During mitosis, cohesin- and condensin-based pericentric chromatin loops function as a spring network to balance spindle microtubule force.
Abstract
The mechanisms by which sister chromatids maintain biorientation on the metaphase spindle are critical to the fidelity of chromosome segregation. Active force interplay exists between predominantly extensional microtubule-based spindle forces and restoring forces from chromatin. These forces regulate tension at the kinetochore that silences the spindle assembly checkpoint to ensure faithful chromosome segregation. Depletion of pericentric cohesin or condensin has been shown to increase the mean and variance of spindle length, which have been attributed to a softening of the linear chromatin spring. Models of the spindle apparatus with linear chromatin springs that match spindle dynamics fail to predict the behavior of pericentromeric chromatin in wild-type and mutant spindles. We demonstrate that a nonlinear spring with a threshold extension to switch between spring states predicts asymmetric chromatin stretching observed in vivo. The addition of cross-links between adjacent springs recapitulates coordination between pericentromeres of neighboring chromosomes.
Introduction
The physical basis of the chromatin spring
The mitotic spindle ensures the equal distribution of chromosomes during cell division. Sister chromatids are bioriented through binding via the kinetochore, to microtubules emanating from opposite spindle poles (Fig. 1 A). The kinetochore is a specialized protein/DNA structure built on centromere DNA that binds to the plus end of dynamically growing and shortening kinetochore microtubules (kMTs). In yeast, each chromosome is tethered to the spindle via a single kMT (Peterson and Ris, 1976; O’Toole et al., 1999). The kinetochore promotes the formation of a pericentric chromatin loop (Yeh et al., 2008), predisposing the kinetochore to protrude from the surface of the chromosome. Cohesin and condensin complexes are enriched in ∼50 kb of the pericentric chromatin (Blat and Kleckner, 1999; Megee et al., 1999; Tanaka et al., 1999; Hagstrom et al., 2002; Wang et al., 2005), which together with DNA constitute the “chromatin spring” to counter microtubule-based forces (Bouck and Bloom, 2007; Stephens et al., 2011). Interpolar microtubules (ipMTs) from opposite spindle poles overlap and are cross-linked by microtubule motor proteins and microtubule-associated proteins. Kinesins act as force production machines that slide the ipMTs apart, generating an extensional force on the spindle pole bodies (Saunders and Hoyt, 1992; Saunders et al., 1997). The balance of microtubule-based extensional force and a chromatin spring contractile force is necessary to produce a steady-state spindle length and tension at the kinetochore that satisfies the spindle checkpoint (Bloom and Yeh, 2010).
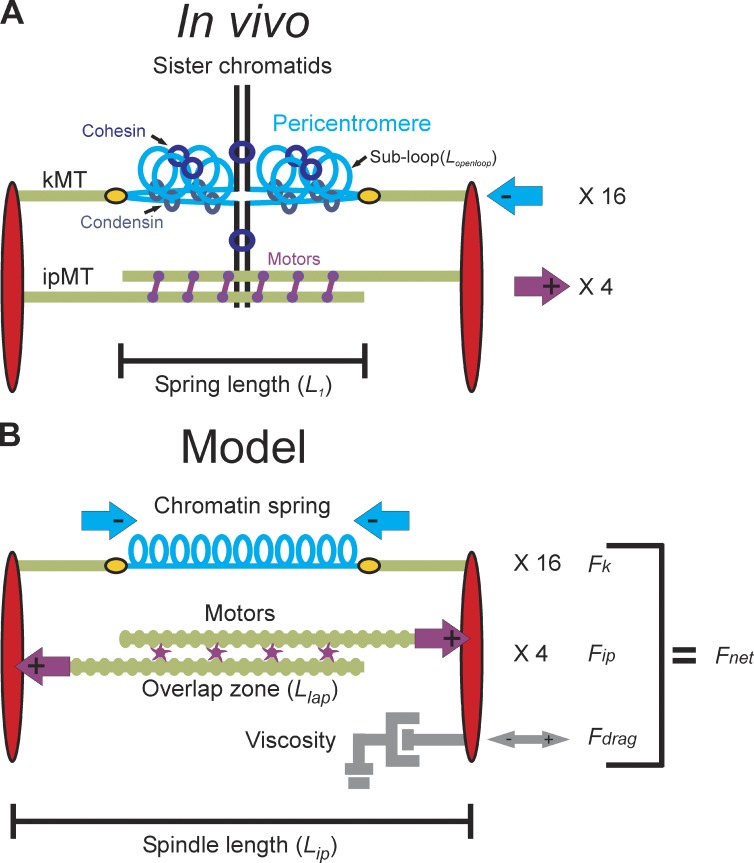
Figure 1.
Structure of the yeast mitotic spindle. (A) Microtubules (kMTs, green) emanating from opposite spindle pole bodies (red) bind to the centromere via the kinetochore (yellow). Sister centromeres are spatially separated in metaphase and reside at the apex of a pericentric chromatin loop (in blue) that extends perpendicularly from the chromosome axis (black; Yeh et al., 2008). The total contour length of the pericentric chromatin loop is split between an axial component (approximated by the distance between the two kinetochores, L1) and subloops (Lopenloop) that extend perpendicular to the spindle axis. Condensin is more proximal to the spindle axis than cohesin. Approximately eight ipMTs overlap (two shown) and are bound by kinesin 5 motor proteins (purple). Although only one replicated chromosome is depicted with two kMTs, there are 16 chromosomes (32 sister chromatids) in budding yeast and ∼32 kMTs. The 16 kinetochores from each pole are clustered in mitosis. The aggregate chromatin spring length is measured by the distance between the two clusters (L1). (B) The model is written as a coupled system of stochastic and deterministic differential equations in which the sum of the forces applied to one spindle pole body is used to numerically solve for velocity at each time step. Spindle length is defined experimentally as the distance between the spindle pole bodies (red) in metaphase (Lip). The pericentric chromatin functions as a spring (blue, Fk). Its length is the distance between two sister kMT plus ends (Lspring). Kinesin motors (purple) bind to and couple ipMTs at the overlap zone (Llap) and slide ipMTs apart, generating an outward extensional force, Fip. The viscous properties of the nucleus are represented as a dashpot and resist movement of the spindle pole bodies in either direction (gray, Fdrag).
The microtubules, microtubule-based motor proteins, and kinetochore components of the segregation apparatus have been explored with biophysical techniques, leading to a detailed understanding of their function (Mogilner and Craig, 2010; Alushin and Nogales, 2011; McIntosh et al., 2012; Umbreit and Davis, 2012; Watanabe, 2012). Mathematical models of the metaphase spindle in several systems aim to account for the distribution and dynamics of spindle microtubules (Sprague et al., 2003; Cytrynbaum et al., 2005; Gardner et al., 2005; Wollman et al., 2005; Civelekoglu-Scholey et al., 2006; Gay et al., 2012). We and others have developed stochastic models of kMT plus-end dynamics in the metaphase spindle that were evaluated by statistical measures of how well simulations predicted experimentally observed distributions of fluorescent kinetochore proteins (Sprague et al., 2003; Gardner et al., 2005, 2007, 2010). In these models, introduction of spatial gradients in dynamic instability across the spindle, as well as tension-mediated regulation of kMT plus-end dynamics, was required to fit experimental data (Gardner et al., 2005). A stochastic model that includes kinetochore attachment and detachment accurately recapitulates chromosome dynamics during metaphase and anaphase as well as the timescale for correction of erroneous attachments in Schizosaccharomyces pombe (Gay et al., 2012). Although, microtubule dynamics were not explicitly modeled, the Gay et al. (2012) model incorporated a spatial gradient in kMT detachment rate (inspired by the Aurora B spatial gradient in vertebrate cells; Liu et al., 2009; Welburn et al., 2010) that is analogous to the spatial gradients of Gardner et al. (2005). Overall, with appropriate tuning of the spatial gradients, these models were able to recapitulate experimentally observed features of microtubule plus-end fluorescence (Gardner et al., 2005), kinetochore separation, and laser ablation (Gay et al., 2012). However, none of the models explicitly consider the physical properties of the chromatin spring (Mogilner and Craig, 2010).
Simple models have assumed that chromatin behaves as a Hookean (i.e. linear force–extension relation) spring. This spring is presumed to be derived via cohesion between sister chromatids (Gay et al., 2012), cohesin and condensin-based chromatin loops (Ribeiro et al., 2009; Stephens et al., 2011), or an entropic wormlike chain (Greulich et al., 1987).
Here, we develop models for the chromatin spring based on experimental observations in budding yeast. Several features of budding yeast make it particularly amenable for a quantitative analysis of the contributions of chromatin to spindle structure and dynamics. The mitotic spindle is streamlined and stereotypic (250-nm-diameter × 1,400–1,600-nm-length cylinder), providing the opportunity for statistical analysis of length and its variation with time (Winey and Bloom, 2012). Condensin and cohesin are enriched in the pericentric chromatin and occupy unique spatial positions relative to the spindle microtubules and kinetochore (Stephens et al., 2011). Condensin lies proximal to the spindle axis and is responsible for axial compaction by looping pericentric chromatin, whereas cohesin is radially displaced from the spindle axis and confines pericentric chromatin loops around the spindle apparatus (Fig. 1 A; Stephens et al., 2011). Through the use of integrated lactose operator (LacO) arrays and LacI-GFP, we can visualize the chromatin at known distances (in base pairs) from the microtubule attachment site defined by a 125-bp DNA sequence. A linear spring in a predictive mathematical model fails to predict spindle length and chromatin dynamics. Cross-linked chromatin loops with nonlinear length changes provide a working model for the nature of the chromatin spring in mitosis.
Results
Force balance within the budding yeast mitotic spindle
The components contributing to force balance in the mitotic spindle are depicted in Fig. 1 B. We consider three main force-generating processes: (1) an extensional force arising from motors sliding ipMT, Fip; (2) an opposing contractile force generated by the chromatin spring, Fk; and (3) a viscous drag force, Fdrag. Other forces acting on the spindle and not considered in the model are described in Materials and methods. At the low Reynolds numbers inside the cell, the Fdrag is proportional to the velocity of the spindle length (denoted Lip) given by Stokes’ law (Batchelor, 1967), Fdrag = −Cdrag Vip, in which Vip = dLip/dt. Holding one spindle pole fixed and summing all forces acting along the primary spindle axis, we obtain the net force on the moveable spindle pole,
Based on a linear force–velocity relation, as explained in the supplemental material, we arrive at a dynamical equation for spindle length,
| (1) |
in which Vmax is the maximum velocity of the spindle motors (Table S1, Spindle velocity). This force balance model is written as a coupled system of stochastic and deterministic differential equations in which the sum of the forces applied to one spindle pole body yields the dynamics of the spindle length (Lip), through integration of Eq. 1.
The basis for a stable spindle length in our model is that a quasi–steady state is reached between inward and outward forces, with fluctuations about a mean spindle length arising from microtubule-based motor activity. This mean length is dictated by the mean number of force-producing motors in the overlap zone and the corresponding extension of the chromatin springs from their rest length, which give Fip and Fk, respectively (Fig. 1). The outward force (Fip) arises from sliding of antiparallel ipMTs caused by plus end–directed motors bound in the overlap region (denoted double-bound motors, D(t)). The forces exerted by each motor, Fm, are additive, so Fip is proportional to the total number of double-bound motors, Fip = D(t) × Fm. The number of productive motors D(t) is given by rates of attachment and detachment, Kon and Koff, so that D(t) fluctuates with a Poisson distribution. The length of the chromatin spring Lsp is the total spindle length Lip minus the length of each kMT , in which and are the length of the left and right kMT, respectively. Thus, the chromatin restoring force is in which Lrest is the rest length of the spring in the absence of force. Lrest is estimated to be ∼200 nm, based on an upper bound given by the diffraction limit ∼250 nm (as sister centromere-linked LacO arrays are not resolved after spindle collapse) and a lower bound given by the chromatin persistence length of 170–220 nm (Bystricky et al., 2004). With these assumptions, Eq. 1 becomes
A linear spring model recapitulates the increase in both the mean and variance of spindle length observed in mutant cells
We first perform numerical simulations with a minimal linear stochastic (MLS) model, in which LkMT(t) and D(t) are described, respectively, by Gaussian and Poisson distributions. The model predicts increases in mean and variance of spindle length upon decreasing the linear chromatin spring constant (Fig. S1, E and F) consistent with experimental observations (Stephens et al., 2011). However, the MLS model fails to capture features such as the spatial distribution of kMTs (Gardner et al., 2005) and asymmetry in the behavior of the chromatin spring (Stephens et al., 2011). Therefore, unlike the situation in certain transcriptional networks (Munsky et al., 2012) or the bimodal response to extracellular signaling (Birtwistle et al., 2012; Kim and Sauro, 2012), our results demonstrate that noise is insufficient to enhance our understanding of mitotic spindle behavior.
We introduced a spatial catastrophe gradient and tension-dependent rescue of kMT plus-end dynamics (Materials and methods; Eqs. 3 and 4). This new model, termed the coupled linear stochastic (CLS) model, couples microtubule and spring dynamics. As the chromatin spring lengthens, tension on the kMT increases, switching the MT from shortening to growth (rescue), whereas the growing kMT allows the chromatin spring to shorten, reducing tension. We incorporated experimentally measured motor on and off rates and distributed the total number of motors among four species (unbound, kMTs, ipMTs, and double bound in the interpolar zone; see Materials and methods). The interpolar overlap zone (Llap) is the region where antiparallel ipMTs from each spindle pole are bundled (Winey et al., 1995; O’Toole et al., 1999). The extensional force (Fip) depends on the number of kinesin motors bound to antiparallel microtubules (Llap). Parameter values are detailed in Table S1. Because Ase1 binds anti- and parallel microtubule bundles (Janson et al., 2007), this provides an upper estimate on overlap zone length. To estimate Llap, we introduced Ase1-GFP in cells with labeled spindle pole bodies (Spc29-RFP; Fig. S3). The distribution of Ase1-GFP increases linearly with spindle length in metaphase (slope of 0.45; Fig. S3 A). Using either constant or length-adjusted Llap does not affect the trends in spindle length and variation (Figs. S3 and S4). We used the constant overlap zone in the model based on the robustness of the sensitivity analysis (Figs. S4 and S5 and Table S3).
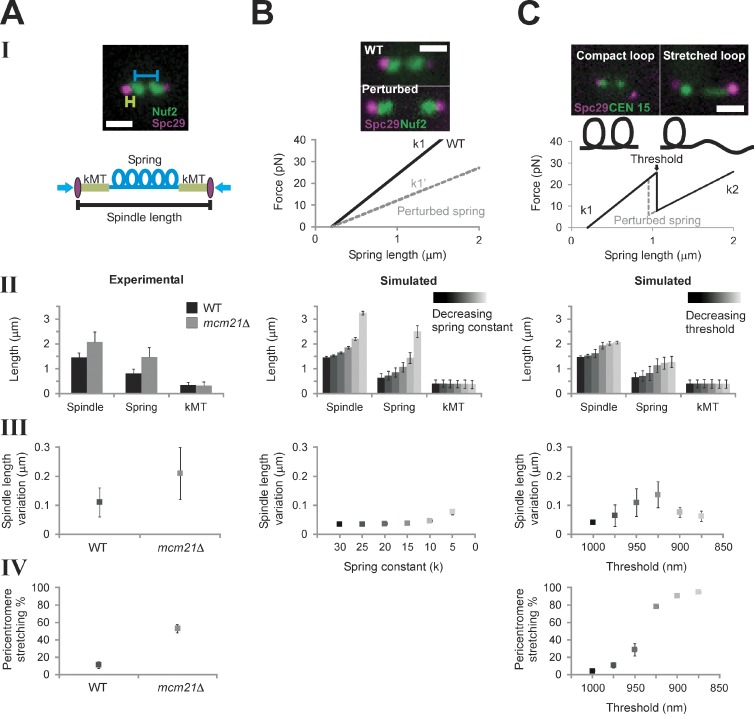
The physical behavior of the pericentric chromatin depends on the concentration of cohesin and condensin complexes. Reduction of pericentric cohesin (mcm21Δ) or condensin (brn1-9) leads to increased spindle length and variation (Fig. 2 A, II and III; Stephens et al., 2011). Variation in spindle length refers to fluctuations about the mean over a time course of observation. In the model, mcm21Δ is captured by decreasing the chromatin spring constant (Fig. 2 B, I; and Table 1). A decreased (softer) spring in the CLS model leads to an increased mean spindle length and greater variance in spindle length, (Fig. 2 B, II and III). Introduction of experimentally measured motor on/off rates and kMT dynamics and 16 individual springs did not alter the trend from the minimal model (Fig. S1, E and F). Increases in spindle length and variation upon depletion of pericentric cohesin or condensin can be recapitulated through decreasing the linear chromatin spring constant in both a minimal (MLS) and coupled model (CLS).
Figure 2.
Model simulations with a piecewise continuous spring recapitulate experimental observations. (A) Experimental data of WT and mcm21Δ cells (pericentromere depleted of cohesin). Spindle length (II) and pericentromere stretching (IV) were measured using population images of cells containing Spc29-RFP spindle poles and LacO at CEN15, respectively. Spindle length variation (III, fluctuation about the mean spindle length) was measured by tracking spindle poles over ∼10 min (Stephens et al., 2011). (B) Numerical simulations of spindle dynamics with a linear chromatin spring F = −k (Lspring − Lrest), in which Lrest = 200 nm (lighter = decreasing spring constant). The spring constant (k) was decreased to simulate perturbation of the spring (see B, I). Experimental images display an increase in the interkinetochore distance (distance between kinetochore clusters, Nuf2) in cohesin/condensin mutants. Simulations using a Hookean spring do not generate asymmetric pericentromere stretching. (C) Simulations of spindle dynamics with the spring defined by a piecewise continuous equation dependent on a threshold (lighter = decreasing threshold). (top) Experimental images reveal two spring states (compact, stretched) of a pericentromere LacO array. Under a force threshold (Lthreshold) the spring is looped (k1 and Lrest1, compact), and above the threshold, a loop stretches, adding length to the spring, which decreases the spring constant and increases the rest length (k2 and Lrest2, stretched). When Lspring < Lthreshold, F = −k1 (Lspring − Lrest). At Lspring ≥ Lthreshold, the spring constant is reduced from k1 to k2 (k2 = k1 (L1/L1 + Lopenloop)) and greater rest length (Lrest2 = Lrest1 + Lopenloop), giving F = −k2 (Lspring − Lrest2), in which the mean experimental aggregate spring length is L1 = 800 nm and Lopenloop = 450 nm (10 kb of nucleosomal chromatin from the stretched loop). (B and C) Simulated population measurements for linear (B) and nonlinear (C) spring models (spindle length [II] and pericentromere stretching [IV]) were generated by running the model and randomly selecting one time step [n = 500, five groups, 100 simulations each]). Simulated time lapses (III) were run for 1,000 s, 50 s for equilibrating, and 950 s measured (n = 25). Bars, 1 µm. Error bars represent standard deviation.
Table 1.
Comparison of model predictions and experimental outcomes
| Attributes | Experimental | Minimal linear stochastic | Coupled linear stochastic | Coupled nonlinear stochastic | Coupled nonlinear stochastic + cross-links |
| Outward motor force | Poisson | On/off rates | On/off rates | On/off rates | |
| Inward spring force | Hookean | Hookean | Piecewise continuous | Piecewise continuous | |
| kMT dynamics | Gaussian | Gardner et al., 2005 | Gardner et al., 2005 | Gardner et al., 2005 | |
| Spring network | Individual | Individual | Individual | Cross-linked network | |
| Measurable attributes | Perturbation of the chromatin spring | ||||
| Spindle length | Increases | Increases | Increases | Increases | Increases |
| Spindle variation | Increases | Increases | Increases | Increases | Increases |
| Chromatin stretching | Increases | NM | NM | Increases | Increases |
| Kinetochore declustering | Increases | NM | Constant | Increases | Increases |
| Coordinated stretching | ~40% WT | Not predicted | Not predicted | Not predicted | Predicted |
NM, not measureable.
A nonlinear spring hypothesis
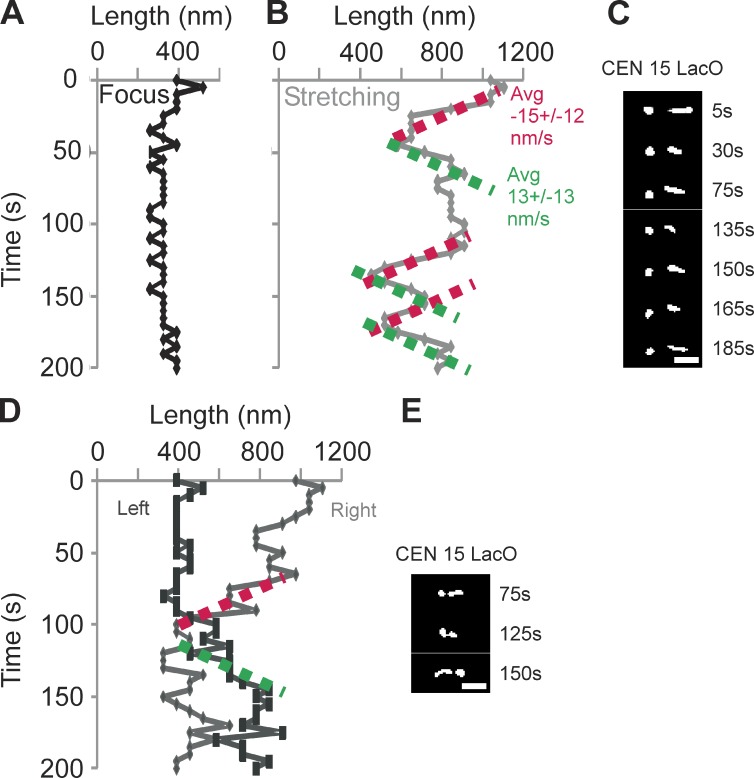
During metaphase, sister LacO-LacI GFP arrays inserted proximal to the centromere appear as diffraction-limited spots that transiently separate into two foci upon biorientation (Goshima and Yanagida, 2000; He et al., 2000; Tanaka et al., 2000; Pearson et al., 2001). These LacO arrays can switch between a focus and a linear filament (∼450-nm length, nucleosome fiber packing) indicative of decompaction and extension (Figs. 2 C and 3; He et al., 2000; Bachant et al., 2002; Warsi et al., 2008). If the pericentric chromatin were a simple linear spring as in the CLS model, sister chromatid LacO arrays would experience the same force and stretch to the same degree. For wild type (WT), 10% of cells exhibit chromatin stretching. However, only one of the sister LacO arrays stretches (asymmetric stretching: WT 100%, n = 102; mcm21Δ 90%, n = 59; brn1-9 95%, n = 43; e.g., Figs. 2 C and 3; Stephens et al., 2011). In the stretched state, LacO arrays occupy a spindle proximal position relative to the compacted foci (Stephens et al., 2011). The changes in compaction and spatial position provide evidence for dynamic chromatin loops not accounted for by linear spring models (MLS or CLS).
Figure 3.
Rate of pericentromere chromatin stretching and recompaction. Time-lapse microscopy of sister pericentric CEN15 LacO arrays in mcm21Δ cells. Images were taken every 5 s for 200 s. Images were deconvolved as described in the Materials and methods, to determine the length of a LacO array along the spindle axis. (A–C) A representative time lapse shows stretching (line) and compaction (foci) of the LacO array over time. (A) Change in length of the left sister LacO array as a function of time in a single cell. The left LacO array appears predominantly as a spot and is slightly larger than the diameter of a diffraction spot. (B) Length of the right sister LacO array as a function of time. To determine a mean compaction and stretching rate, we fit a linear slope to regions that displayed greater than three successive steps in one direction. The mean compaction and stretching rates are −15 ± 12 nm/s (n = 9) and 13 ± 13 nm/s (n = 9), respectively, for six cells. (C) Selective images from the time lapse of sister LacOs A and B. The time point is indicated to the right. (D) Example from a time lapse indicating that sister chromatid stretching can switch from side to side. The right LacO array condenses as the left LacO array commences stretching. (E) Selective images from D. The time point is indicated to the right. Bars, 1 µm.
To account for the asymmetry in chromatin extension, we model chromatin loops (Fig. 2 C, I; Stephens et al., 2011) as a piecewise continuous function that exhibits a sawtooth force versus extension law. At a characteristic threshold length, a loop unravels, switching the spring constant (Fig. 2 C, I). In the stretched state, the increase in available chromatin (L1 + Lopenloop = L2) is compensated by a decrease in spring constant, such that k2 = k1 × (L1/L2) to maintain force (F = −k (L − Lrest)). The aggregate experimental WT spring length is 800 nm (L1; Fig. 1 A). The length of chromatin added upon triggering the threshold condition is estimated to be 450 nm (Lopenloop). The length reflects the nucleosomal contour length of the subloops (10 kb of pericentric chromatin, ∼450 nm of 11-nm fiber, and L2 = 800 + 450 = 1,250 nm; Fig. 1 A and Table S1). Furthermore, we consider an increase in the chromatin rest length L1rest + Lopenloop = L2rest, in agreement with experimental evidence that depletion of pericentric cohesin or condensin affects pericentric sister LacO separation in the absence of tension (Lam et al., 2006; Vas et al., 2007; Ng et al., 2009). Unlike previous models, this coupled nonlinear stochastic (CNLS) model contains a piecewise continuous spring that accounts for dynamically extensible asymmetry in the chromatin spring.
The nonlinear spring model recapitulates the frequency of chromatin stretching and increase in both the mean and variance of spindle length observed in mutant cells
To estimate the threshold at which the spring stretches, we explored model values that best fit the frequency of stretching observed in vivo. The mean distance between paired kMTs is ∼800 nm in WT cells. In simulations, we explored parameter values of spring extension threshold between 800 and 1,100 nm. A threshold of 975 nm reproduces the frequency of ∼10% stretching in WT cells (Fig. 2, A [IV] and C [IV]). Cells depleted of pericentric cohesin or harboring temperature-sensitive mutants in condensin have longer spindles in metaphase, increased spindle length variance, and increased frequency of chromatin stretching (Fig. 2 A). The distance at which chromatin stretching is observed therefore depends on the concentration of cohesin and condensin within the spindle (Stephens et al., 2011). As the spring threshold is reduced, there is an increased frequency of chromatin stretching in the CNLS model (Fig. 2 C, IV). Decreasing the threshold in the model to 925–950 nm best matches the frequency of stretching observed in mutant cells depleted of cohesin or condensin (∼50% stretching).
Simulations with the CNLS model show an increase in spindle length variation with increases in the fraction of stretched chromatin strands (variation 65–135 nm and 10–50% stretching; Fig. 2 C, III and IV). When all 16 springs are in the same conformational state (all k1 or k2), the variance of the spindle length is minimal. Similarly, the maximum variance occurs when roughly half of the springs are in a given state (Fig. 2 C, III and IV). In experiments, populations of WT spindles (∼10% stretching) have reduced length variance relative to mutant populations in which ∼50% of cells show pericentric chromatin stretching (pericentric cohesin and condensin mutants; Fig. 2 A, III and IV). Experimental observations of spindle length distributions in cells with compact versus stretched LacO arrays reveal two distinct populations in both WT and mutant cells (WT, P < 1.5 × 10−11; mcm21Δ, P < 1.3 × 10−10; brn1-9, P < 1 × 10−6; Fig. S2, F–H). This is also evident from fitting the probability distribution functions of spindle lengths in WT, mcm21Δ, and brn1-9 with Gaussian distributions (Fig. S2, C–E; and Table S2). Although fittings to the CLS data with one or two Gaussians were indistinguishable from each other, introducing a second Gaussian consistently improved the goodness of fit for the CNLS data (Table S2, R2 values). The multiplicities of modes in the CNLS model reflect the stretching process through the nonlinear spring law.
Dynamics of chromatin stretching reflect the rate of microtubule depolymerization
The dynamics of chromatin stretching provides a measure of kinetic coupling to microtubule growth and shortening. Chromatin stretching is not instantaneous but occurs within a timescale of 15 s to minutes (Fig. 3). In the model, kMT depolymerization is limited by the shortening rate (1.5 µm/min). Upon an instantaneous change in spring constant, the tension on kMTs is reduced, and the microtubules switch to the shortening state (catastrophe). To determine whether the rate of pericentric chromatin extension and compaction is predicted by kMT dynamics, we imaged cells depleted of pericentric cohesin (mcm21Δ) with LacO 1.8 kb from the centromere at 5-s intervals (Fig. 3). The rate of chromatin extension (13 nm/s or 0.8 ± 0.8 µm/min, n = 9 from six cells) and compaction (15 nm/s or 0.9 ± 0.7 µm/min, n = 9 from six cells) is on the same timescale of microtubule shortening and growth, respectively (∼1–1.5 µm/min; Fig. 3; Carminati and Stearns, 1997; Gardner et al., 2005). The CNLS model recapitulates the observed rate of change in chromatin shape. Although the mechanisms of chromatin extension and compaction are likely to be more complex than stretching and recoil of a random coil described herein, the rates of microtubule growth and shortening are consistent with the observed rate of change from spot to stretched LacO array in the pericentric chromatin. In addition, it is likely that microtubule shortening along the spindle axis pulls the LacO arrays proximal to the spindle axis where the chromatin continues to stretch as kMTs shorten (Stephens et al., 2011).
Decreasing the spring length threshold predicts declustering of kMT plus ends
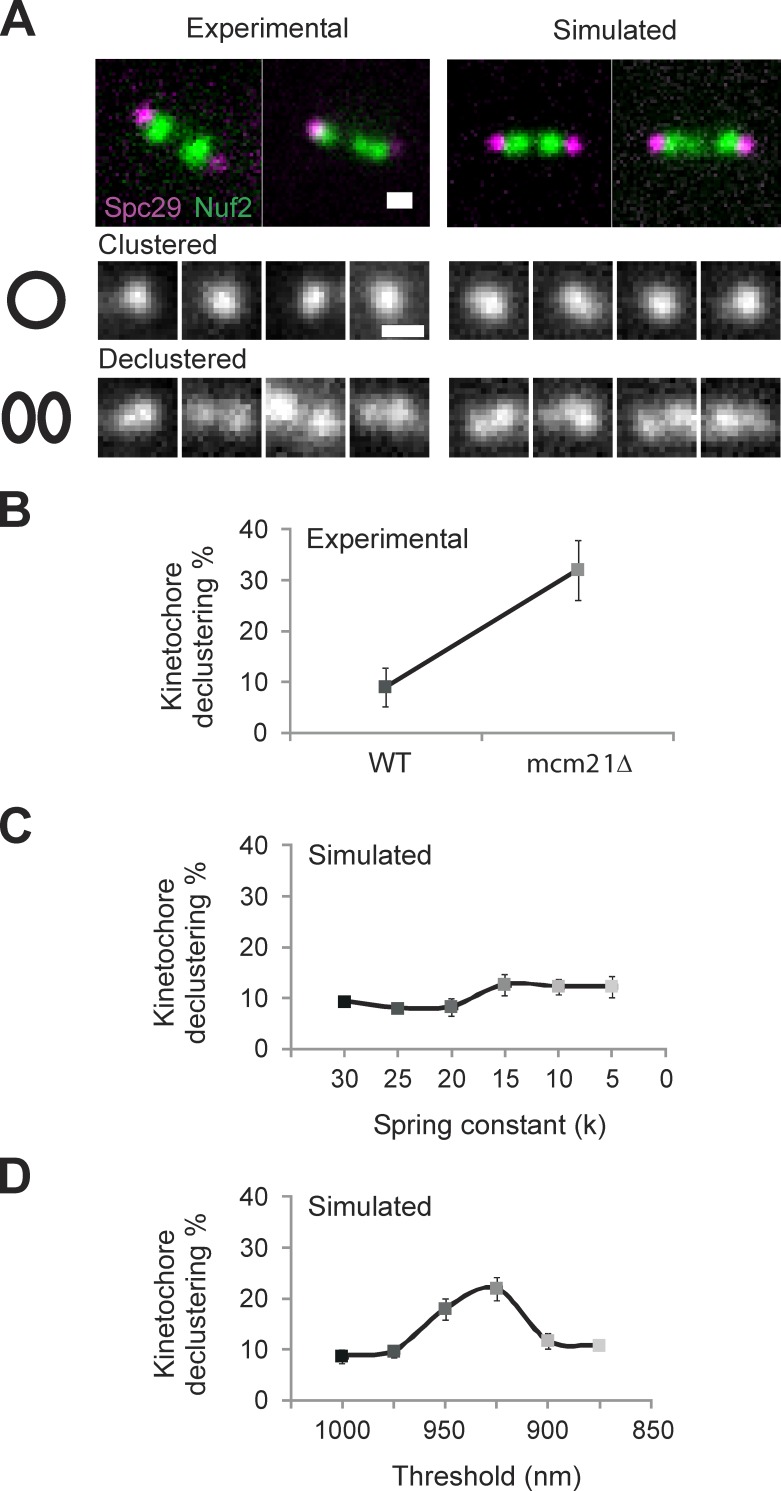
The 32 yeast kinetochores and kMT plus ends, 16 on each side, are clustered into two foci each slightly larger than a diffraction-limited spot in metaphase (Haase et al., 2012). Based on the stochastic growth and shortening of kMTs with no other spatial information, there is no a priori reason to expect that 16 individual microtubules emanating from the spindle pole will have the same length. In previous models, spatial gradients for microtubule dynamics (Gardner et al., 2005) or Aurora gradients for kinetochore detachment (Gay et al., 2012) have been invoked. These gradients provide plausible mechanisms for kMT clustering. Alternatively, mechanisms to cluster individual kinetochore proteins or pericentromeric chromatin could contribute to kMT length control in the spindle. Depletion of histone H3 or pericentric cohesin (mcm21Δ) has been shown to result in the declustering of 16 kinetochores (Ng et al., 2009; Verdaasdonk et al., 2012). Herein, we examined the clustering of the inner (Ame1p-GFP) and outer (Ndc80-GFP or Nuf2p-GFP) kinetochore (Fig. 4 A, top, Nuf2). There is a significant disruption of kinetochore clusters in experimental conditions that increase chromatin stretching (mcm21Δ: 32 ± 6% declustered, n = 230; vs. WT: 9 ± 4%, n = 209; Fig. 4 A). The physical basis for kinetochore declustering in these mutant conditions is unknown.
Figure 4.
Experimental kinetochore declustering is predicted by simulations with a piecewise continuous chromatin spring (CNLS) but not a linear spring (CLS). (A) Experimental and simulated images of WT and mcm21Δ cells with Spc29-RFP (spindle poles) and Nuf2 or Ndc80-GFP (kinetochores) were scored as clustered (kinetochore focus) or declustered in cells having the focus of 16 kinetochores split into multiple foci. Example experimental (left) and simulated (right) images showing a bundle of clustered (middle) and declustered (bottom). (B) Experimental declustering in WT and mcm21Δ cells (WT: 9 ± 4%, n = 209, two experiments; mcm21Δ: 32 ± 6%, n = 230, two experiments). (C and D) Model simulations were used to generate images that match the physical geometry of the mitotic spindle (model convolution; Quammen et al., 2008; Gardner et al., 2010). The position of the spindle poles and plus end of each kMT were convolved with the point-spread function of our microscope objective to produce a simulated image of spindle poles and clusters of kinetochore proteins at the microtubule plus ends. Declustering was scored using the same criteria as in experimental images (n = 300, three groups of 100). (C) Decreasing the linear spring constant by an order of magnitude results in an insignificant increase (9–12%, χ2 > 0.30) in kinetochore declustering. (D) Decreasing the threshold of a piecewise continuous spring results in a significant increase in declustering (10–22%, χ2 < 1 × 10−4) comparable to experimental (B vs. D). Bars, 0.5 µm. Error bars represent standard deviation.
To compare predictions on kinetochore clustering from the coupled linear (CLS) and nonlinear (CNLS) model with respect to experimental observations, we convolved the position of kMT plus ends from the model with the point-spread function of our microscope objective: a process known as model convolution (Sprague et al., 2003; Gardner et al., 2010). The power of model convolution lies in the ability to perform a statistical comparison between multiple runs of the model and multiple experimental images (Gardner et al., 2007, 2010). To determine whether perturbation of a linear chromatin spring influences kMT plus-end clustering, we reduced the spring constant of the chromatin spring in the CLS model. This results in an insignificant change in kinetochore declustering over an order of magnitude change in spring constant (k = 30 pN/µm, 9 ± 1%; vs. k = 5 pN/µm, 12 ± 3%; χ2 > 0.30; Fig. 4 C). This is consistent with observed kinetochore clustering upon removal of the chromatin spring by preventing DNA replication via cdc6 (Gardner et al., 2005). In contrast, when the chromatin stretching threshold is decreased (CNLS model), the incidence of declustering increases to experimentally observed levels (10 ± 1% to 22 ± 4%; χ2 < 1 × 10−4; Fig. 4, B and D). In the CNLS model, declustering arises because of the nonlinear behavior of 16 independent springs in the system. Thus, unlike simpler chromatin spring models, the discontinuous spring predicts kinetochore declustering in WT and mutant strains as well as spindle length variation and asymmetric chromatin stretching (Table 1).
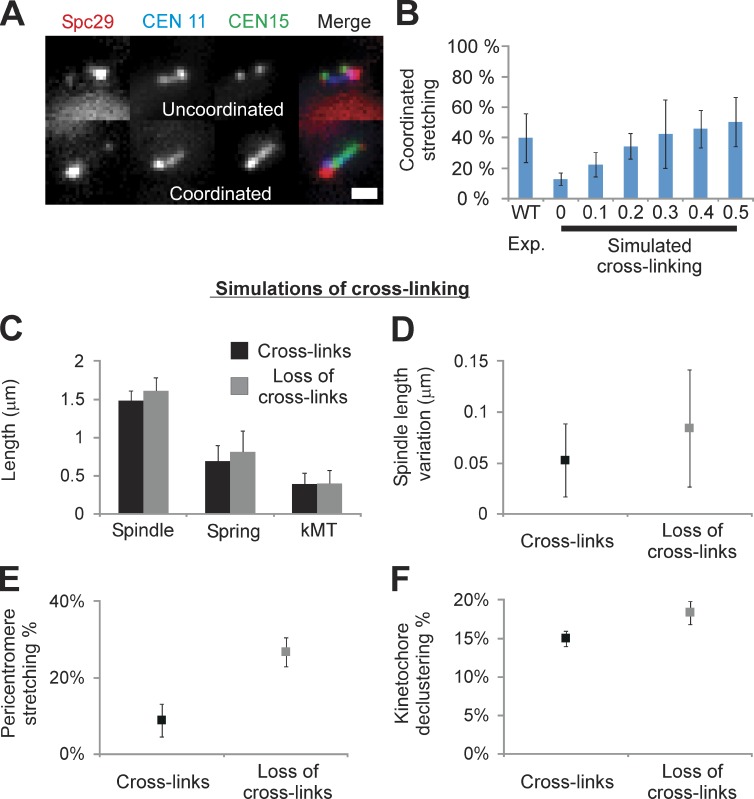
Chromatin springs may be cross-linked
To address whether the pericentromere chromatin springs of each chromosome are independent or coupled, we introduced two centromere-linked LacO arrays on different chromosomes (CEN15 LacO/LacI-GFP and CEN11 tetracycline operator [TetO]/tetracycline repressor [TetR]–CFP; Fig. 5). In WT cells with a single labeled chromosome, the incidence of stretching is 12 ± 3% (n = 261, 11%; Fig. 5; Stephens et al., 2011). LacO and TetO arrays exhibit similar stretching frequencies (13% LacO and 11% TetO). If the springs stretch independently, upon one spring stretching, only 12% of cells should have a second labeled spring stretched. In contrast, coordinated stretching was much more frequent (WT: 40 ± 16% coordinated stretching, n = 45; χ2 < 2 × 10−17; Fig. 5 B). The approximately fourfold increase from the expected probability suggests that the dynamics of chromatin loops on different chromosomes are not independent. With pericentric chromatin cylindrically distributed around the spindle axis (based on the distribution of kMTs), each strand would have two nearest neighbors in the region proximal to the kinetochores. We introduced cross-links in the model by introducing additional springs between nearest-neighbor chromatin strands. In this model, each chromosome is represented by a chromatin spring between the two spindle pole bodies, and one cross-link spring is attached to each of the neighboring strands on either side (see Materials and methods). The cross-link springs apply force proportional to the difference in length between adjacent strands, acting to bring them to the same length.
Figure 5.
Observed coordinated stretching is predicted in simulations with cross-links between adjacent chromatin springs. Cells were labeled with Spc29-RFP (spindle poles), TetO/TetR-CFP at 0.4 kb from CEN11, and LacO/LacI-GFP 1.8 kb from CEN15. WT cells exhibit stretching of a single pericentromere LacO in 11% of cells (Stephens et al., 2011) and recapitulated herein (12 ± 3%, n = 261). (A) Cells with two labeled arrays (CEN11 and CEN15) exhibit one stretching (uncoordinated, top) or both stretching (coordinated, bottom; WT: 40 ± 16%, n = 45 stretching cells). (B) Coordinated stretching was measured in simulations of a piecewise continuous spring, in which asymmetric stretching can be predicted (Fig. 2 C). Chromatin springs were cross-linked with a Hookean spring of increasing strength relative to the chromatin spring constant (0–0.5× = 0–15 pN vs. chromatin spring constant of 30 pN, n = 500, five experiments [Exp.] of 100). For each cross-linking spring constant, the threshold was altered to obtain 12 ± 2% stretching if a single pericentromere was labeled (experimental WT single stretching percentage). Population simulations were then measured for coordinated stretching of any pair of springs. In the absence of cross-linking (0, Simulated) the predicted frequency of coordinated stretching is 13 ± 4%, less than observed experimentally (left, WT). Cross-linking springs with 0.3× the spring constant of the chromatin spring best match experimental (P = 0.88, 42 ± 22%, right; vs. WT 40 ± 16%, left). (C–F) Loss of cross-linking (0 kcross-link; 955 nm Lthreshold) displays increased spindle length (C), spindle variation (D), pericentromere stretching (E), and kinetochore declustering (F; P < 0.05). Bar, 1 µm. Error bars represent standard deviation.
In the CNLS model with no cross-linking, a threshold for stretching of 975 nm yields a spring stretching frequency of 11 ± 3% (n = 500; Fig. 2 C, IV) for any given chromosome and a correlated stretching frequency of 13 ± 4% (Fig. 5 B, simulated cross-linking = 0) for any given pair of chromosomes, consistent with independent probabilities. We investigated cross-linking spring constants (kcross-link) corresponding to 0.1–0.5× the chromatin spring constant (30 pN/µm). Addition of cross-linking provides a mechanism to distribute tension between neighbors, making each chromatin spring less likely to stretch at a given spindle length. For a given kcross-link, the threshold was altered to match the observed frequency of single chromosome stretching in WT cells (12 ± 2%, yielding 965–947-nm thresholds for 0.1–0.5× cross-linking springs). We found that kcross-link = 0.3× yielded the best fit to the observed correlated stretching frequency (P = 0.88; Fig. 5 B).
To determine whether depletion of cross-linking in the CNLS + cross-linking model could account for experimentally observed behavior, we simulated loss of cross-links. We analyzed spindle length, length variation, loop stretching, and declustering in the CNLS with kcross-link = 0.3× (WT proxy) and upon depletion of cross-linking springs kcross-link = 0 (perturbed spring proxy). Upon removing the cross-links, spindle behavior was similar to the CNLS model with a decreased threshold. All simulation outputs increased (P < 0.05; Fig. 5, C–F), recapitulating in vivo perturbation of the chromatin spring via pericentric cohesin or condensin depletion. Ablation of cross-linking serves as another mechanism to increase loop stretching.
Discussion
The mitotic spindle is a force regulatory machine built for chromosome segregation. Using the mitotic spindle in yeast allows us to investigate the core components to avoid complexities that exist in larger spindles found in mammalian cells. The key features of the machine are an extensional microtubule-based motor force and a restoring chromatin spring force. Tension or microtubule attachment at kinetochores silences the spindle checkpoint in preparation for faithful segregation (Bloom and Yeh, 2010). A major question in the field is how the cell measures tension across an attachment site consisting of multiple kMTs. The lack of information regarding the functional properties of the chromatin spring hinders a mechanistic understanding of how tension is distributed between sister kinetochores in mitosis.
To address the biophysical basis of the chromatin spring, we used mathematical models of the yeast spindle. The main experimental features (Table 1) are spindle length and variation, pericentric chromatin stretching, and kinetochore clustering. The simplest models containing a linear chromatin spring (noise, MLS; coupled, CLS) capture spindle length and variation but fail to recapitulate the complexities found in vivo (Table 1).
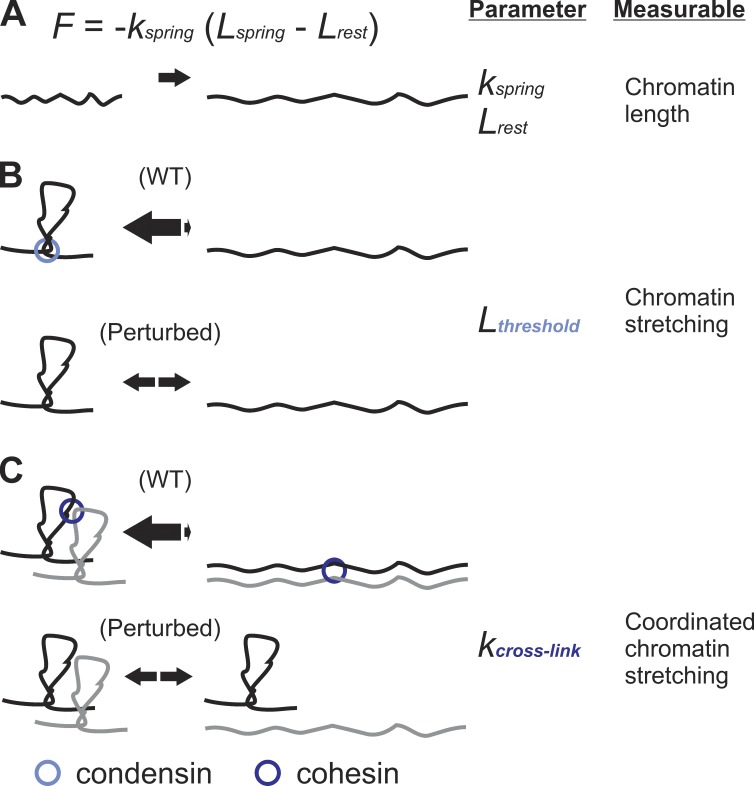
Visualization of a single chromosome using integrated LacO spots revealed asymmetric pericentric chromatin spring behavior during metaphase (Figs. 2 C and 3). LacO spots exhibit dynamic transitions from a focus to an extended state (Fig. 3). When a LacO spot on one sister stretches, the other sister does not (n = 102; Stephens et al., 2011). These data suggest pericentric chromatin does not function as a linear spring. A simple nonlinear spring is a piecewise continuous force–extension rule with a threshold condition for switching between looped and unlooped springs. An alternative form of a nonlinear spring is a wormlike chain, with an exponential force–extension curve. Our model exhibits a sawtooth force–extension curve and recapitulates chromatin stretching and kinetochore declustering (Table 1), not matched by systems with linear springs. A Hookean spring (F = −kspring (Lspring − Lrest)) has two adjustable parameters, rest length (Lrest) and spring constant (kspring; Fig. 6 A). The piecewise continuous spring contains a third additional parameter, a threshold (Lthreshold) for determining the state of chromatin looping that alters the linear spring parameters (Fig. 6 B). An additional fourth parameter is the cross-linking spring constant (kcross-link; Fig. 6 C) invoked to fit experimental data of 40% coordinated stretching (Fig. 5 B, WT). The cross-linking spring parameter affects both the threshold and linear spring parameters. These additional model parameters provide new functional attributes for probing the role of chromatin proteins, such as cohesin and condensin, in organizing pericentric chromatin into higher-order loops that function as a spring.
Figure 6.
Testable parameters of a piecewise continuous spring. (A) The most common form of a spring is given by a Hookean spring equation F = −kspring (Lspring − Lrest), in which k is the spring constant, and Lrest is the spring rest length. Simulations of a linear spring fail to account for behavior of the spindle and the pericentric chromatin upon experimental depletion of pericentric cohesin or condensin (see Table 1). (B) A nonlinear spring with a threshold length (Lthreshold) recapitulates increase in spindle length and fluctuations, asymmetric chromatin stretching, and kinetochore declustering. The threshold represents the length/force at which a compact loop transitions to a stretched loop. Cohesin and condensin increase the threshold of the chromatin loops maintaining compaction (equilibrium arrows shifted toward loops). Perturbation of the chromatin spring through depletion of pericentric cohesin or condensin decreases the length/force the loops can resist, causing the loops to stretch freely (equal amounts of compact and stretched loops; Fig. 2, A and C). The Lthreshold variable is an alternative way to modulate the native linear spring constant (k) and rest length (Lrest). (C) Experimentally observed stretching of two chromosomes could be simulated through the addition of a cross-linking spring between neighboring chromosomes (kcross-link; Fig. 5). Cross-linked chromatin springs can distribute tension, thereby increasing the ability of a single chromatin spring to resist reaching Lthreshold or extreme stretching (equilibrium arrows shifted toward the looped state). Simulation and experimental data suggest condensin and cohesin modulate Lthreshold and kcross-link, respectively.
The physical basis for a system with spring extension thresholds is consonant with chromatin loops in the pericentric region. The evidence for chromatin loops is twofold. One is the transition from spots to linear arrays of pericentric DNA visualized with LacO; two is the spatial redistribution from a spindle distal to proximal position coincident with the spot to linear transition (Stephens et al., 2011). The enrichment of cohesin and condensin and their ability to bring distal regions of chromatin into proximity (Hirano, 2006; Nativio et al., 2009; Mishra et al., 2010) provide additional evidence for pericentric chromatin loops. Pericentric condensin is localized proximal to the spindle axis and is responsible for axial compaction in accord with the SMC (structural maintenance of chromosomes) complex dictating the Lthreshold parameter (Fig. 6 B). Pericentric cohesin’s distal localization from the spindle axis and role in radial confinement of the pericentric chromatin best fit with the depicted kcross-link parameter (Fig. 6 C; Stephens et al., 2011). The depiction of cohesin and condensin in Fig. 6 accounts for their spatial segregation in the pericentromere, different biochemical roles, and common effect on the spindle and spring looping behavior both experimentally and in simulations. Therefore, we propose that cohesin and condensin together with pericentric DNA form a nonlinear spring that is part of a cross-linked network in the pericentromere. A similar cross-linked network mechanism for constraint of mammalian mitotic chromosomes has been attributed to protein-mediated compaction, linking, and DNA entanglements (Kawamura et al., 2010; Sun et al., 2011). Cross-linking between kinetochores and/or kMTs could also contribute to such a mechanism. Condensin and cohesin function to adjust chromatin compaction and provide a mechanism to distribute tension across single and multiple microtubule attachments in the spindle.
The organization of pericentric chromatin loops into a cross-linked network provides new insights into mechanisms of tension sensing in mitosis. The force exerted at the point of microtubule attachment is unlikely to be distributed uniformly between sister microtubule attachment sites. It is unclear how tension is sensed at a multimicrotubule attachment site, such as the mammalian kinetochore (16–20 microtubules), or the clusters of 16 yeast kinetochores, with stochastic microtubule dynamics. Alternatively, the cross-linking model provides a mechanism whereby the sum of forces from a multiattachment site structure can be integrated. When tension is applied to a cross-linked network, the heterogeneous distribution of cross-links can lead to local regions of high stress (Panyukov et al., 2009). Slip rings (or molecular pulleys) provide a mechanism to distribute tension from one location to the entire network (Okumura and Ito, 2001; Granick and Rubinstein, 2004). Cohesin and condensin have the physical attributes to function as slip rings (Glynn et al., 2004; Lengronne et al., 2004; Cuylen et al., 2011; Hu et al., 2011) and provide the chemistry for regulating elasticity both within the centromere and across multiple attachment sites. Upon the loss or depletion of these proteins, pericentric chromatin and tension sensing is impaired in both yeast (Yong-Gonzalez et al., 2007; Ng et al., 2009) and mammals (Ribeiro et al., 2009; Samoshkin et al., 2009; Uchida et al., 2009; Manning et al., 2010). We propose that a chromatin network composed of SMC “tension adjusters” functions to equalize tension across sister kinetochores, whose microtubule attachment sites exhibit stochastic dynamics throughout early mitosis.
Materials and methods
Cell preparation
Cells were incubated in YPD (2% glucose, 2% peptone, and 1% yeast extract) at 24 or 32°C for WT or mcm21Δ strains. Asynchronous cultures were grown to log phase and then imaged. Only metaphase cells were analyzed as outlined in Stephens et al. (2011). Cells were considered in metaphase with separated Nuf2 kinetochore foci and not linearly increasing in spindle length (indicative of anaphase). Cells with pericentric LacO and TetO were considered in metaphase when sister arrays were separated (LacO and TetO) and not linearly increasing in separation distance.
Imaging
Wide-field microscope images were acquired at room temperature (25°C) using a microscope stand (Eclipse TE2000-U; Nikon) with a 100× Plan Apochromat, 1.4 NA digital interference contrast oil immersion lens with a camera (ORCA ER; Hamamatsu Photonics). MetaMorph 7.1 (Molecular Devices) was used to acquire unbinned z-series image stacks with z-step size of 300 nm. Population Imaging was performed in water on Concanavalin A–coated coverslips. Live imaging of cells was performed on 25% gelatin slab with yeast complete 2% glucose media. Image exposure times were between 400 and 800 ms.
Analyzing pericentric LacO array stretching
LacO/LacI-GFP strains were grown in synthetic dextrose-His media to induce LacI-GFP under the HIS promoter as outlined by Goshima and Yanagida (2000) and Pearson et al. (2001). LacO array strains used to determine the rate of chromatin stretching and shrinkage were KBY 9039 (mcm21Δ) LacO 6.8-kb centroid from CEN15 (10-kb array inserted at 1.8 kb from CEN15). Time-lapse images of LacO/LacI-GFP stretching with Spc29-RFP–labeled spindle poles in KBY 9039 were captured using unbinned (65 nm/pixel) single-plane acquisitions at 5-s intervals. Each single-plane image was rotated using MATLAB (MathWorks) to align all spindles axis horizontally along the same y coordinate. Images were deconvolved using Huygens Compute engine 4.1.0 (Scientific Volume Imaging), and background was subtracted using MetaMorph. The length of the focus and stretched LacO/LacI-GFP signal were measured inclusively in MetaMorph and logged into Excel (Microsoft). A compact spot exhibits an isotropic shape, parallel and perpendicular to the spindle axis, and a stretched spot is anisotropic, extended parallel to the spindle axis (Stephens et al., 2011). An aspect ratio (parallel/perpendicular) of >1.2 is a quantitative measure of anisotropy (Haase et al., 2012). Extended arrays are ≥600 nm in length along the spindle axis (aspect ratio of 1.2; Fig. 3 A).
Kinetochore declustering
Population images were acquired of strains containing a kinetochore marker (Nuf2, Ndc80, or Ame1) with labeled spindle poles (Spc29): KBY 8526, 473A Nuf2-GFP-Ura Spc29-RFP-hygromycin B (Hb); KBY 9413, 473A Ndc80-tdtomato-Nat Ame1-GFP-Kan; KBY 9070, 473A mcm21Δ-Nat Nuf2-GFP-Ura Spc29-RFP-Hb; and KBY 9423, 473A mcm21Δ-Nat Nat:Trp Ndc80-tdtomato-Nat Ame1-GFP-Kan. MetaMorph line scans were drawn along the spindle axis through the kinetochores to determine whether each sister kinetochore structure remains clustered as one peak or declustered into multiple peaks.
Simulation outputs of spindle length and the length of each of the 32 kMTs emanating from each pole (16 each) were converted into XML files for generating simulated fluorescent images. A comma separated value to XML converter script was used in MATLAB to import the lengths into a set geometry to be used for the Microscope Simulator 2.1.1 (Center for Computer Integrated Systems for Microscopy and Manipulation, The University of North Carolina-Chapel Hill; Quammen et al., 2008). For each pole, the 16 corresponding kMTs were arranged in a 250-nm diameter around the spindle pole axis. The spindle pole was marked by an oblong sphere and labeled purple. The plus end of each of the kMTs (rods) was marked with a small sphere labeled green. The Microscope Simulator convolved the image using the point-spread function of our microscope as outlined by Sprague et al. (2003). The simulated image was configured to match experimental signal to noise (410 max and 270 min) and background noise standard deviation (±4.8) using gain, offset, and Gaussian noise functions. The simulated images were then analyzed in MetaMorph the same as experimental images outlined in the previous paragraph.
Coordinated stretching
To build a strain with pericentric regions labeled on two different chromosomes, we incorporated a 10-kb TetO array 0.4 kb away from the centromere at the met14 locus of CEN11 using a plasmid-targeting protocol (Rohner et al., 2008) into a strain that already contained a 10-kb LacO array tagged with LacI-GFP 1.8 kb from CEN15. TetR-CFP (pDB49: TetR-CFP-Hb) was used to tag the TetO array. Spindle pole bodies were marked with Spc29-RFP. Images were acquired using MetaMorph and were obtained in z-series stacks of 10 images with a step size of 200 nm. Images were binned two by two (pixel size of 130 nm). A triple pass dichroic was used in conjunction with YFPg, CFP, and RFP excitation filters, no detectable bleed through was found. LacO and TetO array stretching was analyzed in metaphase cells with two separated arrays for both the LacO and TetO. Stretching events were determined as explained in the subsection Analyzing pericentric LacO array stretching. Simulation population outputs were analyzed for uncoordinated or coordinated stretching among all 16 chromosomes.
Mitotic spindle simulation
The Mitotic Spindle Simulation software, written in MATLAB/Simulink, along with an accompanying manual can be downloaded from supplemental materials or the Center for Computer Integrated Systems for Microscopy and Manipulation, The University of North Carolina-Chapel Hill website.
MLS model
This minimal model considers prescribed stochastic cellular forces, a linear chromatin spring, and viscous drag, each of them acting along the spindle axis. Thus, a force balance along the spindle axis includes the extensional motor force from double-bound motors, Fip (+), the chromatin spring restoring force, Fk(−), and the drag force opposing spindle movement, Fdrag:
| (2) |
Force laws.
(a) The drag force is given by Stokes’ drag law: Fdrag(t) = −CdragVip(t), in which Vip is the velocity of the ipMT. (b) The motor force from double-bound motors is additive: Fip(t) = FMD(t), in which FM is the force per motor, and D(t) is the number of double-bound motors. Stall forces determined experimentally are in the range ∼5.5–7.5 pN, depending on ATP concentration (Visscher et al., 1999). In this study, FM is assumed constant and equal to 6 pN. For this minimal model, we posit D(t) as a Poisson distribution with given rates of attachment and detachment, kon and koff. This assumption on the number of motors is equivalent to a population balance of motors, in which the only species are double-bound and unbound motors,
(c) The length of the kMTs is normally distributed with mean Lm and variance σm: (d) The restoring force from chromatin is that of a Hookean (linear) spring: in which Lrest = 200 nm is the rest length of the spring.
Introducing these assumptions into Eq. 2, solving for Vip, and then writing Vip as the discrete rate of change of the spindle length, Lip(t), we find
Finally, solving for Lip(t + Δt) gives
Note that here, we can use values of and D at time t + Δt because these quantities are independent of the spindle length. A typical solution of this equation is plotted in Fig. S1 D, and Table S4 shows the parameters of the MLS model and the default values used in our simulations.
Distribution of Lip(t) calculated using the MLS model follows a log-normal distribution. A variable X is said to be log-normally distributed if log(X) is normally distributed. Although, in a normal distribution, the effects of independent factors are additive, the effects of independent factors acting on a log-normally distributed variable are multiplicative, and log-normal distributions usually refer to variables that remain positive.
CLS model
Motor dynamics.
Rather than simply draw the number of double-bound motors, D(t), from a Poisson distribution parameterized by kon and koff, we now distribute the total number of motors among four species: SipMT for motors single bound to the ipMTs, SkMT for motors single bound to the kMTs, U for unbound or free motors, and D for double-bound motors in the ipMT overlap zone.
The population dynamics is summarized in the following figure, with all the rates given in 1/s.
For each simulation, the total motor pool D + SipMT + U + SkMT is constant. Table S5 summarizes the assumptions made on the different rates of detachment and attachment.
(a) The dynamics of attachment of single-bound motors in ipMTs to become double-bound motors is assumed to follow a binomial process, SipMT → D ∼ B(Slap, 0.12), in which Slap is the number of motors in the overlap region Llap of ipMTs,
in which our simulations assume Llap = 0.8 µm. In addition, the probability of success is constant and equal to 12%. Because the relation between the probability of attachment, pon, and the rate of attachment, kon,1, is We find kon,1 ≈ 0.13.
(b) The rate of attachment of free motors to ipMTs and kMTs is assumed to be proportional to the tubulin concentration (constant) and the percentage of the total length that is available for attachment,
(c) The rate of detachment of bound motors is assumed constant, koff,2 = koff,1 = 0.3 s−1.
KMT length dynamics.
The kMTs grow and shrink stochastically through polymerization and depolymerization, but the process is biased by the state of the kMTs, i.e., their length relative to a threshold length and the tension in the kMTs. The dynamics of this process was studied by Gardner et al. (2005) and probability of switching states is graphed in Fig. S1 A.
Fitting to Gardner et al. (2005) data gives the following relationships for the probabilities of rescue, pr, and catastrophe, pc,
| (3) |
| (4) |
In addition, if LkMT < 50 nm, pr = 1, and if LkMT > 800 nm, pc = 0.389.
In Eqs. 3 and 4, Fk is given in piconormals, and LkMT is given in micrometers. To find whether at a given time step a kMT is growing or shortening, the following procedure is implemented. (a) Two random numbers, rc and rr, are drawn from a uniform distribution. (b) rc and rr are used to compute two logical values, a1 and a2, as
(c) Changes in kMT length are then determined by the following rules: if a1 = 0; a2 = 0, do nothing; if a1 = 0; a2 = 1, rescue (at a rate of 17 nm/s); if a1 = 1; a2 = 0, catastrophe (at a rate of 25 nm/s); and if a1 = 1; a2 = 1, same as the previous time step.
Coupled dynamics of 16 chromatin springs.
In this model, we include the dynamics of all 16 chromatin springs and their corresponding kMTs. Furthermore, we assume the springs are arranged in parallel so that the total spring force, Fk, is the sum of individual spring forces
The consequences of including more than one spring in the spindle dynamics are shown in Fig. S1 (E and F) and discussed in the Results subsection A linear spring model recapitulates the increase in both the mean and variance of spindle length observed in mutant cells.
Force–velocity relationship of the ipMTs.
We impose a linear force–velocity relationship defined by two parameters: a maximum (stall) force, FM = 6 pN, and a maximum speed, Vmax = 50 nm/s. The net force (sum of forces) felt by double-bound motors on ipMTs is
| (5) |
which is then distributed evenly across double-bound motors and gives a mean force per motor,
The ipMT (therefore spindle) velocity can be determined as
| (6) |
From Eq. 6, we see that (a) if Fk + Fdrag = 0, Vip = Vmax. In other words, if the forces from the chromatin spring and fluid drag cancel, the spindle will move at the maximum speed of one motor. (b) If the net force on the ipMTs is 0, Fnet = 0, Vip = 0, and the spindle is stationary. In this quasiequilibrium condition, each motor is at or near stall force (6 pN), and this force arises from the spring and drag forces acting on the motors. (c) The mean force acting on a motor can be found as Fper motor = FM(1−Vip/Vmax) as shown in Fig. S1 B.
Numerical integration.
With these new dynamics, Lip(t + Δt) can no longer be solved explicitly because D(t + Δt) and depend on Lip(t + Δt). Solving Eq. 6 for the spindle velocity gives
To integrate this equation in time and find Lip(t + Δt), we perform a predictor–corrector scheme. The spindle length in the predictor step, Lip(t*), is calculated as Lip(t) = Lip(t) + ΔtVip(t). After this predictor step, we use Lip(t*) to find D(t*) and and perform a corrector step, Lip(t + Δt) = Lip(t) + Δt/2[Vip(t) + Vip(t*)], in which
Finally, from Eq. 5, it is clear that the CLS model no longer imposes a deterministic force balance as in the MLS model (Eq. 2). In contrast, the sum of all forces is a stochastic variable, Fnet, that fluctuates around 0, as shown in Fig. S1 C.
CNLS model
In this model, we assume a nonlinear spring force, as explained in the Results subsection A nonlinear spring hypothesis. In this new spring force, we posit a threshold value (Xthres) for the spring extension. Spring lengths above the threshold value result in a decreased spring constant and increase in rest length. This new spring force can be then written as
| (7) |
Here, we assume xloop and L1 to be constants and equal to 450 and 800 nm, respectively, which gives k2 = 0.64 k1. We recognize that the instantaneous switching between spring states may be more accurately modeled using Kramers’ theory. This theory describes the transition between bistable states as motions over a potential energy barrier. However, for the purposes of this model, the course-grained switching has proven sufficient to capture our experimental observations.
Coupled nonlinear cross-linked stochastic model
The 16 springs are arranged in parallel and linked to their two nearest neighbors. The links are assumed to be soft linear springs with spring constant kcross-link = 0–15 pN/µm. Furthermore, the springs are assumed to be close enough to each other so that the force law in each link is given by
Here, θ is the angle between a cross-link and a spring, and we assumed that adjacent springs are close enough to each other so that θ ≈ 0 and cos(θ) ≈ 1. The force exerted by the spring is then, and in which Fk is calculated using Eq. 7.
Additional spindle forces
Astral microtubules.
In metaphase, there are approximately two to three astral microtubules extending from the spindle pole into the cytoplasm versus the ∼40 spindle microtubules. The astral microtubules are critical for spindle orientation and are acted upon by cytoplasmic dynein (Pearson and Bloom, 2004). Deletion of dynein has little to no effect on the kinetics of spindle elongation or the duration of metaphase (Yeh et al., 1995). Rather, loss of dynein results in misoriented spindles, whose position is monitored by the spindle position checkpoint.
Nuclear membrane forces.
The nuclear membrane has a heterogeneous morphology. Membrane protrusions have been observed to even precede the spindle upon migration into the daughter cell during anaphase (Yeh et al., 1995; Walters et al., 2012). Because spindle elongation is highly stereotypic and independent of changes in nuclear shape, we do not consider the impact of nuclear envelop force on the spindle.
Inward motor forces.
An important component of the spindle machine is the minus-end motor Kar3. KAR3 is a nonessential gene that nonetheless contributes to the fidelity of chromosome segregation in mitosis. Kar3 is found in metaphase along the ipMTs as well as kMTs and microtubule plus ends. Deletion of kar3 suppresses the loss of outward motors, Cin8 and Kip1, giving rise to the model that Kar3 provides an important inward force in metaphase (Saunders and Hoyt, 1992; Saunders et al., 1997). Although it is important to model the role of Kar3 in mitosis, the simplest model is that Kar3 opposes the outward motors Cin8 and Kip1. Incorporation of an additional parameter that opposes Cin8 and Kip1 will not substantively change the model.
Kinetochore forces.
Kinetochore forces are implicit as the mechanism that translates the spatial catastrophe gradient and tension-dependent rescue from the chromatin spring to kMT plus ends. The question of how much force the kinetochore generates has been addressed through elegant in vitro experiments with isolated kinetochores (Akiyoshi et al., 2010). The rupture force to dissociate the kinetochore from a microtubule is ∼9 pN (Akiyoshi et al., 2010). This is equivalent to the force of a single motor protein (∼6 pN). The implicit assumption in the values used in the model is that motors reach their stall force in vivo, and consequently, the spring constant is on the order of motor stalling. An unanswered question in the field is whether motors ever reach their stall force in vivo and how close to the rupture force the system is working. Implementing the microtubule-based force at the kinetochore will change the value of the spring constant but does not substantively change the model.
Variation in chromatin springs is not incorporated in the model, including histone exchange in the pericentric chromatin (Verdaasdonk et al., 2012), the likelihood that spring constants for different chromosomes are not identical nor are the switching thresholds, and variation in chromatin protein number (e.g., cohesin and condensin). These listed sources account for in vivo noise but should not alter the overall trends or behavior of the model.
Online supplemental material
Fig. S1 summarizes the transition from the MLS model to a CLS model. Fig. S2 shows the implementation of a nonlinear spring with two states (histograms of spindle length). Fig. S3 shows that the model is insensitive to overlap dynamics. Fig. S4 shows that model sensitivity analysis reveals robustness of the model. Fig. S5 is a representative single parameter sensitivity analysis that reveals that the threshold for loop stretching disproportionally affects spindle length variation. Table S1 lists the parameters and values used in the model. Table S2 provides the statistics of the Gaussian distributions shown in Fig. S2. Table S3 provides the sensitivity analysis parameters and ranges. Table S4 shows the MLS model values from Fig. S1. Table S5 shows the motor on/off rates of the CLS model from Fig. S1. Supplemental material also includes a PDF file that shows the mitotic spindle simulation manual. A ZIP file is also provided containing the mathematical model. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201208163/DC1.
Supplementary Material
Acknowledgments
We thank Abhilash Guduru (undergraduate, Biomedical Engineering, Department of Applied Mathematics and Statistics, Johns Hopkins University), Robert Baraldi (high school, North Carolina School of Science and Math), Ying Zhou, Patrick Short, Kelly Brannigan (undergraduate, Department of Mathematics and Department of Biology, University of North Carolina-Chapel Hill), and Scott McKinley (University of Florida) for running simulations of the model and participation in fruitful discussions. We thank Dr. Daniel Lew (Duke University) and members of the Bloom laboratory for advice, assistance, and critical readings of the manuscript.
This work was funded by the National Institutes of Health R37 grant GM32238 (to K. Bloom), the National Science Foundation grants DMS-1100281 and DMR-1122483 (to M.G. Forest), and the National Institutes of Health P41 grant EB002025 (to L. Vicci and R. Taylor; Center for Computer Integrated Systems for Microscopy and Manipulation).
Footnotes
Abbreviations used in this paper:
- CLS
- coupled linear stochastic
- CNLS
- coupled nonlinear stochastic
- Hb
- hygromycin B
- ipMT
- interpolar microtubule
- kMT
- kinetochore microtubule
- LacO
- lactose operator
- MLS
- minimal linear stochastic
- TetO
- tetracycline operator
- TetR
- tetracycline repressor
- WT
- wild type
References
- Akiyoshi B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., Biggins S. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 468:576–579 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin G., Nogales E. 2011. Visualizing kinetochore architecture. Curr. Opin. Struct. Biol. 21:661–669 10.1016/j.sbi.2011.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J., Alcasabas A., Blat Y., Kleckner N., Elledge S.J. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 9:1169–1182 10.1016/S1097-2765(02)00543-9 [DOI] [PubMed] [Google Scholar]
- Batchelor G.K. 1967. An Introduction to Fluid Dynamics. Cambridge University Press, Cambridge, UK: 615 pp [Google Scholar]
- Birtwistle M.R., Rauch J., Kiyatkin A., Aksamitiene E., Dobrzyński M., Hoek J.B., Kolch W., Ogunnaike B.A., Kholodenko B.N. 2012. Emergence of bimodal cell population responses from the interplay between analog single-cell signaling and protein expression noise. BMC Syst. Biol. 6:109 10.1186/1752-0509-6-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat Y., Kleckner N. 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 98:249–259 10.1016/S0092-8674(00)81019-3 [DOI] [PubMed] [Google Scholar]
- Bloom K., Yeh E. 2010. Tension management in the kinetochore. Curr. Biol. 20:R1040–R1048 10.1016/j.cub.2010.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D.C., Bloom K. 2007. Pericentric chromatin is an elastic component of the mitotic spindle. Curr. Biol. 17:741–748 10.1016/j.cub.2007.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bystricky K., Heun P., Gehlen L., Langowski J., Gasser S.M. 2004. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl. Acad. Sci. USA. 101:16495–16500 10.1073/pnas.0402766101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati J.L., Stearns T. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629–641 10.1083/jcb.138.3.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G., Sharp D.J., Mogilner A., Scholey J.M. 2006. Model of chromosome motility in Drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys. J. 90:3966–3982 10.1529/biophysj.105.078691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuylen S., Metz J., Haering C.H. 2011. Condensin structures chromosomal DNA through topological links. Nat. Struct. Mol. Biol. 18:894–901 10.1038/nsmb.2087 [DOI] [PubMed] [Google Scholar]
- Cytrynbaum E.N., Sommi P., Brust-Mascher I., Scholey J.M., Mogilner A. 2005. Early spindle assembly in Drosophila embryos: role of a force balance involving cytoskeletal dynamics and nuclear mechanics. Mol. Biol. Cell. 16:4967–4981 10.1091/mbc.E05-02-0154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher J.K., Ballenger M., O’Brien E.T., Haase J., Superfine R., Bloom K. 2009. DNA relaxation dynamics as a probe for the intracellular environment. Proc. Natl. Acad. Sci. USA. 106:9250–9255 10.1073/pnas.0812723106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.K., Pearson C.G., Sprague B.L., Zarzar T.R., Bloom K., Salmon E.D., Odde D.J. 2005. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol. Biol. Cell. 16:3764–3775 10.1091/mbc.E05-04-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.K., Odde D.J., Bloom K. 2007. Hypothesis testing via integrated computer modeling and digital fluorescence microscopy. Methods. 41:232–237 10.1016/j.ymeth.2006.08.002 [DOI] [PubMed] [Google Scholar]
- Gardner M.K., Sprague B.L., Pearson C.G., Cosgrove B.D., Bicek A.D., Bloom K., Salmon E.D., Odde D.J. 2010. Model convolution: A computational approach to digital image interpretation. Cell. Mol. Bioeng. 3:163–170 10.1007/s12195-010-0101-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay G., Courtheoux T., Reyes C., Tournier S., Gachet Y. 2012. A stochastic model of kinetochore–microtubule attachment accurately describes fission yeast chromosome segregation. J. Cell Biol. 196:757–774 10.1083/jcb.201107124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn E.F., Megee P.C., Yu H.G., Mistrot C., Unal E., Koshland D.E., DeRisi J.L., Gerton J.L. 2004. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2:E259 10.1371/journal.pbio.0020259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Yanagida M. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 100:619–633 10.1016/S0092-8674(00)80699-6 [DOI] [PubMed] [Google Scholar]
- Granick S., Rubinstein M. 2004. Polymers: a multitude of macromolecules. Nat. Mater. 3:586–587 10.1038/nmat1199 [DOI] [PubMed] [Google Scholar]
- Greulich K.O., Wachtel E., Ausio J., Seger D., Eisenberg H. 1987. Transition of chromatin from the “10 nm” lower order structure, to the “30 nm” higher order structure as followed by small angle X-ray scattering. J. Mol. Biol. 193:709–721 10.1016/0022-2836(87)90353-6 [DOI] [PubMed] [Google Scholar]
- Haase J., Stephens A., Verdaasdonk J., Yeh E., Bloom K. 2012. Bub1 kinase and Sgo1 modulate pericentric chromatin in response to altered microtubule dynamics. Curr. Biol. 22:471–481 10.1016/j.cub.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K.A., Holmes V.F., Cozzarelli N.R., Meyer B.J. 2002. C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev. 16:729–742 10.1101/gad.968302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison B.D., Hoang M.L., Bloom K. 2009. Persistent mechanical linkage between sister chromatids throughout anaphase. Chromosoma. 118:633–645 10.1007/s00412-009-0224-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P.K. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 101:763–775 10.1016/S0092-8674(00)80888-0 [DOI] [PubMed] [Google Scholar]
- Hirano T. 2006. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7:311–322 10.1038/nrm1909 [DOI] [PubMed] [Google Scholar]
- Hu B., Itoh T., Mishra A., Katoh Y., Chan K.L., Upcher W., Godlee C., Roig M.B., Shirahige K., Nasmyth K. 2011. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr. Biol. 21:12–24 10.1016/j.cub.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A.J., Gittes F., Howard J. 1994. The force exerted by a single kinesin molecule against a viscous load. Biophys. J. 67:766–781 10.1016/S0006-3495(94)80537-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson M.E., Loughlin R., Loïodice I., Fu C., Brunner D., Nédélec F.J., Tran P.T. 2007. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 128:357–368 10.1016/j.cell.2006.12.030 [DOI] [PubMed] [Google Scholar]
- Kawamura R., Pope L.H., Christensen M.O., Sun M., Terekhova K., Boege F., Mielke C., Andersen A.H., Marko J.F. 2010. Mitotic chromosomes are constrained by topoisomerase II–sensitive DNA entanglements. J. Cell Biol. 188:653–663 10.1083/jcb.200910085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Sauro H.M. 2012. In search of noise-induced bimodality. BMC Biol. 10:89 10.1186/1741-7007-10-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam W.W., Peterson E.A., Yeung M., Lavoie B.D. 2006. Condensin is required for chromosome arm cohesion during mitosis. Genes Dev. 20:2973–2984 10.1101/gad.1468806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G.P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F. 2004. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 430:573–578 10.1038/nature02742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., Lens S.M. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning A.L., Longworth M.S., Dyson N.J. 2010. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 24:1364–1376 10.1101/gad.1917310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R., Molodtsov M.I., Ataullakhanov F.I. 2012. Biophysics of mitosis. Q. Rev. Biophys. 45:147–207 10.1017/S0033583512000017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee P.C., Mistrot C., Guacci V., Koshland D. 1999. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell. 4:445–450 10.1016/S1097-2765(00)80347-0 [DOI] [PubMed] [Google Scholar]
- Mishra A., Hu B., Kurze A., Beckouët F., Farcas A.M., Dixon S.E., Katou Y., Khalid S., Shirahige K., Nasmyth K. 2010. Both interaction surfaces within cohesin’s hinge domain are essential for its stable chromosomal association. Curr. Biol. 20:279–289 10.1016/j.cub.2009.12.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilner A., Craig E. 2010. Towards a quantitative understanding of mitotic spindle assembly and mechanics. J. Cell Sci. 123:3435–3445 10.1242/jcs.062208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munsky B., Neuert G., van Oudenaarden A. 2012. Using gene expression noise to understand gene regulation. Science. 336:183–187 10.1126/science.1216379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nativio R., Wendt K.S., Ito Y., Huddleston J.E., Uribe-Lewis S., Woodfine K., Krueger C., Reik W., Peters J.M., Murrell A. 2009. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 5:e1000739 10.1371/journal.pgen.1000739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng T.M., Waples W.G., Lavoie B.D., Biggins S. 2009. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol. Biol. Cell. 20:3818–3827 10.1091/mbc.E09-04-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura Y., Ito K. 2001. The Polyrotaxane gel: A topological gel by figure-of-eight cross-links. Adv. Mater. (Deerfield Beach Fla.). 13:485–487 [DOI] [Google Scholar]
- O’Toole E.T., Winey M., McIntosh J.R. 1999. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 10:2017–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyukov S.V., Sheiko S.S., Rubinstein M. 2009. Amplification of tension in branched macromolecules. Phys. Rev. Lett. 102:148301 10.1103/PhysRevLett.102.148301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Bloom K. 2004. Dynamic microtubules lead the way for spindle positioning. Nat. Rev. Mol. Cell Biol. 5:481–492 10.1038/nrm1402 [DOI] [PubMed] [Google Scholar]
- Pearson C.G., Maddox P.S., Salmon E.D., Bloom K. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255–1266 10.1083/jcb.152.6.1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J.B., Ris H. 1976. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J. Cell Sci. 22:219–242 [DOI] [PubMed] [Google Scholar]
- Quammen C.W., Richardson A.C., Haase J., Harrison B.D., Taylor R.M., II, Bloom K.S. 2008. FluoroSim: A visual problem-solving environment for fluorescence microscopy. Eurographics Workshop Vis. Comput. Biomed. 2008:151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro S.A., Gatlin J.C., Dong Y., Joglekar A., Cameron L., Hudson D.F., Farr C.J., McEwen B.F., Salmon E.D., Earnshaw W.C., Vagnarelli P. 2009. Condensin regulates the stiffness of vertebrate centromeres. Mol. Biol. Cell. 20:2371–2380 10.1091/mbc.E08-11-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner S., Gasser S.M., Meister P. 2008. Modules for cloning-free chromatin tagging in Saccharomyces cerevisiae. Yeast. 25:235–239 10.1002/yea.1580 [DOI] [PubMed] [Google Scholar]
- Samoshkin A., Arnaoutov A., Jansen L.E., Ouspenski I., Dye L., Karpova T., McNally J., Dasso M., Cleveland D.W., Strunnikov A. 2009. Human condensin function is essential for centromeric chromatin assembly and proper sister kinetochore orientation. PLoS ONE. 4:e6831 10.1371/journal.pone.0006831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W.S., Hoyt M.A. 1992. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 70:451–458 10.1016/0092-8674(92)90169-D [DOI] [PubMed] [Google Scholar]
- Saunders W., Lengyel V., Hoyt M.A. 1997. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell. 8:1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler S.C., Liu J.Y., Pellman D. 2003. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. J. Cell Biol. 160:517–528 10.1083/jcb.200210021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague B.L., Pearson C.G., Maddox P.S., Bloom K.S., Salmon E.D., Odde D.J. 2003. Mechanisms of microtubule-based kinetochore positioning in the yeast metaphase spindle. Biophys. J. 84:3529–3546 10.1016/S0006-3495(03)75087-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens A.D., Haase J., Vicci L., Taylor R.M., II, Bloom K. 2011. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J. Cell Biol. 193:1167–1180 10.1083/jcb.201103138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M., Kawamura R., Marko J.F. 2011. Micromechanics of human mitotic chromosomes. Phys. Biol. 8:015003 10.1088/1478-3975/8/1/015003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K., Block S.M. 1994. Force and velocity measured for single kinesin molecules. Cell. 77:773–784 10.1016/0092-8674(94)90060-4 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Cosma M.P., Wirth K., Nasmyth K. 1999. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 98:847–858 10.1016/S0092-8674(00)81518-4 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Fuchs J., Loidl J., Nasmyth K. 2000. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2:492–499 10.1038/35019529 [DOI] [PubMed] [Google Scholar]
- Uchida K.S., Takagaki K., Kumada K., Hirayama Y., Noda T., Hirota T. 2009. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 184:383–390 10.1083/jcb.200811028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbreit N.T., Davis T.N. 2012. Mitosis puts sisters in a strained relationship: force generation at the kinetochore. Exp. Cell Res. 318:1361–1366 10.1016/j.yexcr.2012.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vas A.C., Andrews C.A., Kirkland Matesky K., Clarke D.J. 2007. In vivo analysis of chromosome condensation in Saccharomyces cerevisiae. Mol. Biol. Cell. 18:557–568 10.1091/mbc.E06-05-0454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk J.S., Gardner R., Stephens A.D., Yeh E., Bloom K. 2012. Tension-dependent nucleosome remodeling at the pericentromere in yeast. Mol. Biol. Cell. 23:2560–2570 10.1091/mbc.E11-07-0651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher K., Schnitzer M.J., Block S.M. 1999. Single kinesin molecules studied with a molecular force clamp. Nature. 400:184–189 10.1038/22146 [DOI] [PubMed] [Google Scholar]
- Walters A.D., Bommakanti A., Cohen-Fix O. 2012. Shaping the nucleus: factors and forces. J. Cell. Biochem. 113:2813–2821 10.1002/jcb.24178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.D., Eyre D., Basrai M., Lichten M., Strunnikov A. 2005. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 25:7216–7225 10.1128/MCB.25.16.7216-7225.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warsi T.H., Navarro M.S., Bachant J. 2008. DNA topoisomerase II is a determinant of the tensile properties of yeast centromeric chromatin and the tension checkpoint. Mol. Biol. Cell. 19:4421–4433 10.1091/mbc.E08-05-0547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. 2012. Geometry and force behind kinetochore orientation: lessons from meiosis. Nat. Rev. Mol. Cell Biol. 13:370–382 10.1038/nrm3349 [DOI] [PubMed] [Google Scholar]
- Welburn J.P., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Bloom K. 2012. Mitotic spindle form and function. Genetics. 190:1197–1224 10.1534/genetics.111.128710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Mamay C.L., O’Toole E.T., Mastronarde D.N., Giddings T.H., Jr, McDonald K.L., McIntosh J.R. 1995. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 129:1601–1615 10.1083/jcb.129.6.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman R., Cytrynbaum E.N., Jones J.T., Meyer T., Scholey J.M., Mogilner A. 2005. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr. Biol. 15:828–832 10.1016/j.cub.2005.03.019 [DOI] [PubMed] [Google Scholar]
- Yeh E., Skibbens R.V., Cheng J.W., Salmon E.D., Bloom K. 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130:687–700 10.1083/jcb.130.3.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Haase J., Paliulis L.V., Joglekar A., Bond L., Bouck D., Salmon E.D., Bloom K.S. 2008. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 18:81–90 10.1016/j.cub.2007.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Gonzalez V., Wang B.D., Butylin P., Ouspenski I., Strunnikov A. 2007. Condensin function at centromere chromatin facilitates proper kinetochore tension and ensures correct mitotic segregation of sister chromatids. Genes Cells. 12:1075–1090 10.1111/j.1365-2443.2007.01109.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.