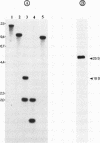
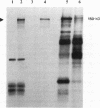
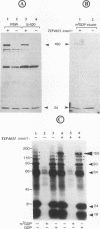
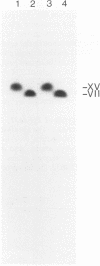
Abstract
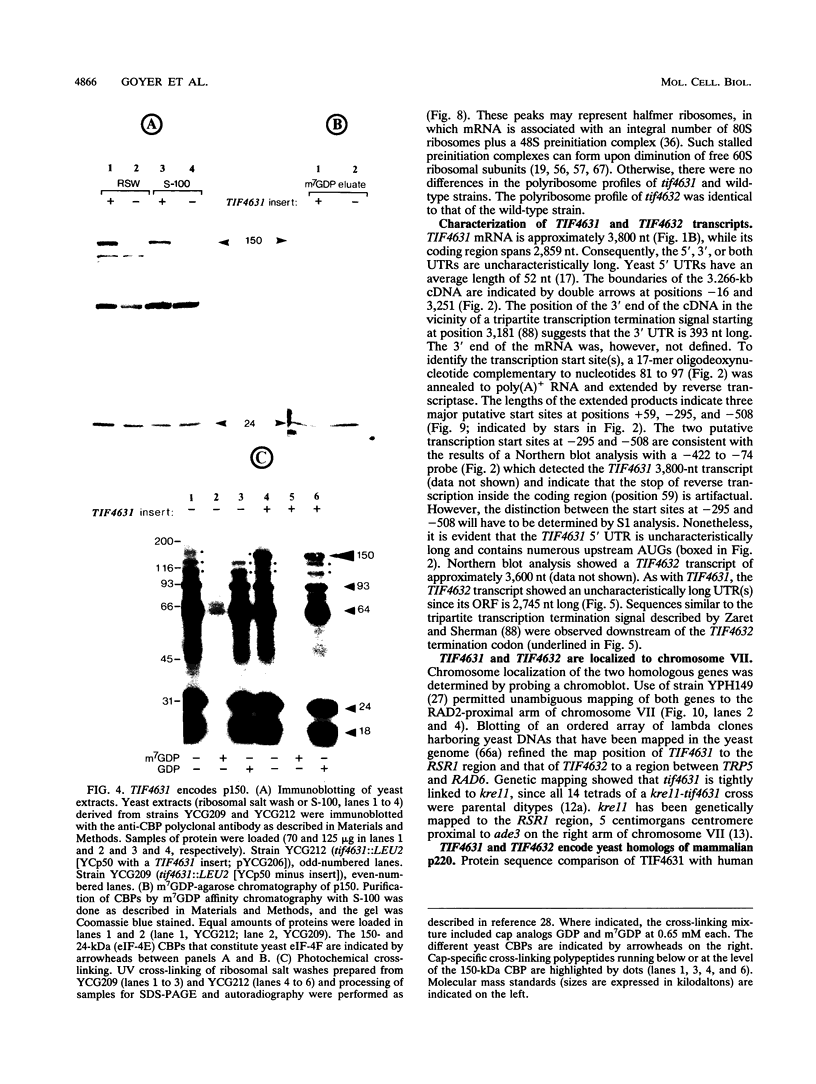
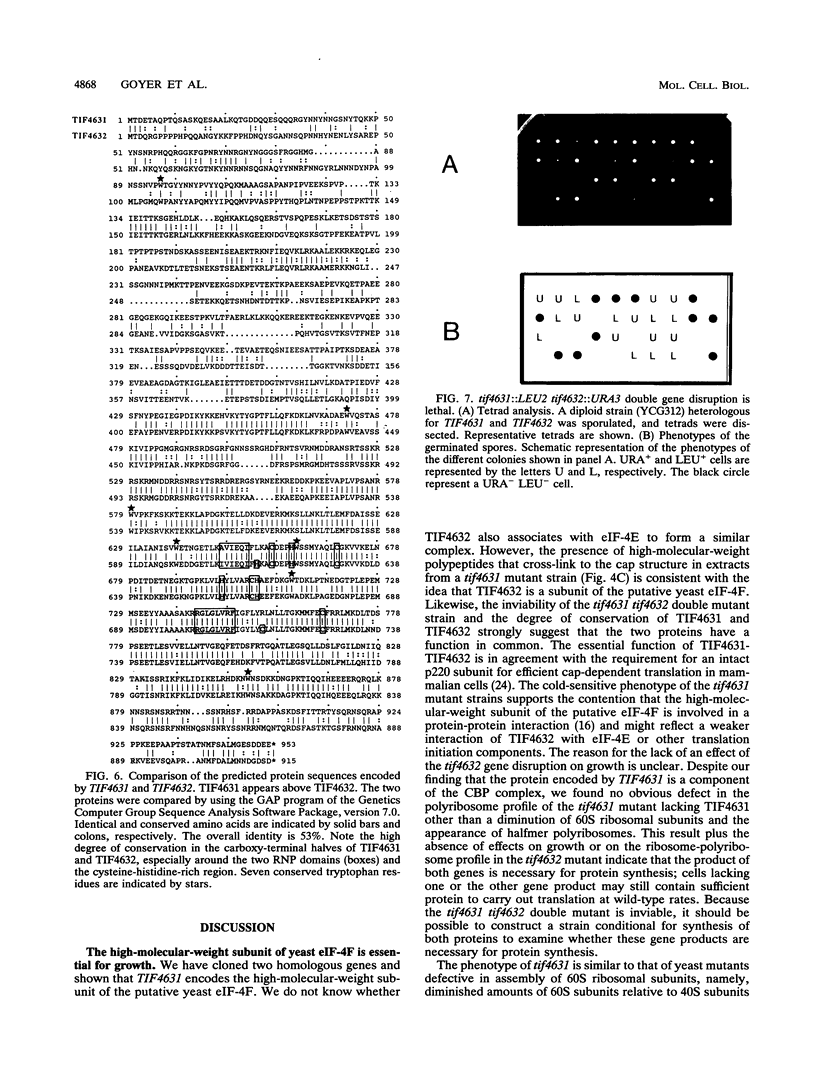
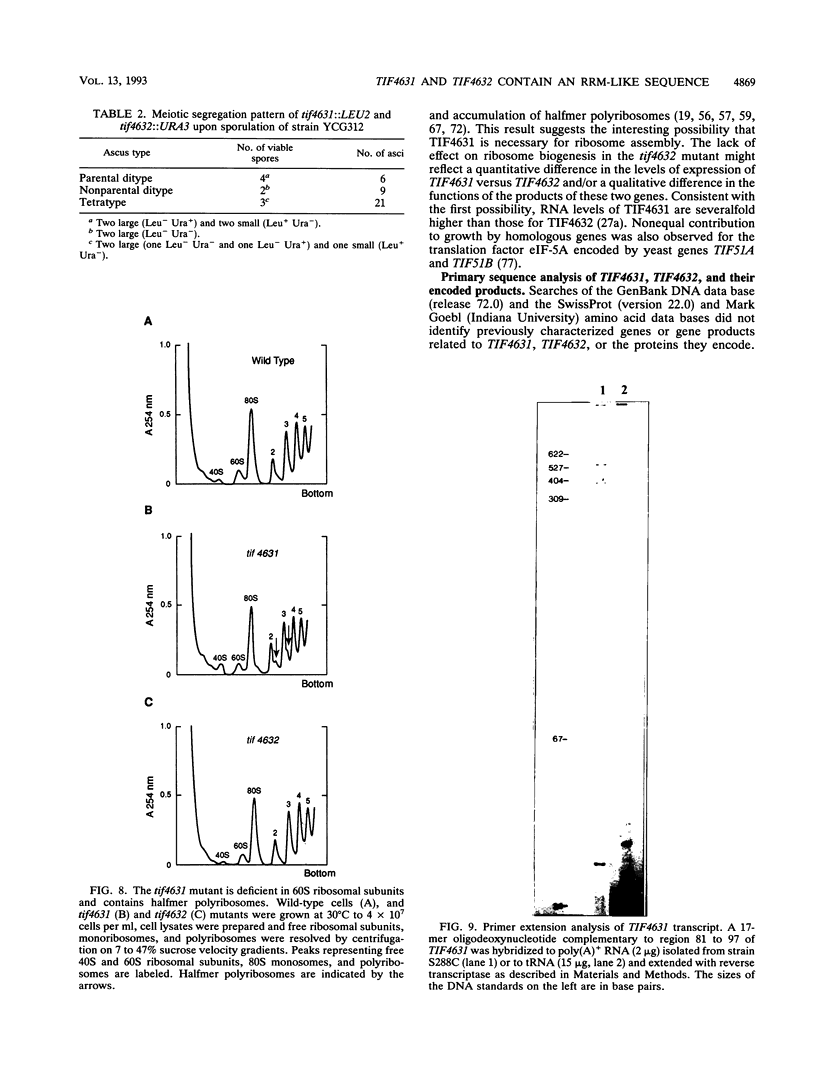
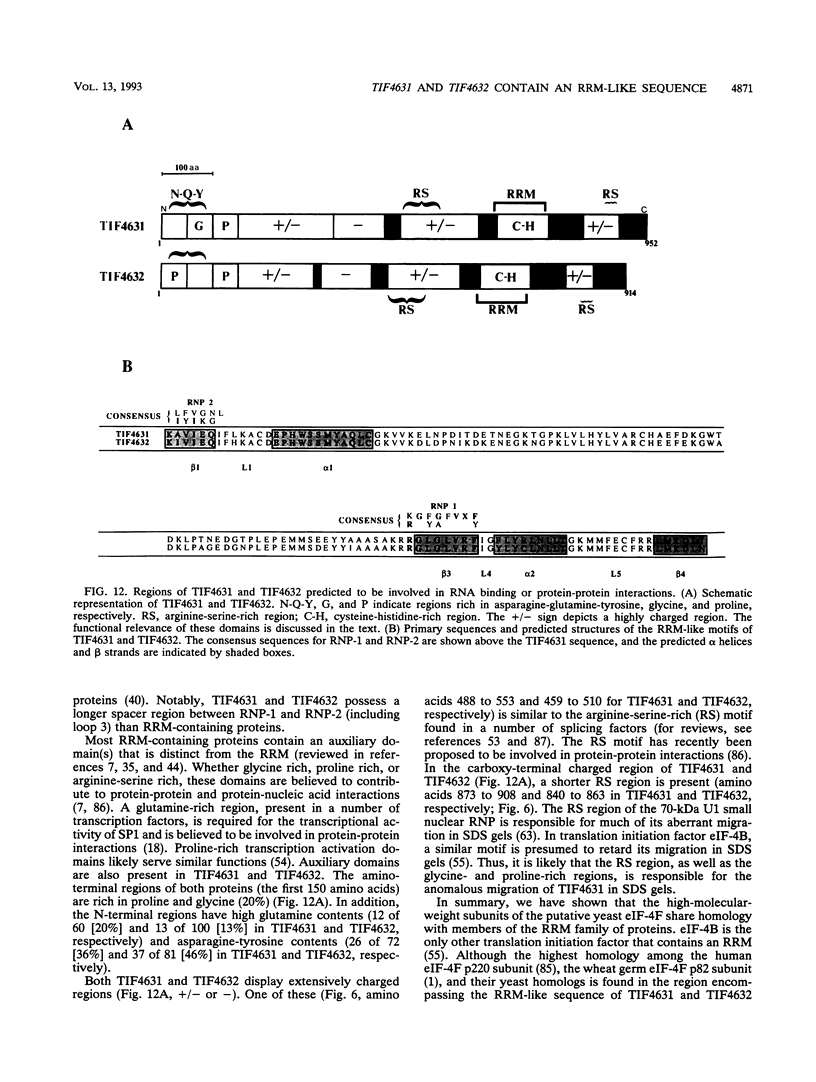
The 5' ends of eukaryotic mRNAs are blocked by a cap structure, m7GpppX (where X is any nucleotide). The interaction of the cap structure with a cap-binding protein complex is required for efficient ribosome binding to the mRNA. In Saccharomyces cerevisiae, the cap-binding protein complex is a heterodimer composed of two subunits with molecular masses of 24 (eIF-4E, CDC33) and 150 (p150) kDa. p150 is presumed to be the yeast homolog of the p220 component of mammalian eIF-4F. In this report, we describe the isolation of yeast gene TIF4631, which encodes p150, and a closely related gene, TIF4632. TIF4631 and TIF4632 are 53% identical overall and 80% identical over a 320-amino-acid stretch in their carboxy-terminal halves. Both proteins contain sequences resembling the RNA recognition motif and auxiliary domains that are characteristic of a large family of RNA-binding proteins. tif4631-disrupted strains exhibited a slow-growth, cold-sensitive phenotype, while disruption of TIF4632 failed to show any phenotype under the conditions assayed. Double gene disruption engendered lethality, suggesting that the two genes are functionally homologous and demonstrating that at least one of them is essential for viability. These data are consistent with a critical role for the high-molecular-weight subunit of putative yeast eIF-4F in translation. Sequence comparison of TIF4631, TIF4632, and the human eIF-4F p220 subunit revealed significant stretches of homology. We have thus cloned two yeast homologs of mammalian p220.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen M. L., Metz A. M., Timmer R. T., Rhoads R. E., Browning K. S. Isolation and sequence of the cDNAs encoding the subunits of the isozyme form of wheat protein synthesis initiation factor 4F. J Biol Chem. 1992 Nov 15;267(32):23232–23236. [PubMed] [Google Scholar]
- Altmann M., Blum S., Pelletier J., Sonenberg N., Wilson T. M., Trachsel H. Translation initiation factor-dependent extracts from Saccharomyces cerevisiae. Biochim Biophys Acta. 1990 Aug 27;1050(1-3):155–159. doi: 10.1016/0167-4781(90)90158-x. [DOI] [PubMed] [Google Scholar]
- Altmann M., Edery I., Sonenberg N., Trachsel H. Purification and characterization of protein synthesis initiation factor eIF-4E from the yeast Saccharomyces cerevisiae. Biochemistry. 1985 Oct 22;24(22):6085–6089. doi: 10.1021/bi00343a009. [DOI] [PubMed] [Google Scholar]
- Altmann M., Handschin C., Trachsel H. mRNA cap-binding protein: cloning of the gene encoding protein synthesis initiation factor eIF-4E from Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):998–1003. doi: 10.1128/mcb.7.3.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M., Krieger M., Trachsel H. Nucleotide sequence of the gene encoding a 20 kDa protein associated with the cap binding protein eIF-4E from Saccharomyces cerevisiae. Nucleic Acids Res. 1989 Sep 25;17(18):7520–7520. doi: 10.1093/nar/17.18.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M., Müller P. P., Pelletier J., Sonenberg N., Trachsel H. A mammalian translation initiation factor can substitute for its yeast homologue in vivo. J Biol Chem. 1989 Jul 25;264(21):12145–12147. [PubMed] [Google Scholar]
- Bandziulis R. J., Swanson M. S., Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989 Apr;3(4):431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Beauchemin N., Benchimol S., Cournoyer D., Fuks A., Stanners C. P. Isolation and characterization of full-length functional cDNA clones for human carcinoembryonic antigen. Mol Cell Biol. 1987 Sep;7(9):3221–3230. doi: 10.1128/mcb.7.9.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein H. D., Sonenberg N., Baltimore D. Poliovirus mutant that does not selectively inhibit host cell protein synthesis. Mol Cell Biol. 1985 Nov;5(11):2913–2923. doi: 10.1128/mcb.5.11.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc A., Goyer C., Sonenberg N. The coat protein of the yeast double-stranded RNA virus L-A attaches covalently to the cap structure of eukaryotic mRNA. Mol Cell Biol. 1992 Aug;12(8):3390–3398. doi: 10.1128/mcb.12.8.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C., Nakayama N., Goebl M., Tanaka K., Toh-e A., Matsumoto K. CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Aug;8(8):3556–3559. doi: 10.1128/mcb.8.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Browning K. S., Lax S. R., Ravel J. M. Identification of two messenger RNA cap binding proteins in wheat germ. Evidence that the 28-kDa subunit of eIF-4B and the 26-kDa subunit of eIF-4F are antigenically distinct polypeptides. J Biol Chem. 1987 Aug 15;262(23):11228–11232. [PubMed] [Google Scholar]
- Browning K. S., Webster C., Roberts J. K., Ravel J. M. Identification of an isozyme form of protein synthesis initiation factor 4F in plants. J Biol Chem. 1992 May 15;267(14):10096–10100. [PubMed] [Google Scholar]
- Cigan A. M., Donahue T. F. Sequence and structural features associated with translational initiator regions in yeast--a review. Gene. 1987;59(1):1–18. doi: 10.1016/0378-1119(87)90261-7. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Deshmukh M., Tsay Y. F., Paulovich A. G., Woolford J. L., Jr Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol. 1993 May;13(5):2835–2845. doi: 10.1128/mcb.13.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I., Hümbelin M., Darveau A., Lee K. A., Milburn S., Hershey J. W., Trachsel H., Sonenberg N. Involvement of eukaryotic initiation factor 4A in the cap recognition process. J Biol Chem. 1983 Sep 25;258(18):11398–11403. [PubMed] [Google Scholar]
- Edery I., Lee K. A., Sonenberg N. Functional characterization of eukaryotic mRNA cap binding protein complex: effects on translation of capped and naturally uncapped RNAs. Biochemistry. 1984 May 22;23(11):2456–2462. doi: 10.1021/bi00306a021. [DOI] [PubMed] [Google Scholar]
- Edery I., Sonenberg N. Cap-dependent RNA splicing in a HeLa nuclear extract. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7590–7594. doi: 10.1073/pnas.82.22.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchison D., Hansen J., Ehrenfeld E., Edery I., Sonenberg N., Milburn S., Hershey J. W. Demonstration in vitro that eucaryotic initiation factor 3 is active but that a cap-binding protein complex is inactive in poliovirus-infected HeLa cells. J Virol. 1984 Sep;51(3):832–837. doi: 10.1128/jvi.51.3.832-837.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y., LaFiandra A., Shatkin A. J. 5'-Terminal structure and mRNA stability. Nature. 1977 Mar 17;266(5599):235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- Georgiev O., Mous J., Birnstiel M. L. Processing and nucleo-cytoplasmic transport of histone gene transcripts. Nucleic Acids Res. 1984 Nov 26;12(22):8539–8551. doi: 10.1093/nar/12.22.8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerring S. L., Connelly C., Hieter P. Positional mapping of genes by chromosome blotting and chromosome fragmentation. Methods Enzymol. 1991;194:57–77. doi: 10.1016/0076-6879(91)94007-y. [DOI] [PubMed] [Google Scholar]
- Goyer C., Altmann M., Trachsel H., Sonenberg N. Identification and characterization of cap-binding proteins from yeast. J Biol Chem. 1989 May 5;264(13):7603–7610. [PubMed] [Google Scholar]
- Goyer C., Lee H. S., Malo D., Sonenberg N. Isolation of a yeast gene encoding a protein homologous to the human Tat-binding protein TBP-1. DNA Cell Biol. 1992 Oct;11(8):579–585. doi: 10.1089/dna.1992.11.579. [DOI] [PubMed] [Google Scholar]
- Green M. R., Maniatis T., Melton D. A. Human beta-globin pre-mRNA synthesized in vitro is accurately spliced in Xenopus oocyte nuclei. Cell. 1983 Mar;32(3):681–694. doi: 10.1016/0092-8674(83)90054-5. [DOI] [PubMed] [Google Scholar]
- Grifo J. A., Tahara S. M., Morgan M. A., Shatkin A. J., Merrick W. C. New initiation factor activity required for globin mRNA translation. J Biol Chem. 1983 May 10;258(9):5804–5810. [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J., Mattaj I. W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990 Oct 5;63(1):109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- Hart R. P., McDevitt M. A., Nevins J. R. Poly(A) site cleavage in a HeLa nuclear extract is dependent on downstream sequences. Cell. 1985 Dec;43(3 Pt 2):677–683. doi: 10.1016/0092-8674(85)90240-5. [DOI] [PubMed] [Google Scholar]
- Haynes S. R. The RNP motif protein family. New Biol. 1992 May;4(5):421–429. [PubMed] [Google Scholar]
- Helser T. L., Baan R. A., Dahlberg A. E. Characterization of a 40S ribosomal subunit complex in polyribosomes of Saccharomyces cerevisiae treated with cycloheximide. Mol Cell Biol. 1981 Jan;1(1):51–57. doi: 10.1128/mcb.1.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey J. W. Translational control in mammalian cells. Annu Rev Biochem. 1991;60:717–755. doi: 10.1146/annurev.bi.60.070191.003441. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Novel mechanisms of translational control in Saccharomyces cerevisiae. Trends Genet. 1988 Jun;4(6):169–174. doi: 10.1016/0168-9525(88)90023-6. [DOI] [PubMed] [Google Scholar]
- Hoffman D. W., Query C. C., Golden B. L., White S. W., Keene J. D. RNA-binding domain of the A protein component of the U1 small nuclear ribonucleoprotein analyzed by NMR spectroscopy is structurally similar to ribosomal proteins. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2495–2499. doi: 10.1073/pnas.88.6.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Horikoshi M., Wang C. K., Schroeder S., Weil P. A., Roeder R. G. Cloning of the Schizosaccharomyces pombe TFIID gene reveals a strong conservation of functional domains present in Saccharomyces cerevisiae TFIID. Genes Dev. 1990 Jul;4(7):1141–1148. doi: 10.1101/gad.4.7.1141. [DOI] [PubMed] [Google Scholar]
- Jackson R. J., Howell M. T., Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990 Dec;15(12):477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- Jagus R., Anderson W. F., Safer B. The regulation of initiation of mammalian protein synthesis. Prog Nucleic Acid Res Mol Biol. 1981;25:127–185. doi: 10.1016/s0079-6603(08)60484-5. [DOI] [PubMed] [Google Scholar]
- Jaramillo M., Dever T. E., Merrick W. C., Sonenberg N. RNA unwinding in translation: assembly of helicase complex intermediates comprising eukaryotic initiation factors eIF-4F and eIF-4B. Mol Cell Biol. 1991 Dec;11(12):5992–5997. doi: 10.1128/mcb.11.12.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J. D., Query C. C. Nuclear RNA-binding proteins. Prog Nucleic Acid Res Mol Biol. 1991;41:179–202. doi: 10.1016/s0079-6603(08)60009-4. [DOI] [PubMed] [Google Scholar]
- Kenan D. J., Query C. C., Keene J. D. RNA recognition: towards identifying determinants of specificity. Trends Biochem Sci. 1991 Jun;16(6):214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- Konarska M. M., Padgett R. A., Sharp P. A. Recognition of cap structure in splicing in vitro of mRNA precursors. Cell. 1984 Oct;38(3):731–736. doi: 10.1016/0092-8674(84)90268-x. [DOI] [PubMed] [Google Scholar]
- Lanker S., Müller P. P., Altmann M., Goyer C., Sonenberg N., Trachsel H. Interactions of the eIF-4F subunits in the yeast Saccharomyces cerevisiae. J Biol Chem. 1992 Oct 15;267(29):21167–21171. [PubMed] [Google Scholar]
- Lax S., Fritz W., Browning K., Ravel J. Isolation and characterization of factors from wheat germ that exhibit eukaryotic initiation factor 4B activity and overcome 7-methylguanosine 5'-triphosphate inhibition of polypeptide synthesis. Proc Natl Acad Sci U S A. 1985 Jan;82(2):330–333. doi: 10.1073/pnas.82.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris-Karatzas A., Montine K. S., Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5' cap. Nature. 1990 Jun 7;345(6275):544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- Lee K. A., Edery I., Sonenberg N. Isolation and structural characterization of cap-binding proteins from poliovirus-infected HeLa cells. J Virol. 1985 May;54(2):515–524. doi: 10.1128/jvi.54.2.515-524.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. J., Newman A. J., Cheng S. C., Abelson J. Yeast mRNA splicing in vitro. J Biol Chem. 1985 Nov 25;260(27):14780–14792. [PubMed] [Google Scholar]
- Maroto F. G., Sierra J. M. Purification and characterization of mRNA cap-binding protein from Drosophila melanogaster embryos. Mol Cell Biol. 1989 May;9(5):2181–2190. doi: 10.1128/mcb.9.5.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W. A binding consensus: RNA-protein interactions in splicing, snRNPs, and sex. Cell. 1989 Apr 7;57(1):1–3. doi: 10.1016/0092-8674(89)90164-5. [DOI] [PubMed] [Google Scholar]
- Mermod N., O'Neill E. A., Kelly T. J., Tjian R. The proline-rich transcriptional activator of CTF/NF-I is distinct from the replication and DNA binding domain. Cell. 1989 Aug 25;58(4):741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- Milburn S. C., Hershey J. W., Davies M. V., Kelleher K., Kaufman R. J. Cloning and expression of eukaryotic initiation factor 4B cDNA: sequence determination identifies a common RNA recognition motif. EMBO J. 1990 Sep;9(9):2783–2790. doi: 10.1002/j.1460-2075.1990.tb07466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Paulovich A. G., Tsay Y. F., Woolford J. L., Jr Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J Cell Biol. 1990 Dec;111(6 Pt 1):2261–2274. doi: 10.1083/jcb.111.6.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M., Pulaski B. A., Woolford J. L., Jr Assembly of 60S ribosomal subunits is perturbed in temperature-sensitive yeast mutants defective in ribosomal protein L16. Mol Cell Biol. 1991 Nov;11(11):5681–5692. doi: 10.1128/mcb.11.11.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P. P., Trachsel H. Translation and regulation of translation in the yeast Saccharomyces cerevisiae. Eur J Biochem. 1990 Jul 31;191(2):257–261. doi: 10.1111/j.1432-1033.1990.tb19118.x. [DOI] [PubMed] [Google Scholar]
- Olmsted J. B. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J Biol Chem. 1981 Dec 10;256(23):11955–11957. [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Photochemical cross-linking of cap binding proteins to eucaryotic mRNAs: effect of mRNA 5' secondary structure. Mol Cell Biol. 1985 Nov;5(11):3222–3230. doi: 10.1128/mcb.5.11.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J., Hunter T. p34cdc2: the S and M kinase? New Biol. 1990 May;2(5):389–401. [PubMed] [Google Scholar]
- Query C. C., Bentley R. C., Keene J. D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989 Apr 7;57(1):89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- Ray B. K., Brendler T. G., Adya S., Daniels-McQueen S., Miller J. K., Hershey J. W., Grifo J. A., Merrick W. C., Thach R. E. Role of mRNA competition in regulating translation: further characterization of mRNA discriminatory initiation factors. Proc Natl Acad Sci U S A. 1983 Feb;80(3):663–667. doi: 10.1073/pnas.80.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray B. K., Lawson T. G., Kramer J. C., Cladaras M. H., Grifo J. A., Abramson R. D., Merrick W. C., Thach R. E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J Biol Chem. 1985 Jun 25;260(12):7651–7658. [PubMed] [Google Scholar]
- Ripmaster T. L., Vaughn G. P., Woolford J. L., Jr A putative ATP-dependent RNA helicase involved in Saccharomyces cerevisiae ribosome assembly. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11131–11135. doi: 10.1073/pnas.89.23.11131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D. Isolation of genes by complementation in yeast. Methods Enzymol. 1987;152:481–504. doi: 10.1016/0076-6879(87)52056-0. [DOI] [PubMed] [Google Scholar]
- Rozen F., Edery I., Meerovitch K., Dever T. E., Merrick W. C., Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990 Mar;10(3):1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen F., Sonenberg N. Identification of nuclear cap specific proteins in HeLa cells. Nucleic Acids Res. 1987 Aug 25;15(16):6489–6500. doi: 10.1093/nar/15.16.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A. B., Davis R. W. Translation initiation and ribosomal biogenesis: involvement of a putative rRNA helicase and RPL46. Science. 1990 Mar 2;247(4946):1077–1079. doi: 10.1126/science.2408148. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar G., Edery I., Gallo R., Sonenberg N. Preferential stimulation of rabbit alpha globin mRNA translation by a cap-binding protein complex. Biochim Biophys Acta. 1984 Nov 22;783(2):122–129. doi: 10.1016/0167-4781(84)90003-4. [DOI] [PubMed] [Google Scholar]
- Schnier J., Schwelberger H. G., Smit-McBride Z., Kang H. A., Hershey J. W. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3105–3114. doi: 10.1128/mcb.11.6.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seal S. N., Schmidt A., Marcus A., Edery I., Sonenberg N. A wheat germ cap-site factor functional in protein chain initiation. Arch Biochem Biophys. 1986 May 1;246(2):710–715. doi: 10.1016/0003-9861(86)90327-9. [DOI] [PubMed] [Google Scholar]
- Sherman F., Wakem P. Mapping yeast genes. Methods Enzymol. 1991;194:38–57. doi: 10.1016/0076-6879(91)94006-x. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. Cap-binding proteins of eukaryotic messenger RNA: functions in initiation and control of translation. Prog Nucleic Acid Res Mol Biol. 1988;35:173–207. doi: 10.1016/s0079-6603(08)60614-5. [DOI] [PubMed] [Google Scholar]
- Sonenberg N., Morgan M. A., Merrick W. C., Shatkin A. J. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5'-terminal cap in mRNA. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4843–4847. doi: 10.1073/pnas.75.10.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N. Picornavirus RNA translation continues to surprise. Trends Genet. 1991 Apr;7(4):105–106. doi: 10.1016/0168-9525(91)90440-2. [DOI] [PubMed] [Google Scholar]
- Sonenberg N. Regulation of translation by poliovirus. Adv Virus Res. 1987;33:175–204. doi: 10.1016/s0065-3527(08)60318-8. [DOI] [PubMed] [Google Scholar]
- Tahara S. M., Morgan M. A., Shatkin A. J. Two forms of purified m7G-cap binding protein with different effects on capped mRNA translation in extracts of uninfected and poliovirus-infected HeLa cells. J Biol Chem. 1981 Aug 10;256(15):7691–7694. [PubMed] [Google Scholar]
- Yan R., Rychlik W., Etchison D., Rhoads R. E. Amino acid sequence of the human protein synthesis initiation factor eIF-4 gamma. J Biol Chem. 1992 Nov 15;267(32):23226–23231. [PubMed] [Google Scholar]
- Zamore P. D., Patton J. G., Green M. R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992 Feb 13;355(6361):609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Zapp M. L., Green M. R. Gene expression. RNA binding: beta s and basics. Nature. 1990 Dec 6;348(6301):485–486. doi: 10.1038/348485a0. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]