Unprecedented proteome plasticity in response to stress in yeast is revealed using a novel screening platform that allows tracking of protein localization and abundance at single-cell resolution.
Abstract
Uncovering the mechanisms underlying robust responses of cells to stress is crucial for our understanding of cellular physiology. Indeed, vast amounts of data have been collected on transcriptional responses in Saccharomyces cerevisiae. However, only a handful of pioneering studies describe the dynamics of proteins in response to external stimuli, despite the fact that regulation of protein levels and localization is an essential part of such responses. Here we characterized unprecedented proteome plasticity by systematically tracking the localization and abundance of 5,330 yeast proteins at single-cell resolution under three different stress conditions (DTT, H2O2, and nitrogen starvation) using the GFP-tagged yeast library. We uncovered a unique “fingerprint” of changes for each stress and elucidated a new response arsenal for adapting to radical environments. These include bet-hedging strategies, organelle rearrangement, and redistribution of protein localizations. All data are available for download through our online database, LOQATE (localization and quantitation atlas of yeast proteome).
Introduction
The budding yeast adapts its physiology to radical external and internal fluctuations in a rapid and robust manner. It survives exposure to toxic substances, consumes a variety of nitrogen and carbon sources, lives both as a haploid or diploid organism, and carries out both mitosis and meiosis. Uncovering the mechanisms underlying its robustness in the face of extreme environments, genetic mutations, and life phases motivates many explorations of this eukaryotic cell. Indeed, much effort has been invested in measuring the transcriptional responses of whole yeast populations under varied conditions over time (Gasch et al., 2000; Hughes et al., 2000; Causton et al., 2001; Nagalakshmi et al., 2008). However, it is now widely appreciated that posttranscriptional, translational, and posttranslational regulation are also required for cell adaptation to stimuli (Stathopoulos-Gerontides et al., 1999; Beyer et al., 2004; Hedbacker et al., 2004; Newman et al., 2006; Sigal et al. 2006, 2007; Erjavec et al., 2007; Aragón et al., 2009; Frenkel-Morgenstern et al., 2010; Eden et al., 2011; Lee et al., 2011; Tkach et al., 2012). Because proteins are the major components that carry out actions within the cell, and are extremely sensitive to their surrounding milieu, even mild changes in protein abundance or subcellular localization may lead to drastic consequences in cellular physiology. We therefore wished to add to previous efforts in studying physiological robustness by characterizing yeast stress responses at the protein level, focusing on two key properties: protein localization and abundance, at a single-cell resolution.
Results
A novel system for measuring genome-wide protein abundance and localization at single-cell resolution
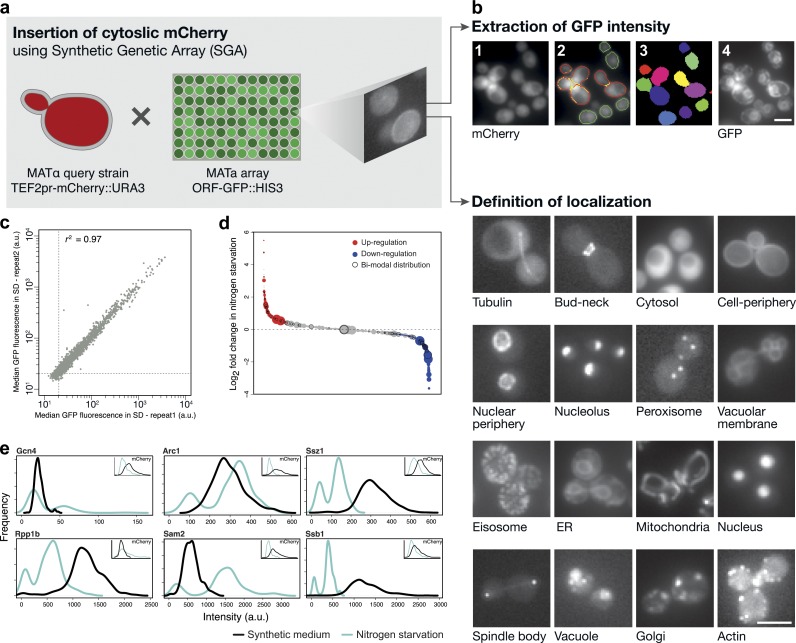
We have constructed a high-throughput microscopy setup to measure the levels of 5,330 yeast proteins and manually assign their localization using strains from the GFP fusion library (Huh et al., 2003). In this collection of strains, a GFP was integrated C-terminally into the genomic locus of each yeast ORF, maintaining the chromosomal context and preserving the natural promoter of each gene (Huh et al., 2003; Newman et al., 2006). To facilitate systematic quantification of GFP fluorescence in these strains, we introduced a constitutively expressed cytosolic marker into the entire library (TEF2pr-mCherry; Fig. 1 a; Tong et al., 2001; Cohen and Schuldiner, 2011). The cytosolic fluorescence allows us to segment the images into hundreds of individual yeast cells for each strain and to extract protein abundance (detected as GFP fluorescence) under the desired condition (Fig. 1 b and Fig. S1).
Figure 1.
Single-cell quantification of GFP-tagged protein levels reveals diverse proteomic changes during nitrogen starvation. (a) To track single cells, we integrated a cassette for expressing cytosolic mCherry into every strain of the yeast GFP collection. (b) We then automatically acquired images of hundreds of individual cells from each strain during growth in control conditions (logarithmic growth in SD) and three different stress stimuli. Using the mCherry (1), we segmented images (2) into single cell objects (3) and extracted GFP intensity in a single-cell resolution (4). Manual inspection of the library enabled the determination of subcellular localization for each protein (bottom). Bars, 5 µm. (c) A scatter plot (r2 = 0.97) of median fluorescence values measured for each of the 5,330 strains of the yeast GFP library in two independent experiments in SD (a.u., arbitrary units). (d) Bubble plot depicting relative abundance (in SD) indicated by the size of the circle and sorted by the log2 ratio of all measured strains under nitrogen starvation relative to SD. Red represents up-regulated events, blue represents down-regulated events, and black circles mark strains that were found to be significantly nonunimodal (i.e., to exhibit two subpopulations) by a Hartigan’s dip test. (e) Single-cell resolution analysis reveals bimodal distribution of expression of ribosome particles and ribosome-associated chaperones during nitrogen starvation relative to SD. Insets shows mCherry expression to remain unimodal in each strain under the same condition. This experiment was performed once on at least 100 cells per sample.
We first tested the reproducibility of our automated microscopy approach for protein abundance. During logarithmic growth in synthetic medium with dextrose (SD), our measurements were highly reproducible (r2 = 0.97, slope = 0.98, P < 0.01) in two biologically independent acquisitions of all strains (Fig. 1 c and Table S1). To assay the accuracy of our system, we compared our data to previously published protein measurements of yeast grown in the same medium. We found high agreement (r2 = 0.79 and r2 = 0.77) between our system and the levels of the 2,153 strains from the same GFP library previously measured by flow cytometry (Newman et al., 2006) and measurements taken for the same library with an independent microscopic setup (Andrews, B., personal communication), respectively (Fig. S2). Our results also correlate with the levels of native, untagged, proteins measured by mass spectrometry (Walther et al., 2010), Western blotting on tandem affinity purification (TAP)-tagged strains (Ghaemmaghami et al., 2003), and ribosomal footprint values (Ingolia et al., 2009; Fig. S2).
In addition to measuring protein abundance, images were used to extract localization information by manual inspection (Fig. 1 b, bottom). We categorized each protein into one of 13 subcellular localizations that can be distinguished without the use of colocalization markers. The main advantage of manual inspection is the miniscule error rate compared with faster automated methods (Rimon and Schuldiner, 2011). To enable accurate determination of a low-abundance cytosolic signal as bona fide expression (in contrast to cellular autofluorescence), we set the threshold of autofluorescence in each condition by measuring a library strain that does not harbor GFP (Fig. S1). Therefore, we could accurately assign expression and novel localizations to 72 proteins that are expressed at low abundance and were not visualized by previous, less sensitive microscopic platforms in SD. We also identified 48 proteins expressed in a condition-specific manner. 21 of the newly visualized proteins are essential, and so reveal new important information on proteins that are vital for yeast cellular life (Fig. S1 and Table S2).
Intensity analysis reveals trends of abundance changes under stress
After establishing the accuracy and reproducibility of this methodology, we used our platform to track proteomic changes during growth in three stress conditions that are known to activate distinct cellular responses: oxidative stress caused by hydrogen peroxide (H2O2), reducing stress caused by DTT, and nitrogen starvation (Gasch et al., 2000; Travers et al., 2000; Giaever et al., 2002; Hillenmeyer et al., 2008; Tyedmers et al., 2008). We found that at a single measured time point each condition evoked hundreds of changes in protein abundance (Fig. S3; for the full lists of up/down-regulated proteins, see Tables S3, S4, and S5). These changes were indicative of the responses required to restore homeostasis in this condition. For example, during nitrogen starvation, 336 proteins were present at a significantly higher level (P < 0.01; Fig. 1 d). These proteins were enriched for involvement in oxidation-reduction processes (P = 2.8 × 10−4) and heterocycle metabolic processes (P = 2.9 × 10−5). In addition, the 360 proteins that were present at significantly lower levels (P < 0.01) were enriched for ribosomal RNA (rRNA) processing (P = 2 × 10−14) and nitrogen compound metabolic process (P = 1.4 × 10−3). Importantly, comparing these direct proteomic measurements to microarray data revealed that over a third of the observed changes in protein levels (in all three stresses) could not be predicted by changes at the mRNA level (Fig. S4 and Tables S3, S4, and S5; Greenbaum et al., 2003; Newman et al., 2006; Walther et al., 2010; Schwanhäusser et al., 2011). This suggests a much more dramatic, proteome-wide, posttranscriptional regulation of protein abundance levels than previously reported.
Measuring protein abundance using single-cell resolution allowed us to explore variations in protein abundance under stress beyond the population level described above and beyond that possible by the pooled samples required for microarray analysis. For example, we detected 57 proteins that acquired a bimodal distribution upon nitrogen starvation (P < 0.05; marked as black circles in Figs. 1 d and 6; representative examples are shown in Fig. 1 e; for the full list of proteins that changed modality under stress see Table S6). This response is not a reflection of cell size (not depicted) or genotype because the mCherry expression in these cells remained unimodal (Fig. 1 e, insets). The bimodal responsive proteins were enriched for subunits of the large ribosomal particle (P < 7 × 10−3) and cellular protein metabolic processes (P < 0.001), which suggests that populations of yeast cells distribute their responses such as to reach two opposite outcomes: cell proliferation versus survival (Joo et al., 2011).
Figure 6.
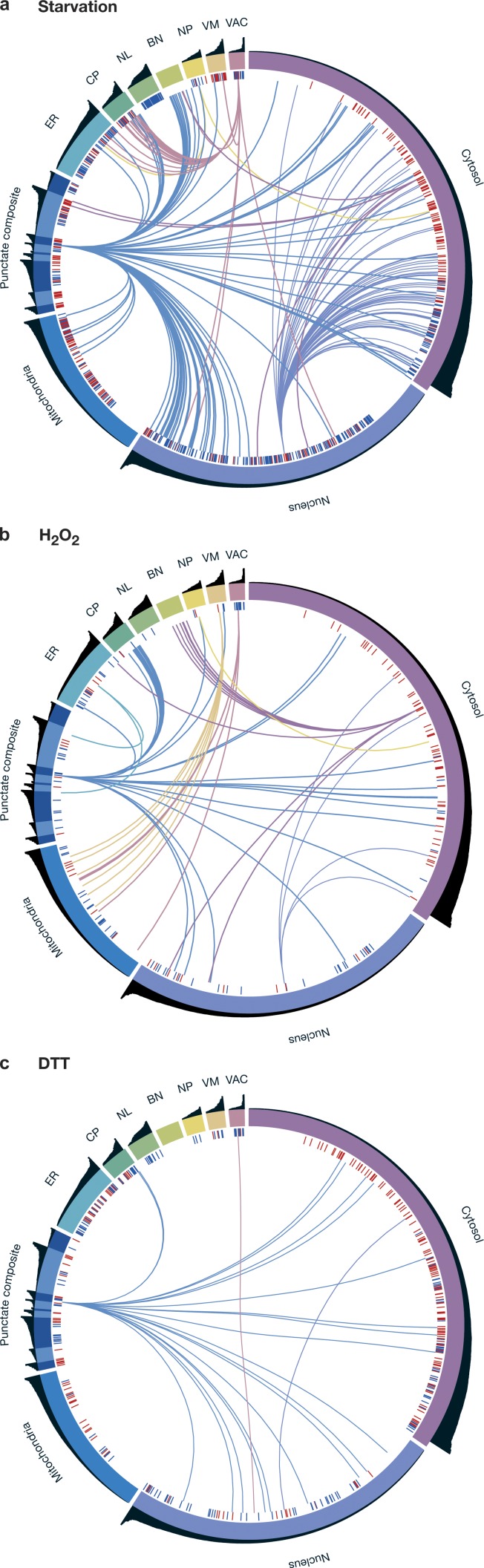
Integration of all proteomic changes gives a “thumbprint” of cellular stress responses. Representative schemes integrating all proteomic changes per each stress (a, starvation; b, H2O2; c, DTT) demonstrate the unique thumbprint of cell remodeling in each of the responses. The entire proteome is divided into organelles represented as colored segments on the circumference. The black bar plot above each organelle gives each protein’s abundance under SD and is sorted in each organelle. Blue (down-regulation) and red (up-regulation) bars demonstrate protein abundance changes. Lines between organelles demonstrate localization changes and are colored according to the destination organelle.
Single cell analysis uncovers a bet-hedging strategy in response to stress
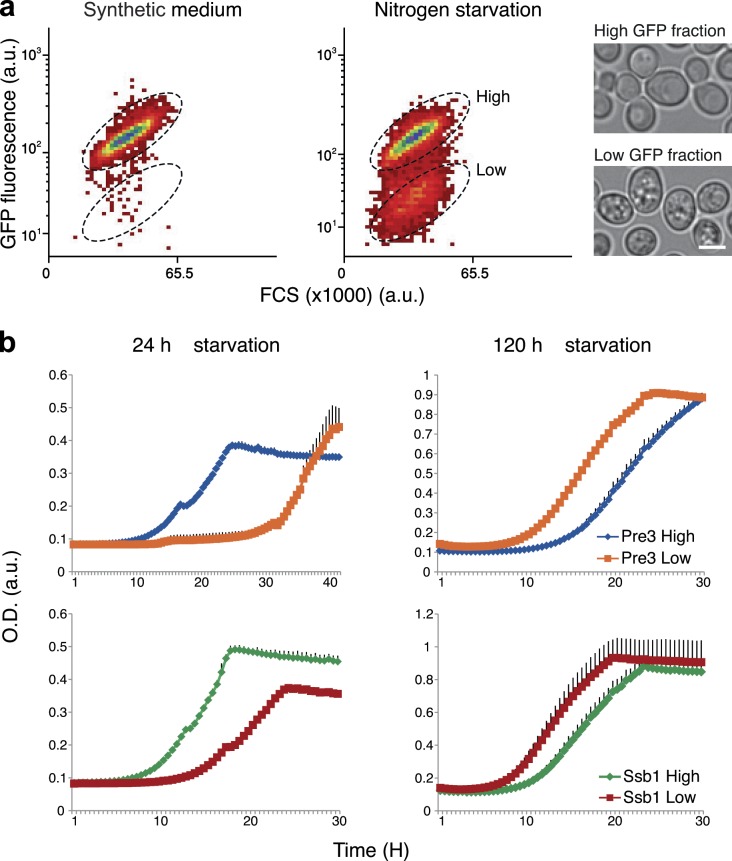
Recent studies observed phenotypic heterogeneity in altered environmental situations displayed by isogenic bacterial and yeast populations (Becskei et al., 2005; Allen et al., 2006; Acar et al., 2008; Gefen et al., 2008; Benbadis et al., 2009; Gefen and Balaban, 2009; Halfmann et al., 2010; Meyerovich et al., 2010; Zhang and Rainey, 2010; Balaban, 2011; Levy and Siegal, 2012; Levy et al., 2012). This phenomenon, termed bet hedging, specifically refers to survival strategies where each subpopulation performs differentially depending on the type of environment that becomes selective. For a population, this provides a long-term fitness advantage (de Jong et al., 2011). Because our observation of bimodality could underlie a “risk-spreading” strategy, either attenuating or augmenting the cell’s ability to survive under starvation, we tested the hypothesis of bet hedging for two representative proteins: Pre3 (the β1 subunit of the 20S proteasome) and Ssb1 (a ribosome-associated molecular chaperone). In strains expressing the GFP-tagged proteins, we sorted out the low and high GFP-expressing fractions after 24 h of nitrogen starvation (Fig. 2 a, middle). By light microscopy we could observe that the cells in each fraction displayed unique morphological characteristics: the high GFP–expressing fraction is similar to logarithmically growing cells, whereas the low GFP–expressing fraction is granular and resembles the quiescent fraction in stationary phase culture (Allen et al., 2006; Benbadis et al., 2009; Fig. 2 a). We then measured the growth rate of the two subpopulations (100,000 cells in each) either after immediate transfer to nutrient-rich conditions (SD) or after prolonged starvation (an additional 4 d) followed by return to nutrient-rich medium. Indeed, we found that the high-GFP fraction had a dramatic advantage over the lower one (high-GFP fractions grew similarly to nonstarved culture; not depicted) if return to rich medium was rapid. However, the low-GFP fraction had clear fitness advantage over the high-GFP fractions if starvation was prolonged (Fig. 2 b).
Figure 2.
A bet-hedging strategy in yeast underlies prolonged survival under starvation. (a) Flow cytometry dot plots show the transition of unimodal distributed population of Pre3-GFP in SD (left) into two distinct subpopulations after 24 h of starvation (right; a.u., arbitrary units). High and low fractions (marked with broken ellipses on the right plot) were sorted accordingly. Microscopic analysis of the two sorted subpopulations demonstrates distinct morphologies. Bars, 5 µm. (b) Growth of the two subpopulations in SD immediately after sorting (24 h starvation, left) versus 4 d after sorting (120 h starvation, right) of both Pre3-GFP and Ssb1-GFP (OD). Each growth well contained 100,000 cells at the starting point. These measurements are the representative means of two repeats; error bars indicate standard deviation in black.
Integrating localization with abundance data reveals stress-specific whole organelle rearrangements
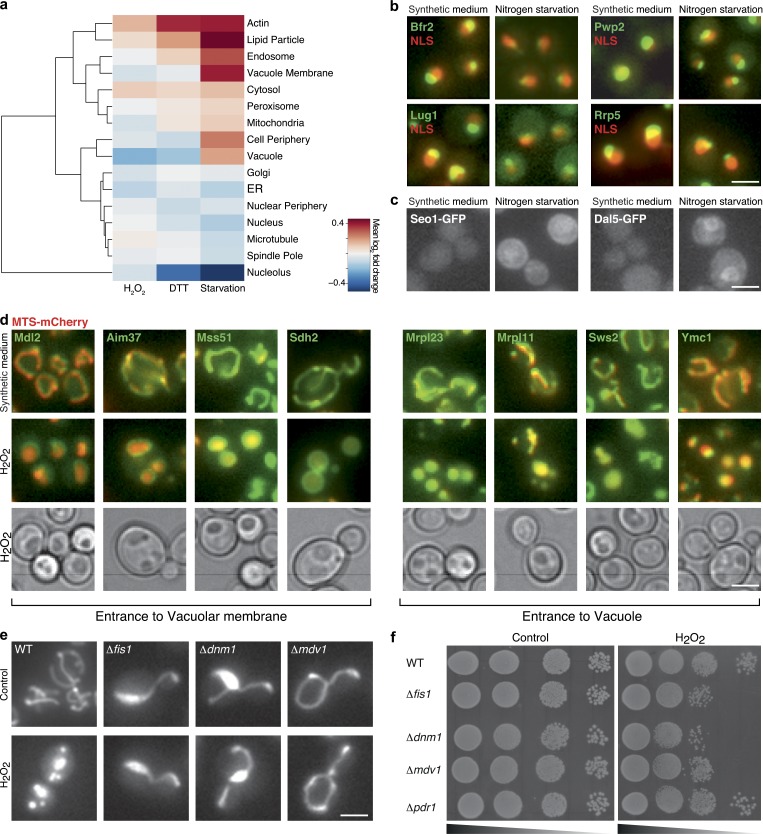
Spatial organization in the organelle-bearing eukaryotic cell is highly regulated and important for cell viability. A fundamental aspect of stress responses is therefore how protein abundance changes in the context of spatial organization. To track the extent of changes in the protein repertoire of each organelle, we calculated the mean log2 fold change of all proteins that were localized to every organelle under each condition (Fig. 3 a). Using this strategy, we observed that each stress condition preferentially affected specific organelles, either increasing or reducing their protein content.
Figure 3.
Integration of proteome abundance and subcellular organization enables characterization of the dynamics of organelle morphology and composition under stress. (a) Hierarchical clustering of mean log2 ratio of the change in abundance of proteins in each organelle under stress relative to SD. (b) Representative images of nucleolus proteins in SD or during growth in starvation medium. Colocalization with a nuclear mCherry (NLS) allows visualization of the massive shrinkage of this compartment relative to the nucleus under these conditions. (c) Representative images of cell periphery proteins that are expressed only under starvation and cause an increase in the protein levels in this organelle. (d) Representative images of mitochondrial proteins tagged with GFP coexpressed with a matrix-targeted dsRed (MTS-dsRed) during growth in SD or H2O2 treatment. During stress these proteins become localized to the vacuole lumen or membrane (as can be seen by the corresponding bright-field image) concomitantly with presence of the mitochondrial dsRed signal in the vacuole. (e) Mitochondrial fragmentation occurs when cells are exposed to medium containing 1 mM H2O2. This fragmentation is blocked in the absence of the fission machinery proteins Fis1, Dnm1, or Mdv1. (f) Fragmentation seems to be required for growth during oxidative stress as loss of fission capacity reduces fitness of cells under 1 mM H2O2 relative to control (YEPD). Bars, 5 µm.
For example, under nitrogen starvation, where protein translation is reduced, we saw dramatic shrinkage of the nucleolus, most likely due to a reduction in rRNA production (Fig. 3, a and b). Under the same condition, plasma membrane content rose, most probably as a result of the cells’ attempt to increase the presence of transporters on the cell surface and thus enhance uptake of sparse nutrients. An example of such an up-regulation is the family of allantoin transporters, which are required for uptake of alternative nitrogen sources (Chisholm et al., 1987; Isnard et al., 1996; Kaur and Bachhawat, 2007), that were below detection threshold under control conditions but became highly expressed when cells were starved for nitrogen (Fig. 3 c). As another example, after DTT treatment we observed up-regulation of actin filament subunits (P = 0.002). This is intriguing in light of previous publications that suggested a major role for cytoskeletal filaments in induction of the secretory pathway stress response: the unfolded protein response (Aragón et al., 2009).
We chose to focus on the changes that occur under H2O2 treatment, where we observed a prominent deflation of the mitochondrial proteome. This was accompanied microscopically by fragmentation of mitochondria (Fig. 3 e). To determine whether these changes are required to combat H2O2-induced stress, we first deleted proteins responsible for mitochondrial fission (Mozdy et al., 2000) and verified that in their absence mitochondria cannot fragment even under oxidative stress. These deletion strains indeed show a growth defect in the presence of H2O2 (Fig. 3 f). To broaden our perspective of essential functions of mitochondrial proteins while combating H2O2-induced stress, we focused on several nonessential mitochondrial proteins that were found to be essential for growth in H2O2 (Hillenmeyer et al., 2008). Following the fluorescent tagged fusions of these proteins by time-lapse resolution, we could find examples of proteins whose fluorescence signal changed dramatically as early as 30 min from initiation of the stress. Overlay with bright-field images suggests that these changes represent presence on the vacuolar membrane (Afg3, Sdh2, Mss51, Cbp3, Mdl2, and Aim37) or in the vacuole lumen (Mrpl11, Pkp1, Sws2, Mrpl23, and Ymc1; Fig. 3 d). Interestingly, this phenomenon occurred for only ∼30% of cells in the population. However, in those cells where it was seen, it was accompanied by accumulation of a matrix-targeted dsRed (MTS-dsRed) in the vacuolar lumen (as can be seen by overlay with bright-field images; Fig. 3 d) and by degradation of the protein over time (Video 1). Important to note is that most mitochondrial proteins (341/353) do not change their subcellular location during oxidative stress, providing a robust and inherent control for overall mitochondrial morphology under these conditions (Video 2). However, it remains formally possible that under oxidative stress mitochondria may swell up in a fashion that is so drastic that they may resemble vacuoles in bright-field images.
Three of the proteins whose localization were altered (Sws2, Mrpl23, and Mrpl11) are components of mitochondrial ribosomes, therefore their elimination would enable a major and rapid down-regulation in mitochondrial proteome production. This sheds new light on how cells clear dysfunctional mitochondria in a timely manner under oxidative stress and ensure rapid reduction of mitochondrial activity. The full list of proteins displaying an altered pattern of their predominant localization under H2O2 is enriched for proteins essential under this condition (35%, P = 0.001; Fig. S5), which suggests that localization change is an integral component of the response arsenal to combat oxidative stress (for the responsive proteins that are essential under starvation but not in reference growth condition, see Fig. S5).
Repatterning of subcellular localization occurs during exposure to stress
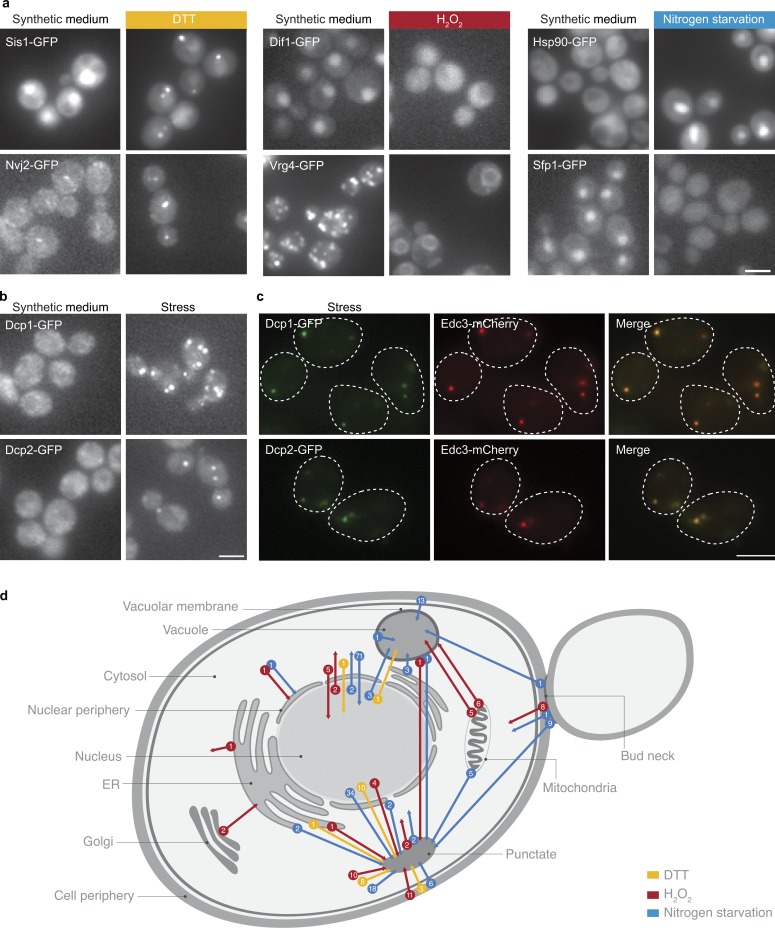
Previous literature demonstrated the importance of localization changes as a regulatory strategy in a handful of specific proteins (Cyert, 2003; Erjavec et al., 2007). Using our platform, we set out to explore these phenomena on a proteomic scale. In addition to expression levels, we systematically determined protein localizations under the same three stress conditions relative to the reference growth condition. Unexpectedly, a large number of proteins (235) displayed a change in their predominant subcellular localization under stress (Figs. 3, 4, and S5). Changes included nuclear proteins that could now be visualized in the cytoplasm and vice versa, plasma membrane proteins that could now be visualized in the vacuole, pan-ER localization that was visualized as nuclear vacuolar junctions, mitochondrial proteins that could be visualized in the vacuole, and proteins from all organelles that could be visualized in punctate foci (for the full scheme of changes observed see Fig. 4 d and for full list of changes see Table S7). Indicative of the functionality of the changes in protein localizations is that at a very high occurrence (48%) we found concerted changes for physically interacting proteins (interacting proteins were determined by Collins et al., 2007; for full details see Table S8). The unprecedented extent and diversity of changes demonstrate the unappreciated capacity of a wide variety of proteins to shift localization.
Figure 4.
Protein localization is dynamic under stress. (a) Two strains per condition (out of a total of 254 unique changes in protein localization) as examples for changes in predominant protein localization between growth in SD and growth under stress. (b) The only localization change that is common under all three stresses is proliferation of P-bodies. Shown are two P-body proteins, Dcp1 and Dcp2, changing localization from the cytosol to punctate foci. Broken lines represent the contour of the cell from bright-field images. (c) Foci of Dcp1 and Dcp2 colocalize with Edc3-mCherry during growth under every stress condition, which shows that these are bona fide P-bodies. (d) A schematic diagram depicting the types and numbers of changes in cellular localization observed during yeast growth in the three different stresses. Bars, 5 µm.
Because protein–protein interactions mediate cellular activity, we hypothesized that the changes that we observed in protein localization may be accompanied by a change in the networks of physical interactions. To test this, we mapped the entire physical interaction network for two proteins that changed their localization under DTT stress using the protein fragment complementation assay (Tarassov et al., 2008). The first, Pin3, is a protein of unknown function whose overproduction induces the appearance of prions (Derkatch et al., 2001). Pin3-GFP shifted from homogenous cytosolic distribution to discrete punctate foci. Indeed, under DTT Pin3 acquired four new significant interactions with the proteins Arc15, Arc18, Las17, and Ths1 (P < 10−12). Arc15, Arc18, and Las17 are all part of cortical actin patches that would give a punctate foci pattern by fluorescent microscopy. In support of these new interactions, the shift of Pin3 to interact with the actin cytoskeleton under stress has been recently demonstrated (Madania et al., 1999; Ganusova et al., 2006; Chernova et al., 2011), validating our hypothesis that changes in protein localization may reflect the acquisition of new cellular functions. The second, Acc1, catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA (Hasslacher et al., 1993). Interestingly, during DTT treatment its predominant localization changed from the cytosol to rod-like foci. In accordance with this movement, Acc1 acquired novel interactors after DTT treatment (for a full list see Table S9). Of note are the new physical interactions with both Pre9 (the α3 subunit of the 20S proteasome) and Rpn8 (the regulatory subunit of the 26S proteasome), which suggests the involvement of Acc1 in proteasomal degradation during exposure to DTT.
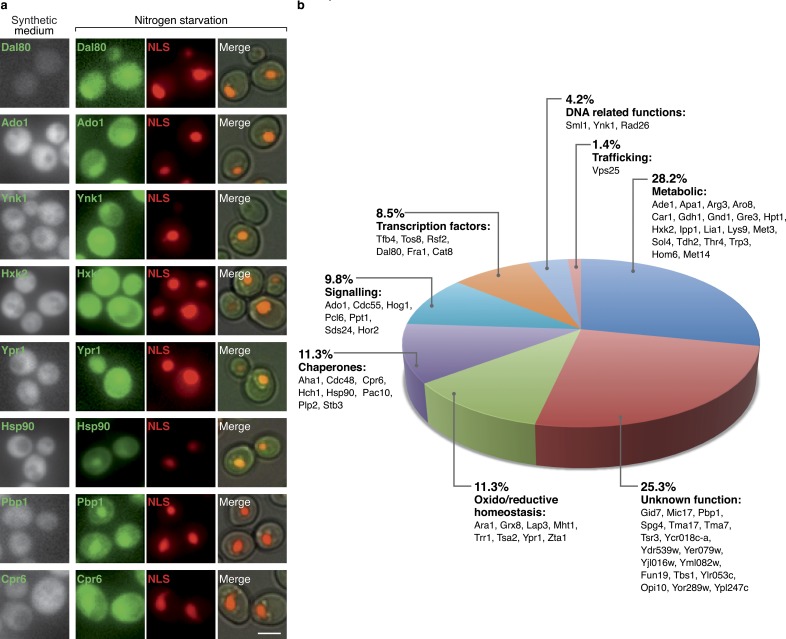
However, by far the most remarkable change in localization during starvation was observed for 71 cytosolic proteins whose predominant subcellular distribution became nuclear (Fig. 5). Although six of these proteins are transcription factors, whose change can be easily rationalized, the vast majority are proteins not previously known to be required in the nucleus under such conditions. For example, eight proteins are chaperones and eight are oxido/reductive homeostasis-related proteins. A striking observation was that 20 metabolic enzymes entered the nucleus (Fig. 5). It has been shown recently that the two mitochondrial enzymes fumarase and aconitase play a minor function in the nucleus that becomes essential under DNA-damaging conditions (Yogev et al., 2010; Ben-Menachem et al., 2011). Our findings would indicate that such cases may be much more prevalent than previously appreciated.
Figure 5.
Examples of relocalizations of proteins from the cytosol into the nucleus during growth in nitrogen starvation medium. 71 cytosolic proteins categorized to eight different cellular functions could be visualized in the nucleus during starvation. (a) A representative protein from each of the categories is shown in SD and under nitrogen starvation, colocalized with a nuclear (NLS)-mCherry. Bars, 5 µm. (b) Pie chart with the relative size of each functional group and information on the genes categorized in it.
Most localization changes were condition-specific (Table S7). However, four proteins changed localization in a concerted manner during all three stresses (Dcp1, Dcp2, Kem1, and Ede1; Fig. 4 b). Three out of the four proteins are localized to P-bodies (Fig. 4 c) and play essential functions in P-body formation, supporting the previously published observation that P-body function and proliferation play a major role during diverse stresses (Nissan et al., 2010).
Collectively, our data open a new porthole to the vast extent of changes exhibited by the cell under stress solely at the level of the proteome. A birds-eye view of these changes can be seen when integrating localization and abundance data into unique “fingerprints” of the yeast cellular response to each stress (Fig. 6).
Discussion
The remodeling of expression patterns that occurs while adapting to new environments was characterized extensively in the past decade using first microarray and now deep-sequencing technologies. This fundamental information has provided significant clues about the way in which we understand stress responses today. Interestingly, previous publications demonstrated only a small overlap between the groups of genes essential to combat a specific stress and the mRNA transcripts that change under the same conditions (Giaever et al., 2002; Hillenmeyer et al., 2008). This strongly suggests that proteomic level changes in stressful environments may be predominant and can give us new insights about the cellular state. Indeed, accumulated literature of proteomic studies promises that there are many more mysteries to be elucidated regarding the cellular response to environmental stress. Using a high-throughput microscopy system, we show here the first nonbiased systematic high-resolution analysis of protein abundance and localization changes at single-cell resolution after three different stress conditions. Moreover, we integrate these two types of data to give a fundamental perspective of abundance changes within the context of spatial eukaryotic organization to detect whole organelle proteomic composition and morphology for the first time. We have observed these stress-specific organelle changes to be crucial for cellular robustness in the face of stress.
One notable example for how such data can be used is in the modeling of transcriptional networks that govern genome expression changes. Our work shows that there are extensive changes in the predominant localization of transcription factors under stress. This strengthens the notion that in many cases transcription factors are controlled not by their levels of expression but rather by their localization. This would suggest that previous analyses that have used only mRNA levels to try and identify transcriptional networks are missing an essential aspect of transcription factor regulation.
There are numerous advantages to probing cellular processes at a single-cell resolution. As an example, it allowed us to uncover the proteins that are regulated in a bi-modal fashion in response to stress. Bet hedging is poorly understood at the molecular level, though it plays a major role in diseases, such as cancer (Levy and Siegal, 2012) and persistence of microbial infections (Levy and Siegal, 2012). Measuring the changes in protein distribution within the population under a variety of conditions should shed novel light on the molecular mechanisms used by species to adapt toward radical environments and promises future solutions to the current failure of chemotherapies and antibiotics on bet-hedging populations.
The platform we present here may be used to measure the proteome under a variety of stress conditions, life phases, genetic backgrounds, and drug treatments. To spearhead these efforts and to facilitate easy access to our data we have created a database of changes in protein levels and localizations (LOQATE, localization and quantitation atlas of the yeast proteome) that will house the information from this current work as well as our future screens (www.weizmann.ac.il/molgen/loqate). Such systems-wide, precise localization and quantitation, directly at the protein level and at single-cell resolution, opens up new avenues in the postgenomic era, which hold the promise to shed light on novel aspects of cell biology that have not been previously characterized.
Materials and methods
Strain management
Insertion of cytosolic mCherry to the GFP library.
Synthetic genetic array (SGA) technique was performed between a MATα haploid strain harboring TEF2pr-mCherry::URA3 integrated into the URA3 locus (plasmid for creation of the strain was a kind gift from D. Breslow, Stanford University, Stanford, CA; Breslow et al., 2008) against the GFP collection (::HIS3; the library was a kind gift from J. Weissman, University of California, San Francisco, San Francisco, CA; Huh et al., 2003). Mating was performed on rich media plates, and selection for diploid cells was performed on plates lacking both HIS and URA. Sporulation was then induced by transferring cells to nitrogen starvation plates for 5 d. Haploid cells containing all desired mutations were selected for by transferring cells to plates containing all selection markers alongside the toxic amino acid derivatives Canavanine and Thialysine (Sigma-Aldrich) to select against remaining diploids and lacking Leucine to select for only spores with an “a” mating type (Tong et al., 2001; Cohen and Schuldiner, 2011). SGA procedure was validated by inspecting representative strains for the presence of the GFP-tagged strains and for the cytosolic mCherry expression. To manipulate the collection in high-density format (384), we used a RoToR bench top colony arrayer (Singer Instruments). Yeast strains used in this study are in Table S10.
Strain growth.
The manipulated 5,330 strains were grown in 50 µl SD (0.67% yeast nitrogen base without amino acids [Conda Pronadisa] and 2% dextrose) containing the appropriate supplements for selection in 384-well plates (catalogue No. 781162; Greiner Bio-One).
Application of stress.
For hydrogen peroxide (H2O2) treatment, cells were grown to early-log phase, at which point H2O2 (catalogue No. 2186–01; J.T.Baker) was added at a final concentration of 1 mM. After 60 min of incubation, plates were taken for microscopic imaging.
For nitrogen starvation, cells were grown to early log phase and centrifuged mildly (1,000 g), then fresh SD (0.67% yeast nitrogen base without amino acids and without ammonium sulfate [Conda Pronadisa] and 2% dextrose) was added. After 15 h of incubation, plates were taken to microscopy imaging.
For DTT treatment, cells grown overnight were back diluted into 2 mM DTT containing SD medium. After 3 h of incubation, plates were taken for microscopic imaging.
Plasmids and deletions.
Deletion strains were prepared by replacement of the ORFs with a pCgMET15 cassette using homologous recombination with 40 bp of homology (Kitada et al., 1995). pRS416 plasmid expressing MTS-dsRed under the ADH1 promoter was provided by J. Nunnari (University of California, Davis, Davis, CA; Meeusen and Nunnari, 2003). pRS426 plasmid expressing NLS-tdTomato under the GPD1 promoter was provided by D. Kaganovich (Hebrew University, Jerusalem, Israel; Kaganovich et al., 2008). Plasmids used in this study are in Table S11 and primers used in this study are in Table S12.
Automated imaging
High-throughput fluorescence microscopy.
Microscopic screening was performed using an automated microscopy setup as described previously (Cohen and Schuldiner, 2011). Cells were moved from agar plates into liquid 384-well polystyrene growth plates using the RoToR arrayer. Liquid cultures were grown overnight in SD medium in a shaking incubator (LiCONiC Instruments) at 30°C. A JANUS liquid handler (PerkinElmer), which is connected to the incubator, was used to back-dilute the strains to ∼0.25 OD into plates containing the same medium. Plates were then transferred back to the incubator and were allowed to grow for 3.5 h at 30°C to reach logarithmic growth phase, as was validated in preliminary calibration. The liquid handler was then used to transfer strains into glass-bottom 384-well microscope plates (Matrical Bioscience) coated with Concanavalin A (Sigma-Aldrich) to allow cell adhesion. Wells were washed twice in medium to remove floating cells and reach cell monolayer. Plates were then transferred into an automated inverted fluorescent microscopic ScanR system (Olympus) using a swap robotic arm (Hamilton). Imaging of plates was performed in 384-well format using a 60× air lens (NA 0.9) in SD medium at 24°C with a cooled charge-coupled device camera (ORCA-ER; Hamamatsu). Images were acquired at GFP (excitation at 490/20 nm, emission at 535/50 nm) and mCherry (excitation at 572/35 nm, emission at 632/60 nm) channels.
Image analysis.
Our screening assay was designed to explore yeast cell biology by assessing two cellular key features of interest: subcellular localization and fluorescence intensity. To analyze these images we have used an in-house script to browse manually and assign localization rapidly and efficiently. To extract proteomic abundance from images, we used the Olympus ScanR analysis software. This allows for the preprocessing of images by background subtraction, and segmentation of images to identify individual cells as separate objects. Specifically, we performed the following steps:
(a) Segmentation on the basis of the edge module of the cytosolic mCherry protein expression.
(b) Background correction using the rolling ball algorithm.
(c) Definition of measured populations. Because several measurements are collected for each cell (e.g., fluorescence intensity, area, shape), we have created a multiparameter gate to ensure that our population was homogenous and that data arise from clearly defined cells only. The mean GFP intensity for each object (cell) of each strain was extracted to Excel files (Microsoft), allowing data processing for single-cell resolution from within a given population.
Data processing
Median measurement.
The median GFP intensity for each strain was measured from the remaining objects for each strain under each condition after removing dead cells. Dead cells are highly fluorescent and must be removed from the analysis because they artificially raise the mean GFP intensity values. Because the fluorescence and shape of dead cells fall within range of those features for normal cells, the software could not gate them out. To automate the removal of dead cells detected as objects by the ScanR software, for each strain we removed any objects that were very high outliers in their mean GFP measurement. Therefore, any objects with mean GFP measurement above UQ + 3 × IQR were removed from analysis, where UQ is the upper quartile and IQR is the interquartile range. Overall, >94% of cells originally screened were classified as alive. On average, 5% of cells per strain were dead under stress. Because we performed two independent measurements under reference conditions (SD), we combined scores from both measurements to get one median and standard deviation value for each strain under reference conditions (summarized in Table S1).
Removing strains from the analysis.
The following strains were removed from the analysis:
(a) Strains in which <25 objects were detected.
(b) Strains whose proper subcellular localization has been demonstrated to be dependent on the C terminus of the protein. A full list of the systematically mislocalized proteins that were removed from analysis is given in Table S13.
(c) Strains whose tagged ORFs are located near the CAN1, LYP1, or URA3 loci. Such strains could not pass the SGA required to make the Tef2-Cherry background. A full list is given in Table S13.
(d) Contaminated strains. Any strains showing localization different than that annotated in SGD or that shown in the original Huh et al. (2003) published dataset were assumed to be the result of contamination and were removed.
(e) Bud neck proteins, as the analysis program could not accurately detect their signal, sometimes out of the detected cell boundaries.
Detection of autofluorescence.
Yeast cells not tagged with GFP emit fluorescence at some baseline intensity. We have found this value to differ significantly across conditions. This may be a result of different cellular conditions under various stresses or of fluctuations in the light source. To account for this, we screened 86 randomly placed wells containing a wild-type strain with no GFP tag in each measured condition. After removing dead cells, the distribution of the intensity values for wild-type objects was not rejected for normality using the Shapiro-Wilk test (P < 0.01). We then approximated the autofluorescence as a normal distribution with mean autofluorescence value Acondition and standard deviation σcondition. Any strain whose median GFP intensity under a given condition was more than Acondition + 2.58σcondition is >99% likely to have biological GFP expression. Strains below this cutoff are marked as “below threshold.”
If a strain falls below this threshold, but localization other than “cytosol” or “ambiguous” was assigned, it is kept in the analysis because we could be certain that we visualized that protein.
Experimental controls.
Testing the reproducibility. To test the stability of our microscopy platform, we plotted two independent measurements of all strains (5,330) in SD (Pearson’s correlation coefficient on logarithmic scaled data tested r2 = 0.97, slope = 0.98, P < 0.01).
Measurement of accuracy. In order to test accuracy of our measurements, we plotted the calculated medians by our method versus two measurements of the same strains by flow cytometry and with the measurements made by a similar high-content screening setup, and found high reproducibility (Fig. S2).
Further testing. In order to show the relevance of these measurements to other proteomic datasets, we plotted the calculated medians by our method versus three measurements of yeast proteomes: native, untagged proteins measured by mass spectrometry, Western blotting on TAP-tagged strains, and ribosomal footprint values, and again found good agreement (see Fig. S2).
Determining abundance change events
For each condition, we determined which strains are significantly up- or down-regulated compared with their reference abundance levels.
Preprocessing.
In addition to the corrections described in “Data processing,” we removed any strains that changed localization under the condition of interest compared with reference medium. Because the GFP signal is sensitive to cellular conditions (such as ionic strength or pH), levels cannot be compared between two proteins showing different localizations.
Some proteins are only expressed under certain stress conditions and are not detected under reference conditions. To not miss these proteins, any strains that fell below the autofluorescence threshold in the reference condition and therefore were not assigned localization, but were detected under the stress condition, were left in the analysis. Similarly, strains detected under reference but below the autofluorescence threshold under treatment were also included.
Normalization of fluorescence signal.
To make fluorescence values comparable across reference and stress conditions, we normalized median fluorescence values for each strain using the normalize.quantiles function in the R preprocessCore library (Bolstad et al., 2003). This method adjusts fluorescence values such that the median values for each strain follow the same distribution under each condition. The fluorescence values of objects for each strain under each condition are scaled to have the corrected median value.
Definition of significant abundance change.
All strains under SD in two independent experiments were sorted by their log10 abundance values and binned such that each bin contained 5% of strains. For each bin, we plotted the distance from the diagonal for each strain in the scatterplot of replicate 1 versus replicate 2. The distances within each bin were normally distributed (Shapiro-Wilk test, P < 0.01). For each stress condition, we plotted the log10 abundance of each strain versus the average of the log10 abundance across the two reference conditions. We then determined the distance of each strain from the diagonal. Based on the distribution of distances between the two SD experiments in the corresponding abundance bin for each strain, we determined an empirical P-value. Strains with P < 0.01 were marked as showing a significant abundance change (Fig. S3), and these events are marked above (red) and below (blue) the broken lines in Fig. S3.
Fold change values.
For each stress condition, we calculated the ratio of median GFP intensity measured under stress to the median GFP intensity under reference conditions. Ratios are given in Table S1.
Testing multimodality.
In the case that not all cells are uniformly affected by a stress condition, the resulting distribution of fluorescence intensities could be multimodal. The significance test described is not informative in this situation, and gives no indication that there might be multiple levels of response throughout a population of cells. To test each strain under each condition for multimodality, we used Hartigan’s dip test (Hartigan and Hartigan, 1985), which tests how well the distribution of fluorescence for the objects fits the tightest fitting unimodal distribution. Strains in which unimodality was rejected with P < 0.05 were marked as multimodal. This test was implemented using the dip.test command in the R dip test library. (The list of strains predicted to have multimodal distributions is given in Table S6). Manual inspection confirmed that all strains rejecting unimodality were indeed bimodal.
Comparison to published datasets
Comparison to essentiality data.
We used published essentiality datasets (Hillenmeyer et al., 2008) to determine which ORFs are essential under each condition tested. We downloaded the homozygous fitness defect scores from the web supplement to Hillenmeyer et al. (2008) and created a list of ORFs for each condition shown to be essential with P < 0.05. We used the conditions 5 mM H2O2 and SD for H2O2 and starvation, respectively.
Comparison to mRNA data.
For each ORF under each condition tested, we determined whether it was up-regulated, down-regulated, or showed no change at both the protein and mRNA level. Proteins were determined to be up or down-regulated based on criteria described in “Definition of significant abundance change.” To determine if an ORF is up- or down-regulated at the mRNA level, we used published expression datasets. For DTT, we compared with the DTT time points at 15, 30, 60, and 120 min (Travers et al., 2000). For starvation and H2O2, we compared with the nitrogen depletion and 0.32 mM H2O2 time points from 30 min to 1 d and 10 min to 160 min, respectively (Gasch et al., 2000). An ORF was defined as up-regulated if any time point showed a twofold increase in expression level compared with reference and as down-regulated if any time point showed a twofold decrease compared with reference. ORFs that had time points showing both a twofold increase and twofold decrease were marked as ambiguous and were removed from the analysis.
We then used these up/down/no change classifications to find ORFs marked as no change in mRNA levels but that did significantly change at the protein level according to our analysis.
Protein–protein interactions analysis
Dihydrofolate reductase–based protein fragment complementation assay (DHFR PCA).
The assay for the yeast DHFR PCA followed entirely the published protocol (Tarassov et al., 2008). In brief, MATa strains with the ORFs of Acc1 and Pin3 fused C-terminally to F[1,2] were mated to the entire MATα collection of ORFs tagged with F[3]. The resulting diploids were subsequently selected for growth in the presence of methotrexate for positive DHFR PCA reconstitution with or without the addition of 3 mM DTT for 5 d in 30°C.
Data acquisition, colony quantification, and statistical analysis.
Complete acquisition and analysis of each plate proceeded as follows. First, images of the diploid methotrexate selection with or without DTT were taken after 120 h of growth. Plate images were saved in JPG format at a resolution of 300 dpi. Using the freely available Balony software (http://code.google.com/p/balony/downloads/list), the first step of the image analysis was to determine expected centers of the colonies arrayed in 48 columns and 32 rows. To adjust for variation in plating and possible rotation of an image during image acquisition, we manually defined the coordinate center of a first colony in a first row of the array and of the last colony of the last row. Results of positional array adjustment and detection of colony centers were also validated manually for all images. We then extracted the area for each colony and set a threshold of a positive interaction to be >150. This threshold was chosen accordingly: in the negative interactions, distribution concentrated normally with a mean = 41, standard deviation = 11.1, whereas positive interactions with controls distributed with a mean = 400, standard deviation = 208. Therefore, each colony got a z-score according to its area and distance from the negative controls mean, and a P-value was calculated following the multiple hypothesis correction.
Online supplemental material
Fig. S1 provides insight into the image analysis process (segmentation, threshold of detection, and newly visualized strains). Fig. S2 shows the comparison of our intensity measurements to published datasets. Fig. S3 shows abundance change events for each stress and provides the statistical analysis for detection of significance. Fig. S4 shows detection of potential posttranscriptional regulation events by comparison to published microarray datasets. Fig. S5 provides examples for proteins that change abundance or localization during growth in H2O2 or nitrogen starvation and are essential during these growth conditions. Table S1 summarizes intensity measurements and localization assignments under all conditions. Table S2 summarizes new assignments for protein localization. Table S3 summarizes significantly changed proteins during starvation. Table S4 summarizes significantly changed proteins in DTT. Table S5 summarizes significantly changed proteins in H2O2. Table S6 summarizes bimodality distribution under stress. Table S7 summarizes localization changes under stress. Table S8 summarizes concerted movements of physically interacting proteins under stress. Table S9 summarizes newly acquired interactions of Acc1 in DTT. Table S10 summarizes yeast strains used in this study. Table S11 summarizes plasmids used in this study. Table S12 summarizes primers used in this study. Table S13 summarizes systematically removed proteins. Video 1 shows a fraction of the population of cells expressing mitochondrial protein Aim37-GFP in which it changes its mitochondrial localization to vacuole membrane under H2O2. Video 2 shows a fraction of the population of cells expressing mitochondrial protein Aim37-GFP in which it did not change its localization under H2O2. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201301120/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201301120.dv.
Supplementary Material
Acknowledgments
We would like to thank Ron Milo, Tobias Walther, Ophry Pines, Michal Sharon, Oren Schuldiner, Yaniv Erlich, and Nir Fridman for helpful discussions and critical reading of the manuscript. We would like to thank Keren Katzav for her graphical designs and support and Anastasia Zarankin for constructing the LOQATE database.
This work was funded by the Minerva Minna James Heineman Stiftung grant and by a European Research Council StG (260395). The robotic setup was purchased through the generous support of the Israeli Science Foundation (ISF) Legacy Heritage fund (grant No. 1995/08) and the Human Frontiers Science Program (CDA0006/2008-C) as well as through the generous donation of Mr. James Nathan, Beverly Hills, CA, and the Estate of Lela London. M. Gymrek was supported by the MIT International Science and Technology Initiatives (MISTI). The authors declare that they have no competing financial interests.
Footnotes
Abbreviations used in this paper:
- MTS-dsRed
- matrix-targeted dsRed
- SD
- synthetic medium with dextrose
- SGA
- synthetic genetic array
- TAP
- tandem affinity purification
References
- Acar M., Mettetal J.T., van Oudenaarden A. 2008. Stochastic switching as a survival strategy in fluctuating environments. Nat. Genet. 40:471–475 10.1038/ng.110 [DOI] [PubMed] [Google Scholar]
- Allen C., Büttner S., Aragon A.D., Thomas J.A., Meirelles O., Jaetao J.E., Benn D., Ruby S.W., Veenhuis M., Madeo F., Werner-Washburne M. 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 174:89–100 10.1083/jcb.200604072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón T., van Anken E., Pincus D., Serafimova I.M., Korennykh A.V., Rubio C.A., Walter P. 2009. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature. 457:736–740 10.1038/nature07641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban N.Q. 2011. Persistence: mechanisms for triggering and enhancing phenotypic variability. Curr. Opin. Genet. Dev. 21:768–775 10.1016/j.gde.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Becskei A., Kaufmann B.B., van Oudenaarden A. 2005. Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nat. Genet. 37:937–944 10.1038/ng1616 [DOI] [PubMed] [Google Scholar]
- Ben-Menachem R., Regev-Rudzki N., Pines O. 2011. The aconitase C-terminal domain is an independent dual targeting element. J. Mol. Biol. 409:113–123 10.1016/j.jmb.2011.03.045 [DOI] [PubMed] [Google Scholar]
- Benbadis L., Cot M., Rigoulet M., Francois J. 2009. Isolation of two cell populations from yeast during high-level alcoholic fermentation that resemble quiescent and nonquiescent cells from the stationary phase on glucose. FEM. Yeast Res. 9:1172–1186 10.1111/j.1567-1364.2009.00553.x [DOI] [PubMed] [Google Scholar]
- Beyer A., Hollunder J., Nasheuer H.P., Wilhelm T. 2004. Post-transcriptional expression regulation in the yeast Saccharomyces cerevisiae on a genomic scale. Mol. Cell. Proteomics. 3:1083–1092 10.1074/mcp.M400099-MCP200 [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 19:185–193 10.1093/bioinformatics/19.2.185 [DOI] [PubMed] [Google Scholar]
- Breslow D.K., Cameron D.M., Collins S.R., Schuldiner M., Stewart-Ornstein J., Newman H.W., Braun S., Madhani H.D., Krogan N.J., Weissman J.S. 2008. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods. 5:711–718 10.1038/nmeth.1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causton H.C., Ren B., Koh S.S., Harbison C.T., Kanin E., Jennings E.G., Lee T.I., True H.L., Lander E.S., Young R.A. 2001. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 12:323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernova T.A., Romanyuk A.V., Karpova T.S., Shanks J.R., Ali M., Moffatt N., Howie R.L., O’Dell A., McNally J.G., Liebman S.W., et al. 2011. Prion induction by the short-lived, stress-induced protein Lsb2 is regulated by ubiquitination and association with the actin cytoskeleton. Mol. Cell. 43:242–252 10.1016/j.molcel.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm V.T., Lea H.Z., Rai R., Cooper T.G. 1987. Regulation of allantoate transport in wild-type and mutant strains of Saccharomyces cerevisiae. J. Bacteriol. 169:1684–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y., Schuldiner M. 2011. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol. Biol. 781:127–159 10.1007/978-1-61779-276-2_8 [DOI] [PubMed] [Google Scholar]
- Collins S.R., Miller K.M., Maas N.L., Roguev A., Fillingham J., Chu C.S., Schuldiner M., Gebbia M., Recht J., Shales M., et al. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 446:806–810 10.1038/nature05649 [DOI] [PubMed] [Google Scholar]
- Cyert M.S. 2003. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311:1143–1150 10.1016/S0006-291X(03)01552-3 [DOI] [PubMed] [Google Scholar]
- de Jong I.G., Haccou P., Kuipers O.P. 2011. Bet hedging or not? A guide to proper classification of microbial survival strategies. Bioessays. 33:215–223 10.1002/bies.201000127 [DOI] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley M.E., Hong J.Y., Liebman S.W. 2001. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell. 106:171–182 10.1016/S0092-8674(01)00427-5 [DOI] [PubMed] [Google Scholar]
- Eden E., Geva-Zatorsky N., Issaeva I., Cohen A., Dekel E., Danon T., Cohen L., Mayo A., Alon U. 2011. Proteome half-life dynamics in living human cells. Science. 331:764–768 10.1126/science.1199784 [DOI] [PubMed] [Google Scholar]
- Erjavec N., Larsson L., Grantham J., Nyström T. 2007. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 21:2410–2421 10.1101/gad.439307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel-Morgenstern M., Cohen A.A., Geva-Zatorsky N., Eden E., Prilusky J., Issaeva I., Sigal A., Cohen-Saidon C., Liron Y., Cohen L., et al. 2010. Dynamic Proteomics: a database for dynamics and localizations of endogenous fluorescently-tagged proteins in living human cells. Nucleic Acids Res. 38(Suppl 1):D508–D512 10.1093/nar/gkp808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusova E.E., Ozolins L.N., Bhagat S., Newnam G.P., Wegrzyn R.D., Sherman M.Y., Chernoff Y.O. 2006. Modulation of prion formation, aggregation, and toxicity by the actin cytoskeleton in yeast. Mol. Cell. Biol. 26:617–629 10.1128/MCB.26.2.617-629.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 11:4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefen O., Balaban N.Q. 2009. The importance of being persistent: heterogeneity of bacterial populations under antibiotic stress. FEMS Microbiol. Rev. 33:704–717 10.1111/j.1574-6976.2008.00156.x [DOI] [PubMed] [Google Scholar]
- Gefen O., Gabay C., Mumcuoglu M., Engel G., Balaban N.Q. 2008. Single-cell protein induction dynamics reveals a period of vulnerability to antibiotics in persister bacteria. Proc. Natl. Acad. Sci. USA. 105:6145–6149 10.1073/pnas.0711712105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 418:387–391 10.1038/nature00935 [DOI] [PubMed] [Google Scholar]
- Greenbaum D., Colangelo C., Williams K., Gerstein M. 2003. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4:117 10.1186/gb-2003-4-9-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R., Alberti S., Lindquist S. 2010. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 20:125–133 10.1016/j.tcb.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartigan J.A., Hartigan P.M. 1985. The Dip Test of Unimodality. Ann. Stat. 13:70–84 10.1214/aos/1176346577 [DOI] [Google Scholar]
- Hasslacher M., Ivessa A.S., Paltauf F., Kohlwein S.D. 1993. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J. Biol. Chem. 268:10946–10952 [PubMed] [Google Scholar]
- Hedbacker K., Townley R., Carlson M. 2004. Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 24:1836–1843 10.1128/MCB.24.5.1836-1843.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M.E., Fung E., Wildenhain J., Pierce S.E., Hoon S., Lee W., Proctor M., St Onge R.P., Tyers M., Koller D., et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 320:362–365 10.1126/science.1150021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T.R., Marton M.J., Jones A.R., Roberts C.J., Stoughton R., Armour C.D., Bennett H.A., Coffey E., Dai H., He Y.D., et al. 2000. Functional discovery via a compendium of expression profiles. Cell. 102:109–126 10.1016/S0092-8674(00)00015-5 [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 324:218–223 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isnard A.D., Thomas D., Surdin-Kerjan Y. 1996. The study of methionine uptake in Saccharomyces cerevisiae reveals a new family of amino acid permeases. J. Mol. Biol. 262:473–484 10.1006/jmbi.1996.0529 [DOI] [PubMed] [Google Scholar]
- Joo Y.J., Kim J.H., Kang U.B., Yu M.H., Kim J. 2011. Gcn4p-mediated transcriptional repression of ribosomal protein genes under amino-acid starvation. EMBO J. 30:859–872 10.1038/emboj.2010.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., Frydman J. 2008. Misfolded proteins partition between two distinct quality control compartments. Nature. 454:1088–1095 10.1038/nature07195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J., Bachhawat A.K. 2007. Yct1p, a novel, high-affinity, cysteine-specific transporter from the yeast Saccharomyces cerevisiae. Genetics. 176:877–890 10.1534/genetics.107.070342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K., Yamaguchi E., Arisawa M. 1995. Cloning of the Candida glabrata TRP1 and HIS3 genes, and construction of their disruptant strains by sequential integrative transformation. Gene. 165:203–206 10.1016/0378-1119(95)00552-H [DOI] [PubMed] [Google Scholar]
- Lee M.V., Topper S.E., Hubler S.L., Hose J., Wenger C.D., Coon J.J., Gasch A.P. 2011. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7:514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S.F., Siegal M.L. 2012. The robustness continuum. Adv. Exp. Med. Biol. 751:431–452 10.1007/978-1-4614-3567-9_20 [DOI] [PubMed] [Google Scholar]
- Levy S.F., Ziv N., Siegal M.L. 2012. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 10:e1001325 10.1371/journal.pbio.1001325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madania A., Dumoulin P., Grava S., Kitamoto H., Schärer-Brodbeck C., Soulard A., Moreau V., Winsor B. 1999. The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell. 10:3521–3538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., Nunnari J. 2003. Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 163:503–510 10.1083/jcb.200304040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerovich M., Mamou G., Ben-Yehuda S. 2010. Visualizing high error levels during gene expression in living bacterial cells. Proc. Natl. Acad. Sci. USA. 107:11543–11548 10.1073/pnas.0912989107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy A.D., McCaffery J.M., Shaw J.M. 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151:367–380 10.1083/jcb.151.2.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U., Wang Z., Waern K., Shou C., Raha D., Gerstein M., Snyder M. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 320:1344–1349 10.1126/science.1158441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J.R., Ghaemmaghami S., Ihmels J., Breslow D.K., Noble M., DeRisi J.L., Weissman J.S. 2006. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 441:840–846 10.1038/nature04785 [DOI] [PubMed] [Google Scholar]
- Nissan T., Rajyaguru P., She M., Song H., Parker R. 2010. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell. 39:773–783 10.1016/j.molcel.2010.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimon N., Schuldiner M. 2011. Getting the whole picture: combining throughput with content in microscopy. J. Cell Sci. 124:3743–3751 10.1242/jcs.087486 [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. 2011. Global quantification of mammalian gene expression control. Nature. 473:337–342 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Sigal A., Milo R., Cohen A., Geva-Zatorsky N., Klein Y., Alaluf I., Swerdlin N., Perzov N., Danon T., Liron Y., et al. 2006. Dynamic proteomics in individual human cells uncovers widespread cell-cycle dependence of nuclear proteins. Nat. Methods. 3:525–531 10.1038/nmeth892 [DOI] [PubMed] [Google Scholar]
- Sigal A., Danon T., Cohen A., Milo R., Geva-Zatorsky N., Lustig G., Liron Y., Alon U., Perzov N. 2007. Generation of a fluorescently labeled endogenous protein library in living human cells. Nat. Protoc. 2:1515–1527 10.1038/nprot.2007.197 [DOI] [PubMed] [Google Scholar]
- Stathopoulos-Gerontides A., Guo J.J., Cyert M.S. 1999. Yeast calcineurin regulates nuclear localization of the Crz1p transcription factor through dephosphorylation. Genes Dev. 13:798–803 10.1101/gad.13.7.798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K., Messier V., Landry C.R., Radinovic S., Serna Molina M.M., Shames I., Malitskaya Y., Vogel J., Bussey H., Michnick S.W. 2008. An in vivo map of the yeast protein interactome. Science. 320:1465–1470 10.1126/science.1153878 [DOI] [PubMed] [Google Scholar]
- Tkach J.M., Yimit A., Lee A.Y., Riffle M., Costanzo M., Jaschob D., Hendry J.A., Ou J., Moffat J., Boone C., et al. 2012. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 14:966–976 10.1038/ncb2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Pagé N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 294:2364–2368 10.1126/science.1065810 [DOI] [PubMed] [Google Scholar]
- Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101:249–258 10.1016/S0092-8674(00)80835-1 [DOI] [PubMed] [Google Scholar]
- Tyedmers J., Madariaga M.L., Lindquist S. 2008. Prion switching in response to environmental stress. PLoS Biol. 6:e294 10.1371/journal.pbio.0060294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Olsen J.V., Mann M. 2010. Yeast expression proteomics by high-resolution mass spectrometry. Methods Enzymol. 470:259–280 10.1016/S0076-6879(10)70011-2 [DOI] [PubMed] [Google Scholar]
- Yogev O., Yogev O., Singer E., Shaulian E., Goldberg M., Fox T.D., Pines O. 2010. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 8:e1000328 10.1371/journal.pbio.1000328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.X., Rainey P.B. 2010. Bet hedging in the underworld. Genome Biol. 11:137 10.1186/gb-2010-11-10-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.