz MD, et al. American Heart Association Advocacy Coordinating Committee. Stroke Council. Council on Cardiovascular Radiology and Intervention. Council on Clinical Cardiology. Council on Epidemiology and Prevention. Council on Arteriosclerosis. Thrombosis and Vascular Biology. Council on Cardiopulmonary. Critical Care. Perioperative and Resuscitation. Council on Cardiovascular Nursing. Council on the Kidney in Cardiovascular Disease. Council on Cardiovascular Surgery and Anesthesia, and Interdisciplinary Council on Quality of Care and Outcomes Research Forecasting the future of cardiovascular disease in the United States: a policy statement from the American Heart Association. Circulation. 2011;123:933–44. doi: 10.1161/CIR.0b013e31820a55f5. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt GM, Horak DA, Niland JC, Duncan SR, Forman SJ, Zaia JA. A randomized, controlled trial of prophylactic ganciclovir for cytomegalovirus pulmonary infection in recipients of allogeneic bone marrow transplants; The City of Hope-Stanford-Syntex CMV Study Group. N Engl J Med. 1991;324:1005–11. doi: 10.1056/NEJM199104113241501. [DOI] [PubMed] [Google Scholar]
- 23.Nieto FJ, Adam E, Sorlie P, Farzadegan H, Melnick JL, Comstock GW, et al. Cohort study of cytomegalovirus infection as a risk factor for carotid intimal-medial thickening, a measure of subclinical atherosclerosis. Circulation. 1996;94:922–7. doi: 10.1161/01.CIR.94.5.922. [DOI] [PubMed] [Google Scholar]
- 24.Speir E, Modali R, Huang ES, Leon MB, Shawl F, Finkel T, et al. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science. 1994;265:391–4. doi: 10.1126/science.8023160. [DOI] [PubMed] [Google Scholar]
- 25.Streblow DN, Soderberg-Naucler C, Vieira J, Smith P, Wakabayashi E, Ruchti F, et al. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell. 1999;99:511–20. doi: 10.1016/S0092-8674(00)81539-1. [DOI] [PubMed] [Google Scholar]
- 26.Cobbs CS, Harkins L, Samanta M, Gillespie GY, Bharara S, King PH, et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347–50. [PubMed] [Google Scholar]
- 27.Huang G, Yan Q, Wang Z, Chen X, Zhang X, Guo Y, et al. Human cytomegalovirus in neoplastic cells of Epstein-Barr virus negative Hodgkin’s disease. Int J Oncol. 2002;21:31–6. [PubMed] [Google Scholar]
- 28.Samanta M, Harkins L, Klemm K, Britt WJ, Cobbs CS. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J Urol. 2003;170:998–1002. doi: 10.1097/01.ju.0000080263.46164.97. [DOI] [PubMed] [Google Scholar]
- 29.Cobbs CS, Soroceanu L, Denham S, Zhang W, Britt WJ, Pieper R, et al. Human cytomegalovirus induces cellular tyrosine kinase signaling and promotes glioma cell invasiveness. J Neurooncol. 2007;85:271–80. doi: 10.1007/s11060-007-9423-2. [DOI] [PubMed] [Google Scholar]
- 30.Rahbar A, Boström L, Lagerstedt U, Magnusson I, Söderberg-Naucler C, Sundqvist VA. Evidence of active cytomegalovirus infection and increased production of IL-6 in tissue specimens obtained from patients with inflammatory bowel diseases. Inflamm Bowel Dis. 2003;9:154–61. doi: 10.1097/00054725-200305000-00002. [DOI] [PubMed] [Google Scholar]
- 31.Institute of Medicine (U.S.). Committee to Study Priorities for Vaccine Development., Stratton KR, Durch J, Lawrence RS. Vaccines for the 21st century: a tool for decisionmaking. Washington, D.C.: National Academy Press, 2000. [PubMed] [Google Scholar]
- 32.Reske A, Pollara G, Krummenacher C, Chain BM, Katz DR. Understanding HSV-1 entry glycoproteins. Rev Med Virol. 2007;17:205–15. doi: 10.1002/rmv.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adler SP, Starr SE, Plotkin SA, Hempfling SH, Buis J, Manning ML, et al. Immunity induced by primary human cytomegalovirus infection protects against secondary infection among women of childbearing age. J Infect Dis. 1995;171:26–32. doi: 10.1093/infdis/171.1.26. [DOI] [PubMed] [Google Scholar]
- 34.Pass RF, Zhang C, Evans A, Simpson T, Andrews W, Huang ML, et al. Vaccine prevention of maternal cytomegalovirus infection. N Engl J Med. 2009;360:1191–9. doi: 10.1056/NEJMoa0804749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schleiss MR. Comparison of vaccine strategies against congenital CMV infection in the guinea pig model. J Clin Virol. 2008;41:224–30. doi: 10.1016/j.jcv.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Schleiss MR, Lacayo JC, Belkaid Y, McGregor A, Stroup G, Rayner J, et al. Preconceptual administration of an alphavirus replicon UL83 (pp65 homolog) vaccine induces humoral and cellular immunity and improves pregnancy outcome in the guinea pig model of congenital cytomegalovirus infection. J Infect Dis. 2007;195:789–98. doi: 10.1086/511982. [DOI] [PubMed] [Google Scholar]
- 37.Wynn KK, Khanna R. Models of CMV infection. Drug Discov Today Dis Models. 2006;3:91–6. doi: 10.1016/j.ddmod.2006.03.011. [DOI] [Google Scholar]
- 38.Frey SE, Harrison C, Pass RF, Yang E, Boken D, Sekulovich RE, et al. Effects of antigen dose and immunization regimens on antibody responses to a cytomegalovirus glycoprotein B subunit vaccine. J Infect Dis. 1999;180:1700–3. doi: 10.1086/315060. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell DK, Holmes SJ, Burke RL, Duliege AM, Adler SP. Immunogenicity of a recombinant human cytomegalovirus gB vaccine in seronegative toddlers. Pediatr Infect Dis J. 2002;21:133–8. doi: 10.1097/00006454-200202000-00009. [DOI] [PubMed] [Google Scholar]
- 40.Pass RF, Duliegè AM, Boppana S, Sekulovich R, Percell S, Britt W, et al. A subunit cytomegalovirus vaccine based on recombinant envelope glycoprotein B and a new adjuvant. J Infect Dis. 1999;180:970–5. doi: 10.1086/315022. [DOI] [PubMed] [Google Scholar]
- 41.Costa C, Astegiano S, Terlizzi ME, Sidoti F, Curtoni A, Solidoro P, et al. Evaluation and significance of cytomegalovirus-specific cellular immune response in lung transplant recipients. Transplant Proc. 2011;43:1159–61. doi: 10.1016/j.transproceed.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 42.Gamadia LE, Remmerswaal EB, Weel JF, Bemelman F, van Lier RA, Ten Berge IJ. Primary immune responses to human CMV: a critical role for IFN-gamma-producing CD4+ T cells in protection against CMV disease. Blood. 2003;101:2686–92. doi: 10.1182/blood-2002-08-2502. [DOI] [PubMed] [Google Scholar]
- 43.Eid AJ, Razonable RR. New developments in the management of cytomegalovirus infection after solid organ transplantation. Drugs. 2010;70:965–81. doi: 10.2165/10898540-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 44.Razonable RR. Immune-based therapies for cytomegalovirus infection. Immunotherapy. 2010;2:117–30. doi: 10.2217/imt.09.82. [DOI] [PubMed] [Google Scholar]
- 45.Komanduri KV, Viswanathan MN, Wieder ED, Schmidt DK, Bredt BM, Jacobson MA, et al. Restoration of cytomegalovirus-specific CD4+ T-lymphocyte responses after ganciclovir and highly active antiretroviral therapy in individuals infected with HIV-1. Nat Med. 1998;4:953–6. doi: 10.1038/nm0898-953. [DOI] [PubMed] [Google Scholar]
- 46.Wang D, Li F, Freed DC, Finnefrock AC, Tang A, Grimes SN, et al. Quantitative analysis of neutralizing antibody response to human cytomegalovirus in natural infection. Vaccine. 2011;29:9075–80. doi: 10.1016/j.vaccine.2011.09.056. [DOI] [PubMed] [Google Scholar]
- 47.Nigro G, Adler SP, La Torre R, Best AM, Congenital Cytomegalovirus Collaborating Group Passive immunization during pregnancy for congenital cytomegalovirus infection. N Engl J Med. 2005;353:1350–62. doi: 10.1056/NEJMoa043337. [DOI] [PubMed] [Google Scholar]
- 48.Britt WJ, Boppana S. Human cytomegalovirus virion proteins. Hum Immunol. 2004;65:395–402. doi: 10.1016/j.humimm.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Cunningham C, Gatherer D, Hilfrich B, Baluchova K, Dargan DJ, Thomson M, et al. Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J Gen Virol. 2010;91:605–15. doi: 10.1099/vir.0.015891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rasmussen L, Geissler A, Cowan C, Chase A, Winters M. The genes encoding the gCIII complex of human cytomegalovirus exist in highly diverse combinations in clinical isolates. J Virol. 2002;76:10841–8. doi: 10.1128/JVI.76.21.10841-10848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pignatelli S, Dal Monte P, Rossini G, Landini MP. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev Med Virol. 2004;14:383–410. doi: 10.1002/rmv.438. [DOI] [PubMed] [Google Scholar]
- 52.Campadelli-Fiume G, Amasio M, Avitabile E, Cerretani A, Forghieri C, Gianni T, et al. The multipartite system that mediates entry of herpes simplex virus into the cell. Rev Med Virol. 2007;17:313–26. doi: 10.1002/rmv.546. [DOI] [PubMed] [Google Scholar]
- 53.Britt WJ, Vugler LG. Processing of the gp55-116 envelope glycoprotein complex (gB) of human cytomegalovirus. J Virol. 1989;63:403–10. doi: 10.1128/jvi.63.1.403-410.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navarro D, Lennette E, Tugizov S, Pereira L. Humoral immune response to functional regions of human cytomegalovirus glycoprotein B. J Med Virol. 1997;52:451–9. doi: 10.1002/(SICI)1096-9071(199708)52:4<451::AID-JMV18>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 55.Tugizov S, Wang Y, Qadri I, Navarro D, Maidji E, Pereira L. Mutated forms of human cytomegalovirus glycoprotein B are impaired in inducing syncytium formation. Virology. 1995;209:580–91. doi: 10.1006/viro.1995.1290. [DOI] [PubMed] [Google Scholar]
- 56.Cui X, Meza BP, Adler SP, McVoy MA. Cytomegalovirus vaccines fail to induce epithelial entry neutralizing antibodies comparable to natural infection. Vaccine. 2008;26:5760–6. doi: 10.1016/j.vaccine.2008.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimamura M, Mach M, Britt WJ. Human cytomegalovirus infection elicits a glycoprotein M (gM)/gN-specific virus-neutralizing antibody response. J Virol. 2006;80:4591–600. doi: 10.1128/JVI.80.9.4591-4600.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Genini E, Percivalle E, Sarasini A, Revello MG, Baldanti F, Gerna G. Serum antibody response to the gH/gL/pUL128-131 five-protein complex of human cytomegalovirus (HCMV) in primary and reactivated HCMV infections. J Clin Virol. 2011;52:113–8. doi: 10.1016/j.jcv.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 59.Fouts AE, Chan P, Stephan JP, Vandlen R, Feierbach B. Antibodies against the gH/gL/UL128/UL130/UL131 complex comprise the majority of the anti-cytomegalovirus (anti-CMV) neutralizing antibody response in CMV hyperimmune globulin. J Virol. 2012;86:7444–7. doi: 10.1128/JVI.00467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J Virol. 2000;74:7720–9. doi: 10.1128/JVI.74.17.7720-7729.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1996;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, et al. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci USA. 2003;100:14976–81. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laddy DJ, Yan J, Kutzler M, Kobasa D, Kobinger GP, Khan AS, et al. Heterosubtypic protection against pathogenic human and avian influenza viruses via in vivo electroporation of synthetic consensus DNA antigens. PLoS ONE. 2008;3:e2517. doi: 10.1371/journal.pone.0002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan J, Yoon H, Kumar S, Ramanathan MP, Corbitt N, Kutzler M, et al. Enhanced cellular immune responses elicited by an engineered HIV-1 subtype B consensus-based envelope DNA vaccine. Mol Ther. 2007;15:411–21. doi: 10.1038/sj.mt.6300036. [DOI] [PubMed] [Google Scholar]
- 65.Chou SW, Dennison KM. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J Infect Dis. 1991;163:1229–34. doi: 10.1093/infdis/163.6.1229. [DOI] [PubMed] [Google Scholar]
- 66.Haberland M, Meyer-König U, Hufert FT. Variation within the glycoprotein B gene of human cytomegalovirus is due to homologous recombination. J Gen Virol. 1999;80:1495–500. doi: 10.1099/0022-1317-80-6-1495. [DOI] [PubMed] [Google Scholar]
- 67.Rasmussen L, Hong C, Zipeto D, Morris S, Sherman D, Chou S, et al. Cytomegalovirus gB genotype distribution differs in human immunodeficiency virus-infected patients and immunocompromised allograft recipients. J Infect Dis. 1997;175:179–84. doi: 10.1093/infdis/175.1.179. [DOI] [PubMed] [Google Scholar]
- 68.Shepp DH, Match ME, Lipson SM, Pergolizzi RG. A fifth human cytomegalovirus glycoprotein B genotype. Res Virol. 1998;149:109–14. doi: 10.1016/S0923-2516(98)80086-1. [DOI] [PubMed] [Google Scholar]
- 69.Wu X, Wang Y, Xu Y, Wu D, Sun A, Zhu Z, et al. Cytomegalovirus glycoprotein B genotype in hematopoietic stem cell transplant patients from China. Biol Blood Marrow Transplant. 2010;16:647–52. doi: 10.1016/j.bbmt.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 70.Bergallo M, Costa C, Gambarino S, Tornicelli A, Astegiano S, Terlizzi ME, et al. Human cytomegalovirus glycoprotein B genotyping from bronchoalveolar lavage specimens. Can J Microbiol. 2011;57:273–7. doi: 10.1139/w11-014. [DOI] [PubMed] [Google Scholar]
- 71.Görzer I, Kerschner H, Jaksch P, Bauer C, Seebacher G, Klepetko W, et al. Virus load dynamics of individual CMV-genotypes in lung transplant recipients with mixed-genotype infections. J Med Virol. 2008;80:1405–14. doi: 10.1002/jmv.21225. [DOI] [PubMed] [Google Scholar]
- 72.Kashiwagi Y, Kawashima H, Matsuura K, Sasamoto M, Takekuma K, Hoshika A, et al. Clinical characteristics and gB genotype of cytomegalovirus infection in Japan. In Vivo. 2002;16:447–50. [PubMed] [Google Scholar]
- 73.Jung GS, Kim YY, Kim JI, Ji GY, Jeon JS, Yoon HW, et al. Full genome sequencing and analysis of human cytomegalovirus strain JHC isolated from a Korean patient. Virus Res. 2011;156:113–20. doi: 10.1016/j.virusres.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 74.Simpson JA, Chow JC, Baker J, Avdalovic N, Yuan S, Au D, et al. Neutralizing monoclonal antibodies that distinguish three antigenic sites on human cytomegalovirus glycoprotein H have conformationally distinct binding sites. J Virol. 1993;67:489–96. doi: 10.1128/jvi.67.1.489-496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pignatelli S, Dal Monte P, Rossini G, Lazzarotto T, Gatto MR, Landini MP. Intrauterine cytomegalovirus infection and glycoprotein N (gN) genotypes. J Clin Virol. 2003;28:38–43. doi: 10.1016/S1386-6532(02)00236-6. [DOI] [PubMed] [Google Scholar]
- 76.Huber MT, Compton T. Characterization of a novel third member of the human cytomegalovirus glycoprotein H-glycoprotein L complex. J Virol. 1997;71:5391–8. doi: 10.1128/jvi.71.7.5391-5398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li L, Nelson JA, Britt WJ. Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J Virol. 1997;71:3090–7. doi: 10.1128/jvi.71.4.3090-3097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ryckman BJ, Rainish BL, Chase MC, Borton JA, Nelson JA, Jarvis MA, et al. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J Virol. 2008;82:60–70. doi: 10.1128/JVI.01910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci USA. 2005;102:18153–8. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Macagno A, Bernasconi NL, Vanzetta F, Dander E, Sarasini A, Revello MG, et al. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol. 2010;84:1005–13. doi: 10.1128/JVI.01809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, et al. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol. 2004;78:10023–33. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McLaughlin-Taylor E, Pande H, Forman SJ, Tanamachi B, Li CR, Zaia JA, et al. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J Med Virol. 1994;43:103–10. doi: 10.1002/jmv.1890430119. [DOI] [PubMed] [Google Scholar]
- 83.Wills MR, Carmichael AJ, Mynard K, Jin X, Weekes MP, Plachter B, et al. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569–79. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruckwardt TJ, Luongo C, Malloy AM, Liu J, Chen M, Collins PL, et al. Responses against a subdominant CD8+ T cell epitope protect against immunopathology caused by a dominant epitope. J Immunol. 2010;185:4673–80. doi: 10.4049/jimmunol.1001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van der Most RG, Concepcion RJ, Oseroff C, Alexander J, Southwood S, Sidney J, et al. Uncovering subdominant cytotoxic T-lymphocyte responses in lymphocytic choriomeningitis virus-infected BALB/c mice. J Virol. 1997;71:5110–4. doi: 10.1128/jvi.71.7.5110-5114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ebert S, Lemmermann NA, Thomas D, Renzaho A, Reddehase MJ, Holtappels R. Immune control in the absence of immunodominant epitopes: implications for immunotherapy of cytomegalovirus infection with antiviral CD8 T cells. Med Microbiol Immunol. 2012 doi: 10.1007/s00430-012-0268-8. [DOI] [PubMed] [Google Scholar]
- 87.Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat Rev Genet. 2008;9:776–88. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kublin J. Welcome to the HVTNews. HVTNEWS: HIV Vaccine Trials Network, 2011:1.
- 89.Yan H, Koyano S, Inami Y, Yamamoto Y, Suzutani T, Mizuguchi M, et al. Genetic variations in the gB, UL144 and UL149 genes of human cytomegalovirus strains collected from congenitally and postnatally infected Japanese children. Arch Virol. 2008;153:667–74. doi: 10.1007/s00705-008-0044-7. [DOI] [PubMed] [Google Scholar]
- 90.Kee ST, Gehl J. W. LE. Clinical Aspects of Electroporation. Springer, New York, NY, 2011. [Google Scholar]
- 91.Selinsky C, Luke C, Wloch M, Geall A, Hermanson G, Kaslow D, et al. A DNA-based vaccine for the prevention of human cytomegalovirus-associated diseases. Hum Vaccin. 2005;1:16–23. doi: 10.4161/hv.1.1.1335. [DOI] [PubMed] [Google Scholar]
- 92.Khan AS, Pope MA, Draghia-Akli R. Highly efficient constant-current electroporation increases in vivo plasmid expression. DNA Cell Biol. 2005;24:810–8. doi: 10.1089/dna.2005.24.810. [DOI] [PubMed] [Google Scholar]
- 93.Shedlock DJ, Talbott KT, Cress C, Ferraro B, Tuyishme S, Mallilankaraman K, et al. A highly optimized DNA vaccine confers complete protective immunity against high-dose lethal lymphocytic choriomeningitis virus challenge. Vaccine. 2011;29:6755–62. doi: 10.1016/j.vaccine.2010.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kuzushima K, Hayashi N, Kimura H, Tsurumi T. Efficient identification of HLA-A*2402-restricted cytomegalovirus-specific CD8(+) T-cell epitopes by a computer algorithm and an enzyme-linked immunospot assay. Blood. 2001;98:1872–81. doi: 10.1182/blood.V98.6.1872. [DOI] [PubMed] [Google Scholar]
- 95.Liu YN, Curtsinger J, Donahue PR, Klaus A, Optiz G, Cooper J, et al. Molecular analysis of the immune response to human cytomegalovirus glycoprotein B. I. Mapping of HLA-restricted helper T cell epitopes on gp93. J Gen Virol. 1993;74:2207–14. doi: 10.1099/0022-1317-74-10-2207. [DOI] [PubMed] [Google Scholar]
- 96.Nastke MD, Herrgen L, Walter S, Wernet D, Rammensee HG, Stevanović S. Major contribution of codominant CD8 and CD4 T cell epitopes to the human cytomegalovirus-specific T cell repertoire. Cell Mol Life Sci. 2005;62:77–86. doi: 10.1007/s00018-004-4363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lim JB, Provenzano M, Kwon OH, Bettinotti M, Caruccio L, Nagorsen D, et al. Identification of HLA-A33-restricted CMV pp65 epitopes as common targets for CD8(+) CMV-specific cytotoxic T lymphocytes. Exp Hematol. 2006;34:296–307. doi: 10.1016/j.exphem.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 98.Beninga J, Kalbacher H, Mach M. Analysis of T helper cell response to glycoprotein H (gpUL75) of human cytomegalovirus: evidence for strain-specific T cell determinants. J Infect Dis. 1996;173:1051–61. doi: 10.1093/infdis/173.5.1051. [DOI] [PubMed] [Google Scholar]
- 99.Elkington R, Walker S, Crough T, Menzies M, Tellam J, Bharadwaj M, et al. Ex vivo profiling of CD8+-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J Virol. 2003;77:5226–40. doi: 10.1128/JVI.77.9.5226-5240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]