Abstract
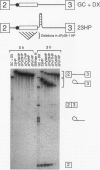
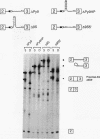
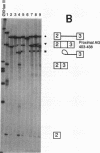
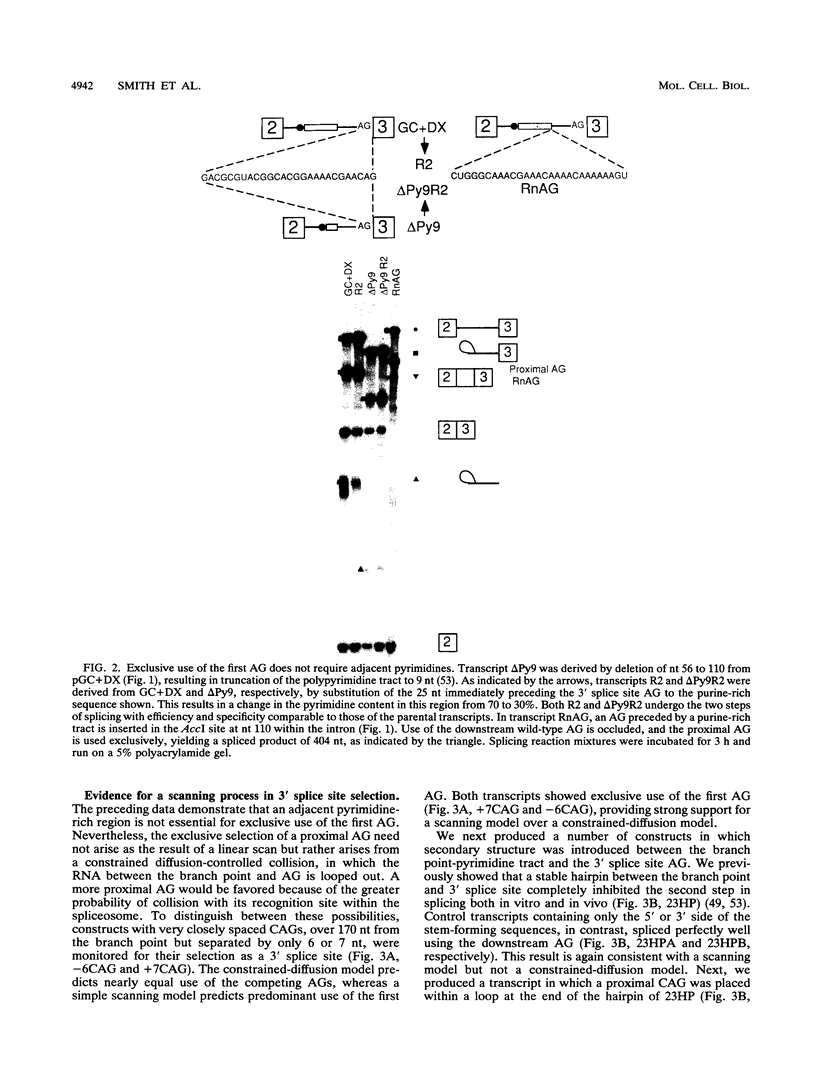
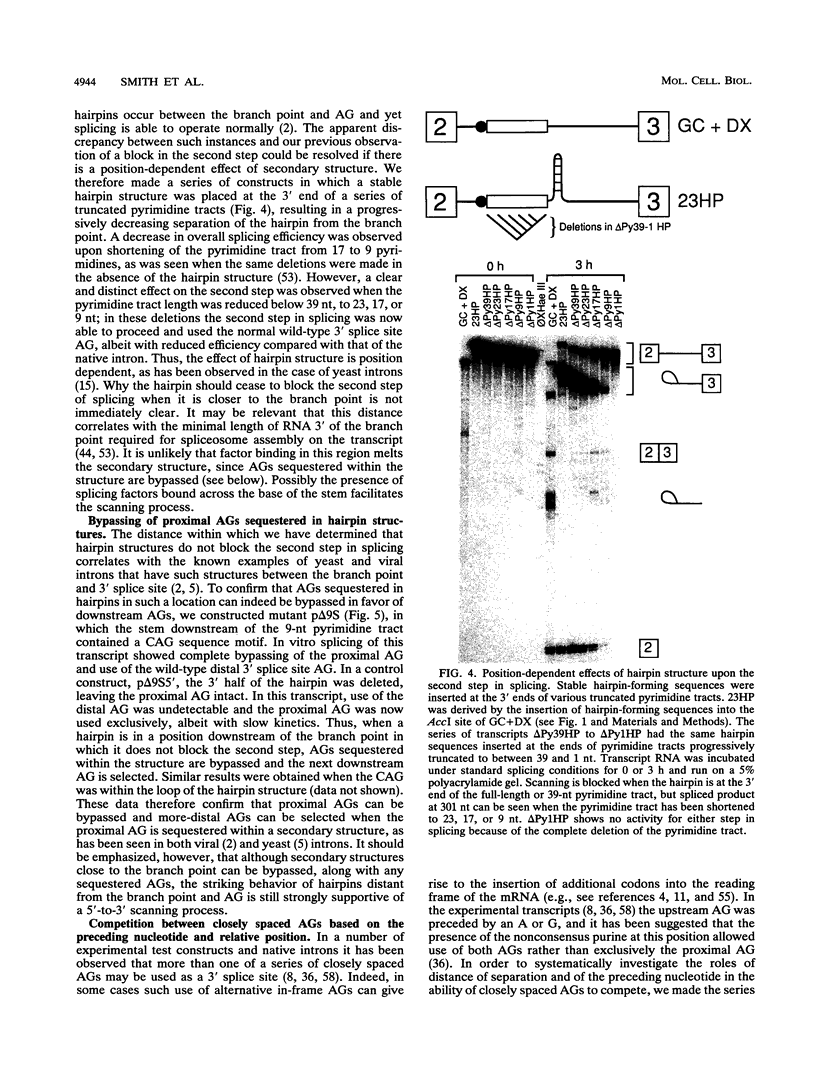
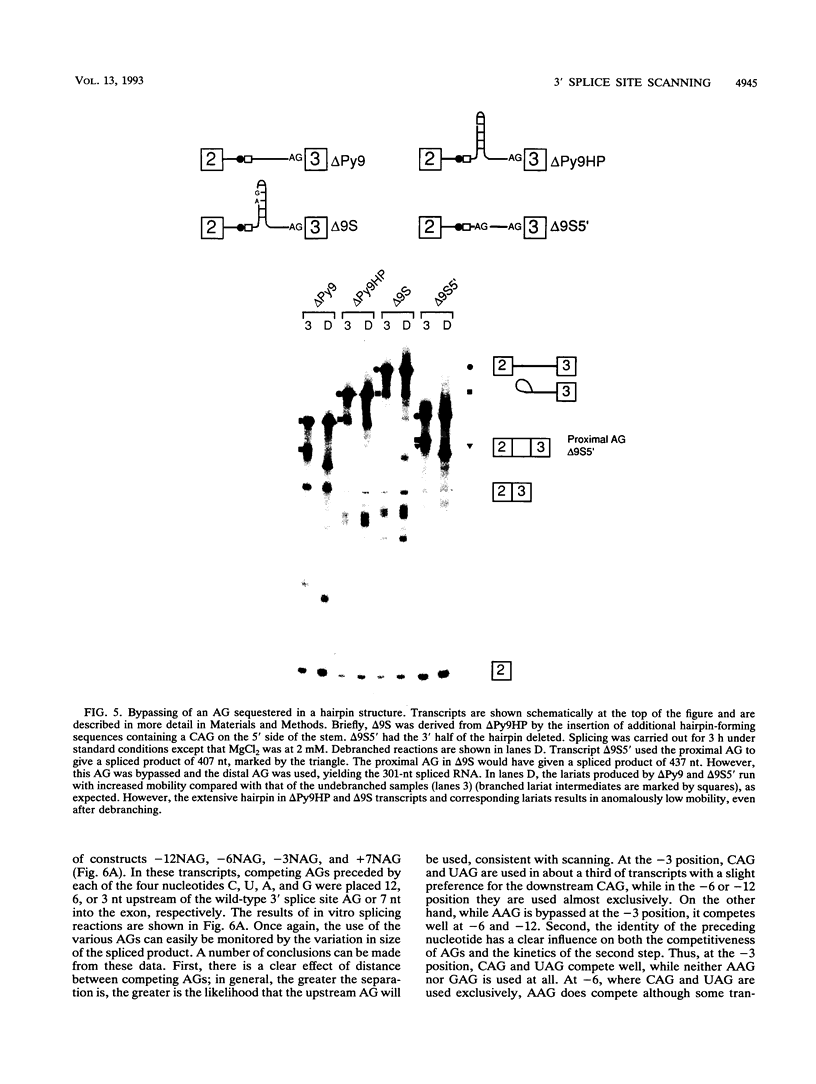
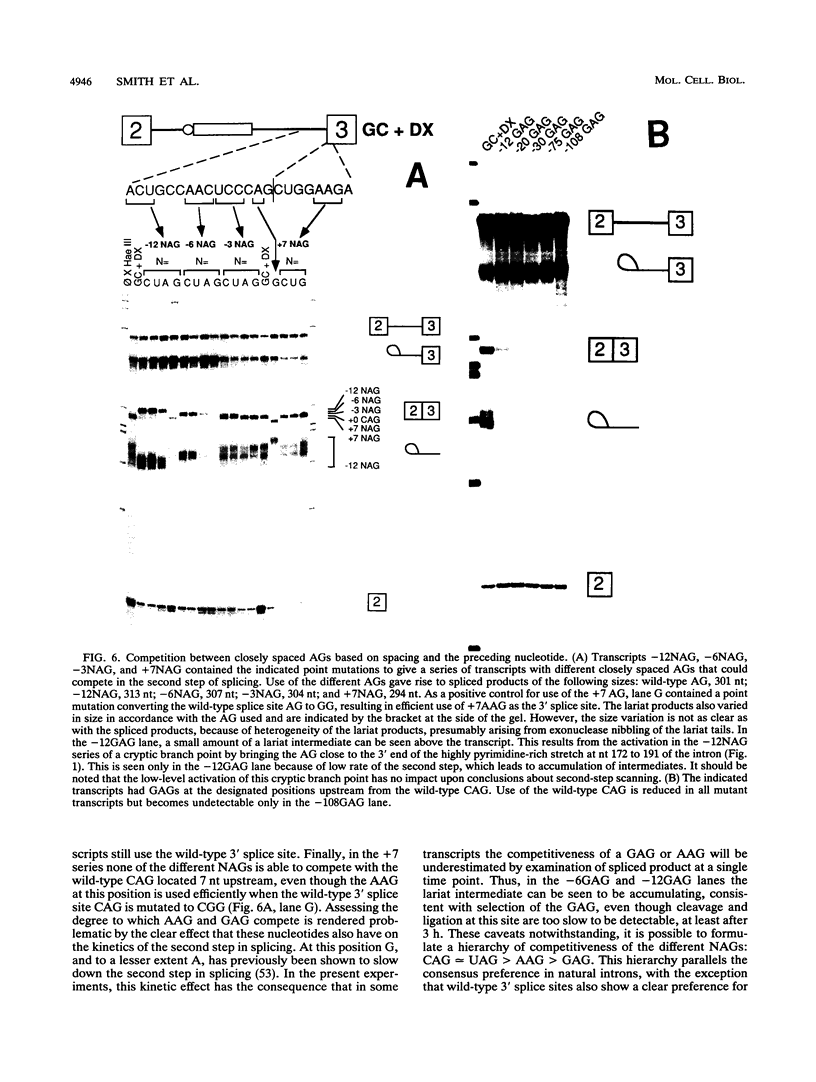
In mammalian intron splicing, the mechanism by which the 3' splice site AG is accurately and efficiently identified has remained unresolved. We have previously proposed that the 3' splice site in mammalian introns is located by a scanning mechanism for the first AG downstream of the branch point-polypyrimidine tract. We now present experiments that lend further support to this model while identifying conditions under which competition can occur between adjacent AGs. The data show that the 3' splice site is identified as the first AG downstream from the branch point by a mechanism that has all the characteristics expected of a 5'-to-3' scanning process that starts from the branch point rather than the pyrimidine tract. Failure to recognize the proximal AG may arise, however, from extreme proximity to the branch point or sequestration within a hairpin. Once an AG has been encountered, the spliceosome can still see a limited stretch of downstream RNA within which an AG more competitive than the proximal one may be selected. Proximity to the branch point is a major determinant of competition, although steric effects render an AG less competitive in close proximity (approximately 12 nucleotides). In addition, the nucleotide preceding the AG has a striking influence upon competition between closely spaced AGs. The order of competitiveness, CAG congruent to UAG > AAG > GAG, is similar to the nucleotide preference at this position in wild-type 3' splice sites. Thus, 3' splice site selection displays properties of both a scanning process and competition between AGs based on immediate sequence context. This refined scanning model, incorporating elements of competition, is the simplest interpretation that is consistent with all of the available data.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abmayr S. M., Reed R., Maniatis T. Identification of a functional mammalian spliceosome containing unspliced pre-mRNA. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7216–7220. doi: 10.1073/pnas.85.19.7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebli K., Gattoni R., Schmitt P., Hildwein G., Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989 Nov;9(11):4852–4861. doi: 10.1128/mcb.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Feng L., Donahue T. F. tRNAi(met) functions in directing the scanning ribosome to the start site of translation. Science. 1988 Oct 7;242(4875):93–97. doi: 10.1126/science.3051379. [DOI] [PubMed] [Google Scholar]
- Cook K. S., Groves D. L., Min H. Y., Spiegelman B. M. A developmentally regulated mRNA from 3T3 adipocytes encodes a novel serine protease homologue. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6480–6484. doi: 10.1073/pnas.82.19.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler J. O., Rossi J. J. Unexpected point mutations activate cryptic 3' splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 1991 Jul;5(7):1252–1263. doi: 10.1101/gad.5.7.1252. [DOI] [PubMed] [Google Scholar]
- Frank D., Patterson B., Guthrie C. Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol Cell Biol. 1992 Nov;12(11):5197–5205. doi: 10.1128/mcb.12.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Y., Colgan J. D., Manley J. L. Multiple cis-acting sequence elements are required for efficient splicing of simian virus 40 small-t antigen pre-mRNA. Mol Cell Biol. 1988 Sep;8(9):3582–3590. doi: 10.1128/mcb.8.9.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X. Y., Ge H., Manley J. L. The role of the polypyrimidine stretch at the SV40 early pre-mRNA 3' splice site in alternative splicing. EMBO J. 1988 Mar;7(3):809–817. doi: 10.1002/j.1460-2075.1988.tb02879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Blanco M. A., Jamison S. F., Sharp P. A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989 Dec;3(12A):1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- Ge H., Noble J., Colgan J., Manley J. L. Polyoma virus small tumor antigen pre-mRNA splicing requires cooperation between two 3' splice sites. Proc Natl Acad Sci U S A. 1990 May;87(9):3338–3342. doi: 10.1073/pnas.87.9.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H., Zuo P., Manley J. L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991 Jul 26;66(2):373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Gerke V., Steitz J. A. A protein associated with small nuclear ribonucleoprotein particles recognizes the 3' splice site of premessenger RNA. Cell. 1986 Dec 26;47(6):973–984. doi: 10.1016/0092-8674(86)90812-3. [DOI] [PubMed] [Google Scholar]
- Goux-Pelletan M., Libri D., d'Aubenton-Carafa Y., Fiszman M., Brody E., Marie J. In vitro splicing of mutually exclusive exons from the chicken beta-tropomyosin gene: role of the branch point location and very long pyrimidine stretch. EMBO J. 1990 Jan;9(1):241–249. doi: 10.1002/j.1460-2075.1990.tb08101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Pre-mRNA splicing. Annu Rev Genet. 1986;20:671–708. doi: 10.1146/annurev.ge.20.120186.003323. [DOI] [PubMed] [Google Scholar]
- Halfter H., Gallwitz D. Impairment of yeast pre-mRNA splicing by potential secondary structure-forming sequences near the conserved branchpoint sequence. Nucleic Acids Res. 1988 Nov 25;16(22):10413–10423. doi: 10.1093/nar/16.22.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M. Branch point selection in alternative splicing of tropomyosin pre-mRNAs. Nucleic Acids Res. 1989 Jul 25;17(14):5633–5650. doi: 10.1093/nar/17.14.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Roscigno R. F., Mulligan G. J., Finn L. A., Weber K. S. Identification of two distinct intron elements involved in alternative splicing of beta-tropomyosin pre-mRNA. Genes Dev. 1990 Jan;4(1):98–110. doi: 10.1101/gad.4.1.98. [DOI] [PubMed] [Google Scholar]
- Jackson R. J., Howell M. T., Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990 Dec;15(12):477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1986 May;83(9):2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. The scanning model for translation: an update. J Cell Biol. 1989 Feb;108(2):229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Metherall J. E., Collins F. S., Pan J., Weissman S. M., Forget B. G. Beta zero thalassemia caused by a base substitution that creates an alternative splice acceptor site in an intron. EMBO J. 1986 Oct;5(10):2551–2557. doi: 10.1002/j.1460-2075.1986.tb04534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen M. P., Smith C. W., Patton J. G., Nadal-Ginard B. Alpha-tropomyosin mutually exclusive exon selection: competition between branchpoint/polypyrimidine tracts determines default exon choice. Genes Dev. 1991 Apr;5(4):642–655. doi: 10.1101/gad.5.4.642. [DOI] [PubMed] [Google Scholar]
- Nakano T., Suzuki K. Genetic cause of a juvenile form of Sandhoff disease. Abnormal splicing of beta-hexosaminidase beta chain gene transcript due to a point mutation within intron 12. J Biol Chem. 1989 Mar 25;264(9):5155–5158. [PubMed] [Google Scholar]
- Newman A. J., Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992 Feb 21;68(4):743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Ohshima Y., Gotoh Y. Signals for the selection of a splice site in pre-mRNA. Computer analysis of splice junction sequences and like sequences. J Mol Biol. 1987 May 20;195(2):247–259. doi: 10.1016/0022-2836(87)90647-4. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G., Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987 Apr 24;49(2):229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- Patterson B., Guthrie C. A U-rich tract enhances usage of an alternative 3' splice site in yeast. Cell. 1991 Jan 11;64(1):181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- Patton J. G., Mayer S. A., Tempst P., Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991 Jul;5(7):1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5' noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985 Mar;40(3):515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Reed R. The organization of 3' splice-site sequences in mammalian introns. Genes Dev. 1989 Dec;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- Reich C. I., VanHoy R. W., Porter G. L., Wise J. A. Mutations at the 3' splice site can be suppressed by compensatory base changes in U1 snRNA in fission yeast. Cell. 1992 Jun 26;69(7):1159–1169. doi: 10.1016/0092-8674(92)90637-r. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Abelson J. Pre-mRNA splicing in yeast. Trends Genet. 1991 Mar;7(3):79–85. doi: 10.1016/0168-9525(91)90276-V. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Green M. R. Role of the 3' splice site consensus sequence in mammalian pre-mRNA splicing. Nature. 1985 Oct 24;317(6039):732–734. doi: 10.1038/317732a0. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Greene J. M., Green M. R. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985 Jul;41(3):833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Zamore P. D., Green M. R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988 Jan 29;52(2):207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Rosbash M. Cleavage of 5' splice site and lariat formation are independent of 3' splice site in yeast mRNA splicing. Nature. 1985 Oct 24;317(6039):735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- Rymond B. C., Torrey D. D., Rosbash M. A novel role for the 3' region of introns in pre-mRNA splicing of Saccharomyces cerevisiae. Genes Dev. 1987 May;1(3):238–246. doi: 10.1101/gad.1.3.238. [DOI] [PubMed] [Google Scholar]
- Schwer B., Guthrie C. PRP16 is an RNA-dependent ATPase that interacts transiently with the spliceosome. Nature. 1991 Feb 7;349(6309):494–499. doi: 10.1038/349494a0. [DOI] [PubMed] [Google Scholar]
- Shapiro M. B., Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987 Sep 11;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano P. G., Guthrie C. 5' splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev. 1988 Oct;2(10):1258–1267. doi: 10.1101/gad.2.10.1258. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Porro E. B., Patton J. G., Nadal-Ginard B. Scanning from an independently specified branch point defines the 3' splice site of mammalian introns. Nature. 1989 Nov 16;342(6247):243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- Spritz R. A., Jagadeeswaran P., Choudary P. V., Biro P. A., Elder J. T., deRiel J. K., Manley J. L., Gefter M. L., Forget B. G., Weissman S. M. Base substitution in an intervening sequence of a beta+-thalassemic human globin gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2455–2459. doi: 10.1073/pnas.78.4.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. H., Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991 Jan 25;64(2):459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- Swanson M. S., Dreyfuss G. RNA binding specificity of hnRNP proteins: a subset bind to the 3' end of introns. EMBO J. 1988 Nov;7(11):3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Kretzner L., Rosbash M. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5' cleavage site. EMBO J. 1988 Aug;7(8):2533–2538. doi: 10.1002/j.1460-2075.1988.tb03101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi J., Alibert C., Temsamani J., Reveillaud I., Cathala G., Brunel C., Jeanteur P. A protein that specifically recognizes the 3' splice site of mammalian pre-mRNA introns is associated with a small nuclear ribonucleoprotein. Cell. 1986 Dec 5;47(5):755–766. doi: 10.1016/0092-8674(86)90518-0. [DOI] [PubMed] [Google Scholar]
- Ulfendahl P. J., Kreivi J. P., Akusjärvi G. Role of the branch site/3'-splice site region in adenovirus-2 E1A pre-mRNA alternative splicing: evidence for 5'- and 3'-splice site co-operation. Nucleic Acids Res. 1989 Feb 11;17(3):925–938. doi: 10.1093/nar/17.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U., Abelson J. PRP18, a protein required for the second reaction in pre-mRNA splicing. Mol Cell Biol. 1990 Jan;10(1):324–332. doi: 10.1128/mcb.10.1.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman D. A., Steitz J. A. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science. 1992 Sep 25;257(5078):1918–1925. doi: 10.1126/science.1411506. [DOI] [PubMed] [Google Scholar]
- Wu J., Manley J. L. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev. 1989 Oct;3(10):1553–1561. doi: 10.1101/gad.3.10.1553. [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Green M. R. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in U1 snRNA suppresses a 5' splice site mutation. Cell. 1986 Sep 12;46(6):827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Weiner A. M. A compensatory base change in human U2 snRNA can suppress a branch site mutation. Genes Dev. 1989 Oct;3(10):1545–1552. doi: 10.1101/gad.3.10.1545. [DOI] [PubMed] [Google Scholar]