Abstract
Double-strand RNA-mediated interference (RNAi) is an effective strategy to knock down target gene expression1-3. It has been applied to many model systems including plants, invertebrates and vertebrates. There are various methods to achieve RNAi in vivo4,5. For example, the target gene may be transformed into an RNAi vector, and then either permanently or transiently transformed into cell lines or primary cells to achieve gene knockdown effects; alternatively synthesized double-strand oligonucleotides from specific target genes (RNAi oligos) may be transiently transformed into cell lines or primary cells to silence target genes; or synthesized double-strand RNA molecules may be microinjected into an organism. Since the nematode C. elegans uses bacteria as a food source, feeding the animals with bacteria expressing double-strand RNA against target genes provides a viable strategy6. Here we present an RNAi feeding method to score body size phenotype. Body size in C. elegans is regulated primarily by the TGF- β - like ligand DBL-1, so this assay is appropriate for identification of TGF-β signaling components7. We used different strains including two RNAi hypersensitive strains to repeat the RNAi feeding experiments. Our results showed that rrf-3 strain gave us the best expected RNAi phenotype. The method is easy to perform, reproducible, and easily quantified. Furthermore, our protocol minimizes the use of specialized equipment, so it is suitable for smaller laboratories or those at predominantly undergraduate institutions.
Keywords: Developmental Biology, Issue 72, Genetics, Cellular Biology, Molecular Biology, Biochemistry, Basic Protocols, RNAi feeding technique, genetic screen, TGF-beta, body size, C. elegans, Caenorhabditis elegans, RNA-mediated Interference, RNAi, RNA, DNA, gene expression knock down, animal model
Protocol
1. Preparing RNAi Feeding Plates Carrying Target Gene Sequence
If using commercially available RNAi libraries (e.g. from Source BioScience LifeSciences), proceed to step 1.4. Alternatively, clone target gene sequences into vector L4440, a commonly used worm RNAi plasmid8, by standard cloning protocol.
Transform the recombinant plasmid into bacterial strain HT115(DE3), culture them on LB agar plates with 25 μg/ml carbenicillin and 12.5 μg/ml tetracycline, and pick a single clone for future use.
Meanwhile, transform the empty vector L4440 into bacterial strain HT115 to use as a control for the experiments.
Culture both recombinant and empty L4440 -carrying bacteria in 1 ml LB broth with 100 μg/ml ampicillin overnight at 37 °C. Tetracycline is not added due to the fact that it decreases RNAi efficiency.
Add another 5 ml LB broth with 100 μg/ml ampicillin into the overnight culture, incubate for another 4-6 hr at 37 °C.
Seed 0.5 ml recombinant or empty L4440 -carrying bacterial culture onto the RNAi worm plates. Label all plates with clone name.
Mark the plates and incubate overnight at 37 °C to grow bacterial lawn; these are the RNAi plates with or without target gene sequence. At least 2 plates should be prepared for each condition.
2. Culture Worms on the RNAi Feeding Plates
Flame sterilize the tip of a platinum wire worm pick. Use the worm pick to pick 6-10 fourth larval stage (L4) hermaphrodites to each RNAi plate that contains either recombinant or empty L4440; the animals will use the bacteria as a food source.
Let hermaphrodites grow at 20 °C (a standard culture condition for C. elegans) overnight, they will become young adults the next day.
Transfer 6-10 young adults into a new correspondingly labelled and prepared RNAi plate.
Let the adults lay eggs for 4-6 hr, then remove all the adults from the plate; this step is to synchronize the progeny. Once the mothers are removed, start to count time. This is time zero.
Incubate the plates at 20 °C to let animals grow to specific developmental stage; in this protocol, we choose young adults for phenotypic analysis. For knockdowns which develop at a normal rate, 72 hr incubation is sufficient for animals to become young adults. However, various genes affect animal development differently. If RNAi causes animals to grow at a different rate, young adults can be identified as those no more than 24 hr past L4 with completed vulval development and 2-6 embryos in the uterus (if fertile). To identify other developmental stages, the investigator should use gonadal and vulval development as a guide.
3. Score Body Size Phenotypes of RNAi Treated Worms
Place two layers of colored label tapes on a glass slide. Make two of these glass slides. Then, place a new glass slide in between the two slides with tape. Melt 2% agarose in water; apply one drop to the center of the new glass slide; then press the second glass slide on the top to make a thin layer of 2% agarose as described previously9.
Once agarose is solidified, remove the top glass slide; the slide with agarose pad is ready to load worms. Label slide with clone name.
Add 10 μl 25 mM NaN3 to the agarose pad; pick 30-40 animals into the NaN3 solution to immobilize them; then put coverslip on the top; repeat for both recombinant and empty L4440-carrying RNAi-treated animals.
Image the worms under dissecting microscope using 2.5x objective lens; here we use Leica digital camera with supporting software Qcapture.
For calibration, first capture an image of a standard micrometer ruler. Then open the image in Image-Pro software, Click on "measure" and choose "calibration" then choose "spatial calibration wizard". With "calibrate the active image" selected, click on "next". Input the name for the calibration and ensure that the spatial reference units are set to micrometer. Then, check the "create a reference calibration" button and click "next". Click on "draw reference line." A reference scale bar will appear. Reposition the scale bar to match the ends of the micrometer, then indicate that the reference indicates 1,000 units. Click "OK", "Next", and "Finish".
Once the software is calibrated, open a worm image. Then, select under the "measure" menu, select "calibration" then click on "select spatial". In the pull down menu, select the file name created for the micrometer calibration. Then, under the "measure" menu, select "measurements." In the window that opens, select the free-draw tool and trace a line through the center of the animal body from head to tail with computer mouse (Figure 1). The length will be reported in the window. Now you have two groups of data: body length of animals treated with target gene RNAi and control animals treated with empty L4440 vector.
Export the length measurements to a Microsoft Excel file, or other suitable statistical software. Analyze the data by calculating the mean and standard deviation for each sample. Then compare samples using Student's t-test.
Representative Results
In our research, we focus on body size regulation by the DBL-1/TGF-β pathway. The loss of function of the DBL-1 pathway results in small body size, including a shorter body length compared with wild-type animals7,10,11. Thus, screens for C. elegans body size mutants are capable of identifying TGF-β signaling components and modifiers. DBL-1 signaling is mediated by a conserved TGF-β signal transduction pathway that includes cell surface receptors and intracellular Smad signal transducers12. To test the effectiveness of RNAi by feeding for the identification of body size mutants, we used this technique to knock down DBL-1 pathway components: dbl-1/ligand; sma-2, sma-3, sma-4/Smads; and sma-6/receptor. For our study, we used two different RNAi hypersensitive C. elegans strains to perform the experiment: eri-1;lin-15b and rrf-313,14. Meanwhile, we also used N2 (standard wild-type) strain and lin-36, a component in the lin-15 pathway whose RNAi sensitivity is nevertheless similar to N213. Our results (Figure 2) show that all of these strains displayed the short body size phenotype as expected (p<0.001), except sma-6 RNAi in lin-36 background. In rrf-3 background, body lengths of young adults after RNAi treatment were 84~95% of vector alone. In eri-1;lin-15b background, body lengths of young adults after RNAi were 68~86% of control animals. Compared with lin-36 and N2 strains, both of the RNAi hypersensitive strains rrf-3 and eri-1;lin-15b were more sensitive.
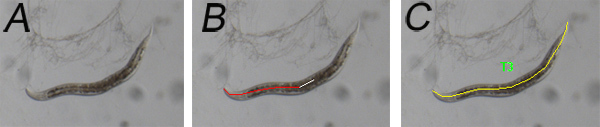 Figure 1. Representative images of animal body length being measured.A. animal image before measurement; B. the center of animal body was traced during the measurement; C. a tracing line along the animal body from head to tail, is measured by software.
Figure 1. Representative images of animal body length being measured.A. animal image before measurement; B. the center of animal body was traced during the measurement; C. a tracing line along the animal body from head to tail, is measured by software.
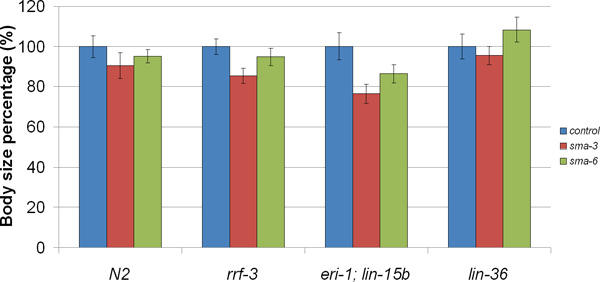 Figure 2. Body length of animals in which DBL-1 pathway components have been inactivated by RNAi feeding method. RNAi animals in all of these background strains displayed the short body length phenotype as expected (p<0.001), except sma-6 RNAi in lin-36 background. RNAi of rrf-3 and eri-1;lin-15b strains demonstrated a stronger phenotype than that of lin-36 and N2 backgrounds.
Figure 2. Body length of animals in which DBL-1 pathway components have been inactivated by RNAi feeding method. RNAi animals in all of these background strains displayed the short body length phenotype as expected (p<0.001), except sma-6 RNAi in lin-36 background. RNAi of rrf-3 and eri-1;lin-15b strains demonstrated a stronger phenotype than that of lin-36 and N2 backgrounds.
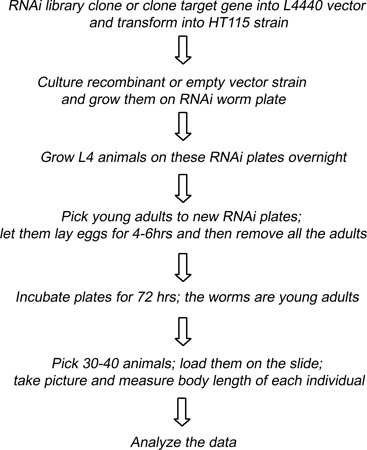 Figure 3. Schematic description of using RNAi feeding strategy to screen for candidate genes.
Figure 3. Schematic description of using RNAi feeding strategy to screen for candidate genes.
Discussion
In this protocol, we describe our method for the identification of body size defective mutants of C. elegans by RNAi feeding. This method is applicable to the identification of TGF-β signaling components. Since such components are highly conserved through evolution15, such screens are relevant to elucidating the molecular mechanisms of TGF-β signaling in all metazoans. An important consideration in designing such a screen is the starting strain. We have demonstrated that both eri-1;lin-15b and rrf-3 are more sensitive to RNAi feeding than the control strains, which is consistent with previous studies12,13. However, even though eri-l;lin-15b demonstrated the strongest phenotype, the animals did not grow well and produced few progeny. Thus, in a large-scale screen, we would not have enough animals for scoring phenotypes from this strain. As a result, we favor the rrf-3 strain to apply the RNAi feeding technique to body size regulation. A second choice to consider in the starting strain is whether to initiate a screen with the DBL-1 pathway intact or to begin with a sensitized strain in which the DBL-1 pathway is either compromised or overactive. These alternative starting points are likely to lead to the identification of overlapping but distinct sets of signaling components. Our protocol can also be modified to the study of other postembryonic phenotypes. In this case, the stage of the animals to be scored for the phenotype of interest may need to be modified by altering the duration of growth in step 2.5. Prior to initiating a large scale screen for other phenotypes, we would recommend a pilot screen with genes known to produce the phenotype of interest such as we describe here for dbl-1, sma-2, sma-3 sma-4, and sma-6.
By using this protocol, knocking down DBL-1 pathway components showed expected body length phenotype. Those RNAi animals were about 68-95% in length compared with control animals. In previous studies, DBL-1 pathway loss of function genetic mutants in adulthood are about 50% shorter than wild-type animals7,10,11. Therefore, the RNAi feeding technique presented here did not fully eliminate gene activity. Thus, the method may be limited in its ability to identify pathway components that have a weak phenotype. Meanwhile, since there are many genes that regulate body length, genes identified from the screen also might not necessarily fall in DBL-1 pathway. Further experiments should be performed to place candidates in pathways, as is true for any other screen method.
In summary, RNAi feeding screens in C. elegans have proven useful in identifying genes involved in a process of interest. In this protocol, we have optimized existing protocols to be highly effective in the identification of body size defects. The optimized protocol can be further modified for the study of other postembryonic phenotypes.
Disclosures
No conflicts of interest declared.
Acknowledgments
We thank James Clark for providing figures of animals at various developmental time points. C. elegans strains in this study were obtained from the Caenorhabditis Genetics Center, which is supported by the NIH National Center for Research Resources (NCRR). This work was supported by CIRG 1817 from CUNY to J.L. and C.S.D.; and by NIH 1R15GM073678-01 and 1R15GM097692-01 to C.S.D. We thank Dr. William J Rice, Dr. Nathalia Holtzman, and Melissa Silvestrini for comments on the manuscript. This work was carried out in partial fulfillment of the requirements for a PhD degree from the Graduate Center of the City University of New York (S. Xiong).
References
- Melnyk CW, Molnar A, Baulcombe DC. Intercellular and systemic movement of RNA silencing signals. Embo J. 2011;30:3553–3563. doi: 10.1038/emboj.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, Shi Y. Small RNA: can RNA interference be exploited for therapy. Lancet. 2003;362:1401–1403. doi: 10.1016/S0140-6736(03)14637-5. [DOI] [PubMed] [Google Scholar]
- Kalantidis K, Schumacher HT, Alexiadis T, Helm JM. RNA silencing movement in plants. Biol. Cell. 2008;100:13–26. doi: 10.1042/BC20070079. [DOI] [PubMed] [Google Scholar]
- Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA in vivo. Febs. J. 2010;277:4814–4827. doi: 10.1111/j.1742-4658.2010.07904.x. [DOI] [PubMed] [Google Scholar]
- Amarzguioui M, Rossi JJ, Kim D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett. 2005;579:5974–5981. doi: 10.1016/j.febslet.2005.08.070. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Savage-Dunn C, et al. Genetic screen for small body size mutants in C. elegans reveals many TGFb pathway components. Genesis. 2003;35:239–247. doi: 10.1002/gene.10184. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Wang J, Tokarz R, Savage-Dunn C. The expression of TGFβ signal transducers in the hypodermis regulates body size in C. elegans. Development. 2002;129:4989–4998. doi: 10.1242/dev.129.21.4989. [DOI] [PubMed] [Google Scholar]
- Liang J, et al. The Caenorhabditis elegans schnurri homolog sma-9 mediates stage- and cell type-specific responses to DBL-1 BMP-related signaling. Development. 2003;130:6453–6464. doi: 10.1242/dev.00863. [DOI] [PubMed] [Google Scholar]
- Savage-Dunn C. TGF-β signaling. WormBook. 2005. pp. 1–12. [DOI] [PMC free article] [PubMed]
- Simmer F, et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Wang D, et al. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- De Robertis EM. Evo-devo: variations on ancestral themes. Cell. 2008;132:185–195. doi: 10.1016/j.cell.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]


