Abstract
Quantitative analysis of single molecule experiments show that adding either of two natural polyamines, spermine or spermidine, produced more compact plectonemes in DNA in physiological concentrations of monovalent salt. They also promoted plectoneme formation at lower values of torsion in measurements of extension versus twist. Quantifying changes in the plectonemic DNA using some results from simple rod models suggested that exposure to polyamines reduced the radii and increased the densities of plectonemes. Thus, polyamines may limit the twist density by favoring writhe which maintains the B-form. Although polymerases may significantly stretch the double helix, denature DNA, and produce twist instead of writhe, natural polyamines stabilize base-pairing, limit twist to maintain the B-form, and promote supercoiling, which is conducive to replication and transcription and essential for DNA packaging.
Keywords: spermine, spermidine, magnetic tweezers, single-molecule techniques
INTRODUCTION
Polyamines are organic compounds having more than one amino group (NH2). They are synthesized in living cells via highly regulated pathways and are known to regulate cell growth, gene transcription and activation of DNA synthesis 1. High levels of polyamines are associated with certain cancers 2. Furthermore, increased polyamine levels can be diagnostic markers of drug resistance in oncological treatment3. How polyamines act in cellular metabolism to regulate normal or cancerous growth is not entirely understood nor are the structural changes in deoxyribonucleic acid (DNA) caused by polyamines in physiologically relevant conditions.
Inside living cells, DNA is compacted in hierarchical topological structures. Plectonemic, negative supercoils (extruded intertwined loops) form the basis of this compaction in live cells. Plectonemes depend on the topological parameters, writhe and twist, and also play crucial roles in the regulation of DNA transcription and replication5. For instance bacterial plasmids are negatively supercoiled4, which is known to favor local denaturation of DNA near replication origins and transcriptional promoters where polymerases must gain access to the coding strand within a double-stranded DNA molecule (dsDNA)5. Positive supercoiling interferes with mRNA synthesis, arresting the progress of RNA polymerase complexes6. Clearly, DNA supercoiling and its regulation fundamentally impacts cellular functions.
Not surprisingly therefore, a family of enzymes, the topoisomerases, can regulate DNA supercoiling. For example, in bacteria, topoisomerase I relaxes negative supercoils by catalyzing torsion-driven rotation of one single DNA strand about the other in a dsDNA segment, and gyrase catalyzes passage of one dsDNA segment through another to introduce negative and relax positive supercoils5. Topoisomerases are prominent regulators of DNA supercoiling, but their activity might be strongly modulated by small molecules like polyamines that alter the mechanical properties of DNA.
Numerous experiments suggest that polyamines stabilize the right-handed, B-form double helix7, protect it from radiation and oxidation8, 9 and compact and even aggregate DNA in low salt buffers10, 11. The multiple positively charged amine groups can certainly interact electrostatically with the negatively charged phosphates of DNA. However, recent infrared spectroscopy data indicate that natural polyamines can bind in the major and minor grooves of DNA to interact with the DNA bases as well12. Spermidine, which has three amine groups, and spermine, which has four (see Fig. 1), bind to the major and minor grooves of DNA12, 13 and are the two most important naturally occurring polyamines in mammals14. However, little is known about how these two polyamines affect DNA supercoiling in physiological concentrations of monovalent salt.
Figure 1.

Structure of spermidine (top) and spermine (bottom). The basic amino (NH2) groups attract protons at physiological pH to become positively charged.
It has been shown that the presence of spermine and spermidine can enhance the transcription rate despite high levels of positive supercoiling by changing the physical properties of supercoiled DNA15. Moreover, the presence of polyamines inhibited topoisomerase I-catalyzed relaxation of negative supercoils perhaps by stabilization of the DNA double helix15. Both the increase of positive supercoiling during transcription and the inhibition of topoisomerase I were observed with sub-millimolar spermine or millimolar spermidine concentrations, which are physiological14. Systematic regulation of the concentrations of polyamines1 may be a primary mechanism by which a cell regulates DNA supercoiling and its related enzyme activities and cellular processes. Nevertheless, only qualitative explanations have been proposed for these effects15, and there are no quantitative measures of how polyamines alter DNA supercoils.
Previously, single-molecule experiments were performed to investigate DNA compaction as a function of tension16 and twist17 at various polyamine concentrations in buffers containing sub-physiological concentrations of monovalent salt. More specifically, those experiments measured the tensions generated by compaction, which were found to be as high as 3 and 4 pN for spermidine and spermine, respectively. Those data were gathered by measuring the maximum level of tension at which single, torsionally constrained DNA molecules suddenly shortened (into compact forms). The compaction depended on polyamine concentrations, and free energies were reported to be −0.2 kT/base pair for spermidine and −0.33 kT/base pair for spermine.
At higher levels of monovalent salt, DNA molecules are less likely to compact. Indeed high concentrations of KCl prevent DNA aggregation and catenation by enzymes18. The following data were recorded for single DNA molecules in 200 mM KCl to avoid compaction and allow analysis of the structure of plectonemic DNA over a low range of tensions. Magnetic tweezers were used to twist and stretch single DNA molecules at various physiological concentrations of the two polyamines. The observations suggest that polyamines stabilize the double stranded, B-form of DNA and promote plectoneme formation. An analysis of the data using some key results from simple elastic rod theories indicated that both spermine and spermidine changed the form of the supercoiled DNA producing more tightly wrapped plectonemes. An order of magnitude less tetravalent spermine produced the same degree of wrapping as the trivalent spermidine.
MATERIAL AND METHODS
Plasmid pDL231719 was used as a polymerase chain reaction (PCR) template to produce a linear, 3215 base-pair (bp) DNA fragment using the primer pair 5′TAATACGACTCACTATAGGG and 5′TATGCCCGAGAAGATGTT. The amplicon was digested with Bam HI and Xba I to give a final 3096 bp fragment. Biotin- or digoxigenin-labeled fragments approximately 1000 bp were created using PCR with the plasmid pDL1051 (D.A. Lewis, NIH) as a template and the primer pairs 5′-TGTATGGAACAACGCATAAC and 5′-TCCAAACTGGAACAACAC or 5′-AAGGTAACTGGCTTCAGC and 5′-TTATTGTCTCATGAGCGG to incorporate about 5% biotin- or digoxigenin-labeled dUTP. These fragments were digested with BamHI or XbaI to generate complementary ends for ligation to the main DNA fragment20. The ligated construct was attached at one end to the anti-digoxigenin-coated glass surface of a flow-chamber and at the other end to a 2.8 μm streptavidin-coated, super-paramagnetic bead (Invitrogen, Carlsbad, CA, USA) (Figure 2). The DNA had multiple attachments at both ends to prevent torsional relaxation. Note that the biotin- and digoxigenin-labeled DNA ends form multiple attachments to the streptavidin-coated bead and anti-digoxigenin-coated glass, so that the free tether length is the unlabeled, 3096 bp main fragment.
Figure 2.
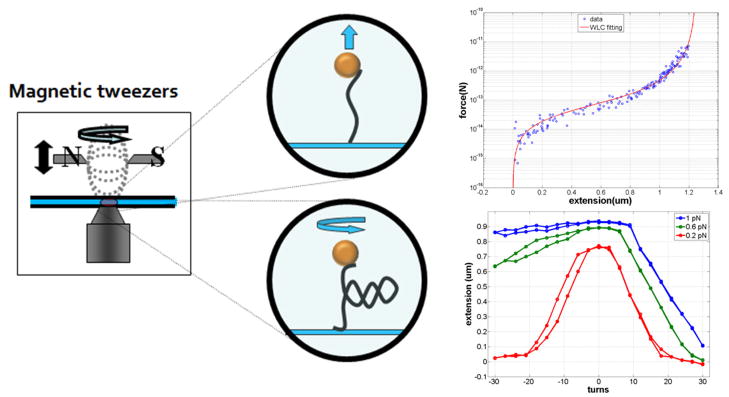
Stretching and twisting DNA with magnetic tweezers. Magnetic tweezers (left) can be used to stretch (upper middle) and twist (lower middle) single DNA molecules tethering para-magnetic beads to a glass surface. Systematically varying the separation between the magnets and the tethered bead generates a range of tensions as shown in a representative force vs. extension curve for a single DNA molecule (upper right). The red curve is a worm-like-chain fit to the data. Rotating the magnets to twist the DNA tether produces extension vs. twist curves for a DNA molecule at different tensions (lower right), from top to bottom: 1, 0.6, and 0.2 pN.
The DNA was incubated in λ buffer, consisting of 10 mM Tis-HCl (pH 7.4), 200 mM KCl, 5% dimethyl sulfoxide (DMSO), 0.1 mM ethylenediaminetetraacetic acid (EDTA), and 0.2 mM dithiothreitol (DTT)),21 with the indicated concentrations of polyamines. Flow chambers of approximately 50 μl were assembled from slides using a continuous bead of silicon grease to form the walls of the chamber before sealing with a coverslip. Spermine was diluted in 200 μl λ buffer to final concentrations of 0, 0.2, 0.5, 0.7, 1 and 2 mM, and spermidine to final concentrations of 0, 1, 2, 5 and 10 mM. Working solutions containing high concentrations of spermine (> 0.5mM) and spermidine (> 2mM) were vortexed before use to dissolve fine particles.
At several polyamine concentrations, magnetic tweezers (MT) were used to stretch and twist the DNA tethers. Tension was varied by changing the separation between the DNA-tethered beads and a pair of neodymium magnets mounted above the microscope stage. Rotation of the magnets caused the oriented, para-magnetic beads to rotate and twist the attached DNA tethers (Fig. 2). Extension versus twist data was recorded at tensions of 0.2, 0.6, and 1.0 pN for several concentrations of spermine and spermidine.
The contour length of tethered DNA under high tension (about 6 pN) agreed well with that predicted from the number of base pairs in the unlabeled fragment. Molecules were stretched and relaxed twice as the force varied from 0.02 to 4 pN and their lengths were recorded. Also, DNA tethers under 0.2, 0.6, and 1 pN of tension were unwound to −10% supercoiling density (σ) (−30 turns for 3kbp DNA), rewound to +10% and unwound back to 0 in steps of 3 turns while recording the tether length.
Data were acquired at a rate of 20 frames/second and analyzed to determine the extension, l = <z>, of the DNA molecule with an error of about 10 nm with 10 sec averaging using video-rate, three-dimensional tracking of the bead22. Measuring the horizontal excursions of the bead <Δx2> allowed the tension in the molecule to be determined via the equipartition theorem, F = kBTl/<Δx2>, with 10% accuracy. Mechanical drift in the data was eliminated using differential tracking of a second bead stuck on the surface. The data were analyzed to determine the probable dimensions of the plectonemes as described (see also appendix I of the supplementary information).
RESULTS AND ANALYSIS
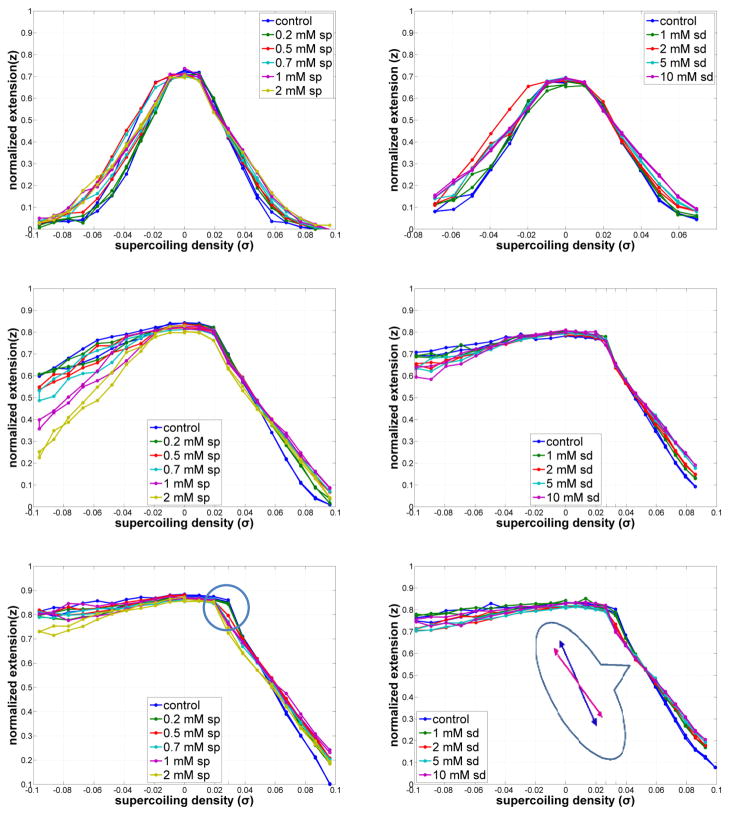
Figure 3 shows the DNA extension, z, normalized by the contour length, L, versus twist for a representative DNA tether at various physiological concentrations of spermine (left panels) or spermidine (right panels). The twist has also been normalized and reported as the “supercoiling density”, σ = (Lk − Lk0)/Lk0 = n/Lk0, where Lk0 is the number of the right-handed helical turns in the torsion-free, B-form dsDNA, and n is the number of mechanically added twists. The effects of polyamines on DNA extension versus twist were investigated at three different tensions (Fig. 3), 0.2 (upper), 0.6 (middle) and 1.0 pN (lower). The blue curves represent the DNA without polyamine.
Figure 3.
DNA extension vs. supercoiling density as a function of spermine (left) or spermidine (right) concentrations at 0.2 (upper), 0.6 (middle), and 1 pN (lower) of tension. “Knee-points” are encircled. The inset emphasizes the change in slope due to added spermidine. Typical measurement errors are shown in supplementary information on a copy of the curves for 0.6 pN with added spermine.
At all tensions, the extension of slightly over-twisted DNA changed very little initially but then contracted sharply beyond a threshold value of twist as the filament “buckled” and formed plectonemes at a critical value of twist, nb. Such buckling has been observed previously in single-molecule experiments with DNA23–25 and modeled and simulated using rod mechanics23, 26. Beyond a threshold value of additional twist, the extension decreased linearly as more and more of the DNA shifted to the plectonemic form.
For 0.2 pN of tension, the extension decreased as a function of either positive or negative supercoiling for control DNA (blue in Fig. 3 top panel) due to the formation of plectonemes. An intermediate tension of 0.6 pN hindered plectoneme formation in unwound molecules. This gentler decrease of the extension upon unwinding corresponded to partitioning between plectoneme formation and some other form of the DNA helix27. Indeed, at a tension of 1 pN, the extension remained constant for negative values of σ, indicating that perhaps plectonemes did not form at all. It is known that under high tension, unwinding partially melts the double helix28 or favors a transition to some other helical form29. However well-defined models of DNA under these conditions are still under development. As a result, only plectonemes formed by positively twisted DNA will be analyzed quantitatively with rod theory in this paper.
Once a plectoneme nucleates in response to positive supercoiling and is sufficiently large, it has been shown from simple rod theories30, 31 that any additional twist is almost completely absorbed as writhe, keeping the torque constant. Thus, over-twisting drives a phase change to a plectonemic form, and a steadily increasing fraction of the DNA adopts the plectonemic phase at a constant torque. The slope of extension versus twist curve reveals the DNA length per writhe in the plectoneme, where more gradual slopes indicate shorter lengths of DNA per writhe, and hence plectonemes of smaller gyres.
Polyamines were found to alter the slope of the extension versus supercoiling density curves for positive σ and presumably the plectoneme structure. Three different effects of polyamines on DNA supercoiling behavior were observed and are described in the following sections.
Spermine and spermidine stabilize B-DNA structure and increase plectoneme formation for negative supercoiling
Spermine and spermidine stabilized the B-form as DNA was unwound at 0.6 pN of tension (middle panels of Fig. 3). When a dsDNA tether is unwound, contraction of the molecule at low tension (0.2 pN) corresponds to the formation of plectoneme(s) as a fraction of the added linking number is absorbed as writhe. However, at higher tension, the torsional stress might disrupt B-DNA and induce the formation of a left-handed winding of unpaired filaments32. Increasing the spermine or spermidine concentration increased the extent of contraction versus unwinding at 0.6 pN, apparently stabilizing the B-form DNA and shifting this torsional equilibrium toward plectoneme formation.
Figure 4 quantitatively summarizes these results in plots of the average magnitude of the slope of the extension versus unwinding data for all the three tensions as a function of increasing concentrations up to 2 mM spermine (left) and 10 mM spermidine (right). At 1 pN, the near-zero slope does not change, because the tension is high enough to oppose any plectoneme formation. Instead at 0.6 pN, increased polyamine concentrations increased the magnitude of the slope indicating more extensive plectoneme formation. At lower tension (0.2 pN), the curves are symmetric even without polyamines. However, note that the slope of the extension versus unwinding curves decreases with increasing polyamine concentrations. This was interpreted as evidence of more compact plectonemes as described below for positive supercoiling.
Figure 4.
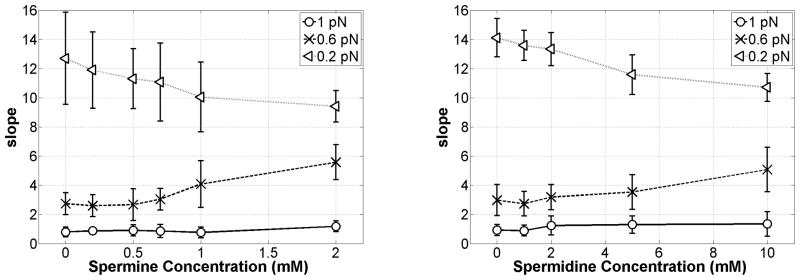
Average slope (dz/dσ, dimensionless) of the DNA extension vs. twist in Fig. 3 for under-wound DNA as a function of spermine (left) or spermidine (right) concentrations at 0.2, 0.6, and 1 pN of tension. Error bars represent standard deviations.
Spermine and spermidine promote plectoneme formation for positive supercoiling
The two polyamines also promote the formation of plectonemes in overwound DNA molecules as shown in Fig. 3, in which they began to form at lower levels of twist as the polyamine concentrations were increased.
This effect was particularly obvious at higher tensions, 0.6 and 1 pN, and the supercoiling density at which writhe initially reduced the extension of the DNA tether is shown as a function of increasing spermine and spermidine in Fig. 5. Large fluctuations of the DNA extension obscured this effect at 0.2 pN of tension. A convenient framework with which to visualize this behavior is to contrast the torsional and flexural stiffness of two rods. If the rods have the same torsional but different flexural stiffness, the more flexible rod will exhibit more writhe at any level of tension to minimize conformational energy. Electrostatic softening of DNA by multivalent cations is well described for polyamine-DNA interactions, but these data indicate that polyamines differentially alter torsional and flexural characteristics of DNA.
Figure 5.
Twist values σ at which writhe first appeared in over-twisted DNA as a function of spermine (left) or spermidine (right) concentrations at 0.6 and 1 pN of tension. The σ values were defined using the linear extrapolation of extension vs. σ curves33. Error bars represent standard deviations.
Spermine and spermidine shrink plectonemic supercoils
Once over-twisted DNA begins to writhe, additional torque is absorbed as writhe and the extension decreases linearly as a function of twist as can be seen in Fig. 3. Figure 6 shows the average magnitude of the slope of these extensions versus twist curves as a function of spermine or spermidine concentration. For all three values of tension, 0.2, 0.6, and 1 pN, the slope decreased with increasing polyamine concentration and somewhat asymptotically approached a constant value. Since the magnitude of the slope indicates the length of DNA consumed in each plectoneme, a lesser slope corresponds to smaller plectonemic gyres. Thus polyamines appeared to promote tightly wrapped plectonemes.
Figure 6.
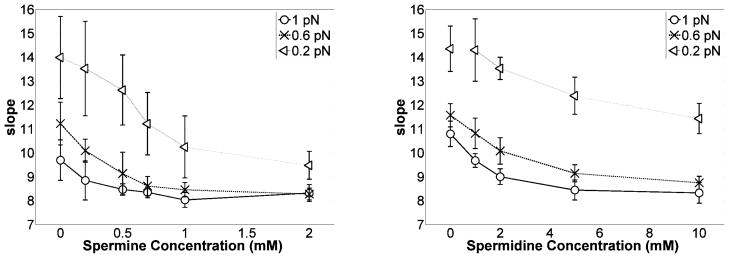
Average magnitude of the slope (dz/dσ, dimensionless) of DNA extension vs. twist for positively supercoiled DNA as a function of spermine (left) or spermidine (right) concentrations at 0.2, 0.6, and 1 pN of tension. Error bars represent standard deviations.
In Fig. 6 it is also clear that the size of plectonemes decreased as tension was increased. At all concentrations the slope of the extension versus twist curves varied inversely with tension. However, the more tightly wrapped plectonemes due to the polyamines were most pronounced at low tension. In order to analyze this plectonemic contraction quantitatively, the writhe density was derived from the slope as described below.
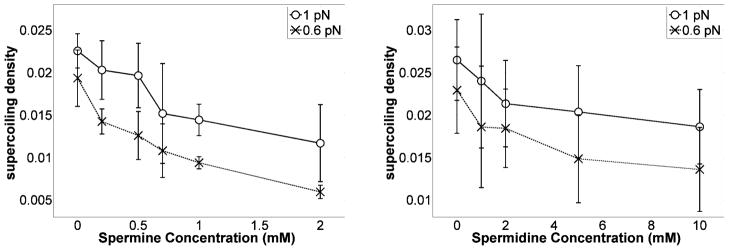
Quantitative analysis of plectonemes
In order to understand how spermine and spermidine affected the topological features of the plectonemic phase in overwound DNA, the writhe per helical turn was determined as a function of increasing polyamine concentrations at 0.6 and 1.0 pN tensions. The plectonemic phase was assumed to consist of two identical, intertwined duplexes with uniform radius, R, and uniform helical angle30, 31, α, and the end loop was ignored30. Furthermore, externally applied twist was assumed to completely convert into writhe, Wr, leaving the torque on the molecule and the size of the plectonemes unchanged24 as new ones were generated. With these assumptions, the writhe density in the plectonemic phase was derived (see Appendix 1 of supplementary information) and found to be inversely proportional to the slope as follows:
| [1] |
where z is the extension normalized by the contour length, L, of the DNA, σ is the number of turns, n, added to DNA normalized by the stress-free linking number Lk0, lp is the length of DNA absorbed into plectonemic phase, Wr is the writhe, and ρth is a contraction factor due to thermal fluctuations that were assumed to occur predominantly in the stretched phase of DNA. This contraction factor ρth was approximated using the Moroz and Nelson approximation for a twisted, fluctuating filament34 under tensions greater than 0.5 pN as follows:
| [2] |
in which,
| [3] |
is the torque exerted by the twisted DNA on the bead, Kb is the bending stiffness, F, is tension, kB is the Boltzmann constant, and T is the absolute room temperature. Note that as plectonemes form, the torque, M3(cr), on the molecule remains constant at a value that depends only on the tension and the bending stiffness.
In order to complete the calculation of ρth, and hence of the writhe density, the bending stiffness was calculated using Kb = kBTPl, for which the bending persistence length, Pl, was determined by fitting the worm-like chain expression34–36 to extension versus force data for torsionally relaxed DNA.
| [4] |
Figure 7 shows the persistence length, which is proportional to the effective bending stiffness, as a function of polyamine concentration. There were no significant changes in the bending persistence length as a function of polyamine concentration in 200 mM KCl. Such invariance is similar to that of persistence lengths previously reported for DNA in 100 mM NaCl which did not change significantly upon addition of 10 mM magnesium37. Both data sets differ from those of Bauman et al. in which millimolar concentrations of multivalent cations in approximately 2 mM NaCl greatly reduced the persistence length of DNA10. The concentration of monovalent salt certainly changes the electrostatic screening environment, but in the case of polyamines, any potential increased flexibility due to charge neutralization by polyamines may also be opposed by polyamine linkages between the two phosphate backbones and the placement of exogenous amines in the minor groove that tend to structurally stiffen the DNA38, 39. Solving Equations [1–4] allowed determinations of the writhe per helical turn as a function of polyamine concentration as shown in Fig. 8.
Figure 7.
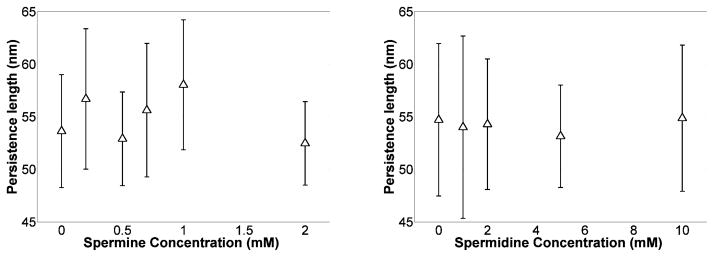
Average persistence length for eight DNA tethers at different spermine (left) or spermidine (left) concentrations. Polyamines did not significantly affect the bending stiffness of DNA. Error bars represent standard deviations.
Figure 8.
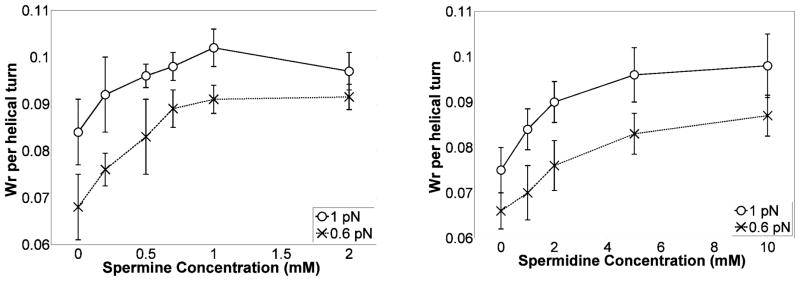
Effect of spermine (left) or spermidine (right) concentrations on the writhe density in the plectonemic phase at 0.6 and 1 pN of tension. The increase in writhe density is a quantitive measure of plectoneme shrinking. Error bars represent standard deviations. Note that the model is valid for small fluctuations at tensions above 0.5 pN.
DISCUSSION
Polyamines are thought to stabilize DNA duplexes, especially AT-rich regions, to prevent their denaturation in response to negative twist. The experiments described above clearly demonstrate that both spermine and spermidine promoted writhe in unwound DNA. Crystal structures and infrared spectra of polyamine-DNA complexes12, 40 show that spermine and spermidine molecules can form hydrogen bonds with bases from opposite strands of DNA duplex. This might reduce denaturation and produce writhe instead of unwinding DNA.
Alternatively to denaturation, negative twist can trigger a right- to left-hand helical transition. Increased unwinding is known to convert small GC repeat DNA insert (about 23 bp) to left-handed Z-DNA at high NaCl concentrations (> 0.7 M)41. Indeed, spermine and spermidine have been shown to induce Z-DNA formation not only in synthetic GC repeat DNA fragment42, 43 but also in calf thymus DNA44. However, investigations using torsionally constrained plasmids (σ = −0.033) without a synthetic GC repeat insert showed that spermine and spermidine were unable to induce the B- to Z-DNA transition45. In the experiments described here, the low tension and degree of unwinding were unlikely to trigger the formation of left-handed DNA 29, and the reduced extensions produced in slightly unwound (less than 1%) DNA were interpreted as plectonemes of right-handed duplexes. Raman and energy minimization studies indicate that spermine and spermidine bind along major and minor grooves of DNA duplex respectively13, 46, and therefore might stabilize the major and minor grooves of B-DNA to favor such writhing in the underwound DNA molecule (Fig. 3).
The effect on over-wound DNA appears to be more subtle. Spermine and spermidine promoted plectoneme formation at lower levels of twist. Once over-wound DNA begins to form plectonemes, the torque and therefore twist per unit length remain constant24. Thereafter plectonemes steadily grow to compensate for further increases in linking number. In the experiments reported here, polyamines increased the writhe density as much as 10% and promoted a commensurate decrease in twist density. A corollary to this idea is that by promoting the formation of compact plectonemes, polyamines tend to decrease the twist density in DNA. Such crowding of writhe to relax twist is a subtle but distinct variation of the conservation of linking number described by the Calugareanu-White-Fuller equation that might be tested as described in Appendix 2 of the supplementary information.
To investigate further the shrinking of plectonemes, if the helical angle is known, one can calculate the radius, R, of the helical plectoneme from the writhe formula of a helix as follows:
| [5] |
The helical angle can be computed from the elastic rod models that account for electrostatic interactions. The computed helical angles are sensitive to the type of model for electrostatic screening31. However, we note that employing either the Ubbink and Odijk approximation of a Poisson-Boltzmann model47 or the Ray and Manning approximation48 of counterion condensation theory49 in the simplistic rod model, the helical angle increases with the tension converging to approximately π/6 for tensions in the range of 0.6 pN and above33. Therefore, assuming a helical angle α≈π/6, Eq. [5] produces a rough estimate of how R may change as a function of the polyamine concentration. The trend is shown in Fig. 9 and the radius was found to shrink substantially as polyamine concentrations increased. Plectoneme formation forces helices into proximity which will increase the binding constant of spermidine and spermine. Enough ions may bind to substantially decrease the repulsion at the distances estimated for the radius of the superhelix (Fig. 9). The accuracy of these results relies on the formation of a uniform helix, which has been analyzed in a simplistic framework for a rod model and appears to be justified30, 31.
Figure 9.
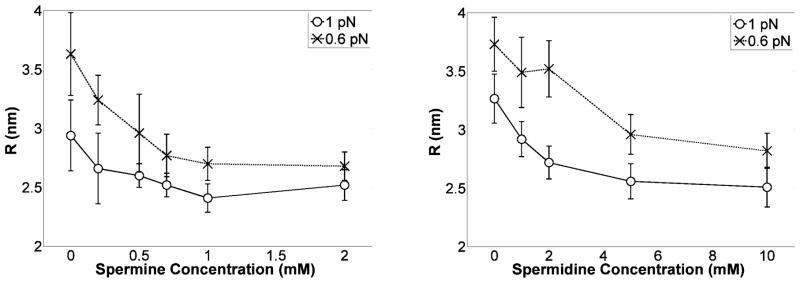
Radius of the helical plectoneme as a function of spermine (left) or spermidine (right) concentrations at 0.6 and 1 pN of tension. The radii of plectonemes decreased substantially as the polyamine concentration was increased, especially at low tension.
While electrostatic neutralization of phosphate repulsions, which would promote more tightly wrapped plectonemes, is a widely accepted phenomenon, a constant helical angle in the plectonemic gyres was assumed for the estimates in Fig. 9. Yet an increase of the helical angle above a threshold polyamine concentration due to repulsions between DNA-bound polyamines, or a drop in the helical angle in response to increasing polyamine concentration due to phosphate charge neutralization are plausible. Nevertheless, since Eq. [5] indicates that the radius of the plectoneme is a direct function of the helical angle, and the radius decreases as polyamine concentration increases in Fig. 9, the radius is likely to be a decreasing function of polyamine concentration.
A more robust description of plectonemic shape will require a model that can dynamically simulate the formation of plectonemes including electrostatic interaction50 and non-homogeneity51 along with accurate modeling of the stress-strain relationship that governs the bending and twisting deformations of DNA as discussed previously52. In general, nonlinear coupling between bending, twisting, and stretching may influence not only plectonemic shapes, but also the buckling of DNA. While the effects of twist-stretch coupling on the buckling and non-uniform twist distribution have been explored53, this coupling and these models need to be systematically developed for application to magnetic tweezer data.
Broadly speaking, polyamines torsionally stiffened either under- or overwound DNA to promote the formation of plectonemes. The fact that polyamines attenuate distortions of helical DNA is likely to be very significant biologically. DNA motor enzymes can produce considerable topological stress in DNA molecules as a result of transcription, replication, and during packaging reactions54. Tuning polyamine levels could limit helical distortions to maintain the binding capacity of proteins that interact with DNA molecules in critical cellular processes. The fact that the levels of polyamines vary and are highly regulated, especially during the cell cycle, may reflect the importance of this regulatory mechanism in living organisms55.
CONCLUSION
In these experiments, both spermine and spermidine stabilized the right-handed, double-stranded B-form of DNA to promote plectoneme formation in both under- and over-wound molecules. Calculations quantitatively revealed increasingly tightly wrapped plectonemes formed by over-winding at physiological concentrations of the two polyamines which differed by a factor of ten. This likely derives from the electrostatic basis of their interactions with DNA and reflects the tetra- and trivalency of the spermine and spermidine cations respectively. The data are consistent with the idea that these two natural polyamines “groom” DNA to limit the twist density and maintain the B-form, despite powerful enzymes that can distort the B-form double helix and produce large topological changes. This would preserve binding by DNA proteins that recognize the B-form, and promote writhe which is critical for DNA packaging.
Supplementary Material
Acknowledgments
FUNDING
This work was supported by National Institutes of Health (5R01GM084070-03 to LF); the Human Frontier Science Program (RGP0051/2009 to DD); and the University Research Council at Emory University (to DD).
Discussions with Prof. Prashant Purohit, Dr. Sebastien Neukirch, and Dr Monica Fernandez were most helpful.
Footnotes
Supporting Information Available
Two appendices describing derivations and a copy of Figure 2b with added error bars are available as supplementary information, which is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Kusano T, Berberich T, Tateda C, Takahashi Y. Planta. 2008;228(3):367–81. doi: 10.1007/s00425-008-0772-7. [DOI] [PubMed] [Google Scholar]
- 2.Wallace HM, Fraser AV, Hughes A. Biochemical Journal. 2003;376:1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grandgirard N, Ly-Sunnaram B, Ferrant D, Gandemer V, Edan C, Le Gall E, Moulinoux JP, Leray E, Goasguen JE. Leukemia Research. 2004;28(5):479–486. doi: 10.1016/j.leukres.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Mcclellan JA, Boublikova P, Palecek E, Lilley DMJ. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(21):8373–8377. doi: 10.1073/pnas.87.21.8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang JC. Nature Reviews Molecular Cell Biology. 2002;3(6):430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 6.Gartenberg MR, Wang JC. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(23):11461–11465. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tabor H. Biochemistry. 1962;1(3):496–501. doi: 10.1021/bi00909a021. [DOI] [PubMed] [Google Scholar]
- 8.Ha HC, Sirisoma NS, Kuppusamy P, Zweier JL, Woster PM, Casero RA. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(19):11140–11145. doi: 10.1073/pnas.95.19.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh TJ, Kim IG. Biotechnology Techniques. 1998;12(10):755–758. [Google Scholar]
- 10.Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Proc Natl Acad Sci U S A. 1997;94(12):6185–90. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosule LC, Schellman JA. Nature. 1976;259(5541):333–335. doi: 10.1038/259333a0. [DOI] [PubMed] [Google Scholar]
- 12.Ouameur AA, Tajmir-Riahi HA. Journal of Biological Chemistry. 2004;279(40):42041–42054. doi: 10.1074/jbc.M406053200. [DOI] [PubMed] [Google Scholar]
- 13.Feuerstein BG, Pattabiraman N, Marton LJ. Nucleic Acids Research. 1990;18(5):1271–1282. doi: 10.1093/nar/18.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis RH, Morris DR, Coffino P. Microbiological Reviews. 1992;56(2):280–290. doi: 10.1128/mr.56.2.280-290.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng HF, Jackson V. Journal of Biological Chemistry. 2000;275(1):657–668. doi: 10.1074/jbc.275.1.657. [DOI] [PubMed] [Google Scholar]
- 16.Todd BA, Rau DC. Nucleic Acids Research. 2008;36(2):501–510. doi: 10.1093/nar/gkm1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Besteman K, Hage S, Dekker NH, Lemay SG. Physical Review Letters. 2007;98(5):058103. doi: 10.1103/PhysRevLett.98.058103. [DOI] [PubMed] [Google Scholar]
- 18.Krasnow MA, Cozzarelli NR. Journal Of Biological Chemistry. 1982;257(5):2687–2693. [PubMed] [Google Scholar]
- 19.Zurla C, Manzo C, Dunlap D, Lewis DE, Adhya S, Finzi L. Nucleic Acids Res. 2009;37(9):2789–95. doi: 10.1093/nar/gkp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finzi L, Dunlap D. Methods Enzymol. 2003;370:369–78. doi: 10.1016/S0076-6879(03)70032-9. [DOI] [PubMed] [Google Scholar]
- 21.Nelson PC, Zurla C, Brogioli D, Beausang JF, Finzi L, Dunlap D. Journal of Physical Chemistry B. 2006;110(34):17260–17267. doi: 10.1021/jp0630673. [DOI] [PubMed] [Google Scholar]
- 22.Strick TR, Charvin G, Dekker NH, Allemand JF, Bensimon D, Croquette V. Comptes Rendus Physique. 2002;3(5):595–618. [Google Scholar]
- 23.Brutzer H, Luzzietti N, Klaue D, Seidel R. Biophysical Journal. 2010;98(7):1267–1276. doi: 10.1016/j.bpj.2009.12.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forth S, Deufel C, Sheinin MY, Daniels B, Sethna JP, Wang MD. Physical Review Letters. 2008;100(14):148301. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. Science. 1996;271(5257):1835–7. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 26.Goyal S, Perkins NC, Lee CL. International Journal of Non-Linear Mechanics. 2008;43(1):65–73. [Google Scholar]
- 27.Leger JF, Romano G, Sarkar A, Robert J, Bourdieu L, Chatenay D, Marko JF. Physical Review Letters. 1999;83(5):1066–1069. [Google Scholar]
- 28.Strick TR, Croquette V, Bensimon D. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(18):10579–10583. doi: 10.1073/pnas.95.18.10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheinin MY, Forth S, Marko JF, Wang MD. Physical Review Letters. 2011;107(10):108102. doi: 10.1103/PhysRevLett.107.108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neukirch S, Marko JF. Phys Rev Lett. 2011;106(13):138104. doi: 10.1103/PhysRevLett.106.138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purohit PK. Shape and energetics of DNA plectonemes. In: Garikipati KA, Ellen M, editors. IUTAM symposium on Cell, Molecular and Tissue Dordrecht, 2010. Springer; Dordrecht: 2010. [Google Scholar]
- 32.Sarkar A, Leger JF, Chatenay D, Marko JF. Phys Rev E Stat Nonlin Soft Matter Phys. 2001;63(5 Pt 1):051903. doi: 10.1103/PhysRevE.63.051903. [DOI] [PubMed] [Google Scholar]
- 33.Clauvelin N, Audoly B, Neukirch S. Biophysical Journal. 2009;96(9):3716–3723. doi: 10.1016/j.bpj.2009.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moroz JD, Nelson P. Macromolecules. 1998;31(18):6333–6347. [Google Scholar]
- 35.Bustamante C, Marko JF, Siggia ED, Smith S. Science. 1994;265(5178):1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 36.Kratky O, Porod G. Recueil Des Travaux Chimiques Des Pays-Bas-Journal of the Royal Netherlands Chemical Society. 1949;68(12):1106–1122. [Google Scholar]
- 37.Porschke D. Biophysical Chemistry. 1991;40(2):169–179. doi: 10.1016/0301-4622(91)87006-q. [DOI] [PubMed] [Google Scholar]
- 38.Bailly C, Mollegaard NE, Nielsen PE, Waring MJ. Embo J. 1995;14(9):2121–31. doi: 10.1002/j.1460-2075.1995.tb07204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouameur AA, Tajmir-Riahi HA. Journal Of Biological Chemistry. 2004;279(40):42041–42054. doi: 10.1074/jbc.M406053200. [DOI] [PubMed] [Google Scholar]
- 40.Jain S, Zon G, Sundaralingam M. Biochemistry. 1989;28(6):2360–2364. doi: 10.1021/bi00432a002. [DOI] [PubMed] [Google Scholar]
- 41.Lee M, Kim SH, Hong SC. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(11):4985–4990. doi: 10.1073/pnas.0911528107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behe M, Felsenfeld G. Proc Natl Acad Sci U S A. 1981;78(3):1619–23. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabor CW, Tabor H. Annu Rev Biochem. 1984;53:749–90. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 44.Hasan R, Alam MK, Ali R. FEBS Lett. 1995;368(1):27–30. doi: 10.1016/0014-5793(95)00591-v. [DOI] [PubMed] [Google Scholar]
- 45.Thomas TJ, Gunnia UB, Thomas T. J Biol Chem. 1991;266(10):6137–41. [PubMed] [Google Scholar]
- 46.Ruiz-Chica J, Medina MA, Sanchez-Jimenez F, Ramirez FJ. Biophysical Journal. 2001;80(1):443–454. doi: 10.1016/S0006-3495(01)76027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ubbink J, Odijk T. Biophysical Journal. 1999;76(5):2502–2519. doi: 10.1016/S0006-3495(99)77405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ray J, Manning GS. Langmuir. 1994;10(7):2450–2461. [Google Scholar]
- 49.Manning GS. Journal of Chemical Physics. 1969;51(3):924. [Google Scholar]
- 50.Lillian TD, Perkins NC. Proceedings of Asme International Design Engineering Technical Conferences and Computers and Information in Engineering Conference; 2010. pp. 1447–1455.pp. 2069 [Google Scholar]
- 51.Goyal S, Perkins NC. International Journal of Non-Linear Mechanics. 2008;43(10):1121–1129. [Google Scholar]
- 52.Palanthandalarn-Madapusi HJ, Goyal S. Automatica. 2011;47(6):1175–1182. [Google Scholar]
- 53.Goyal S, Perkins NC, Lee CL. Journal of Computational Physics. 2005;209(1):371–389. [Google Scholar]
- 54.Bustamante C, Bryant Z, Smith SB. Nature. 2003;421(6921):423–7. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 55.Wallace HM, Fraser AV, Hughes A. Biochem J. 2003;376(Pt 1):1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.