Abstract
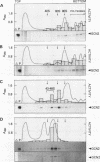
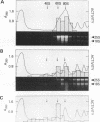
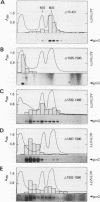
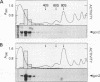
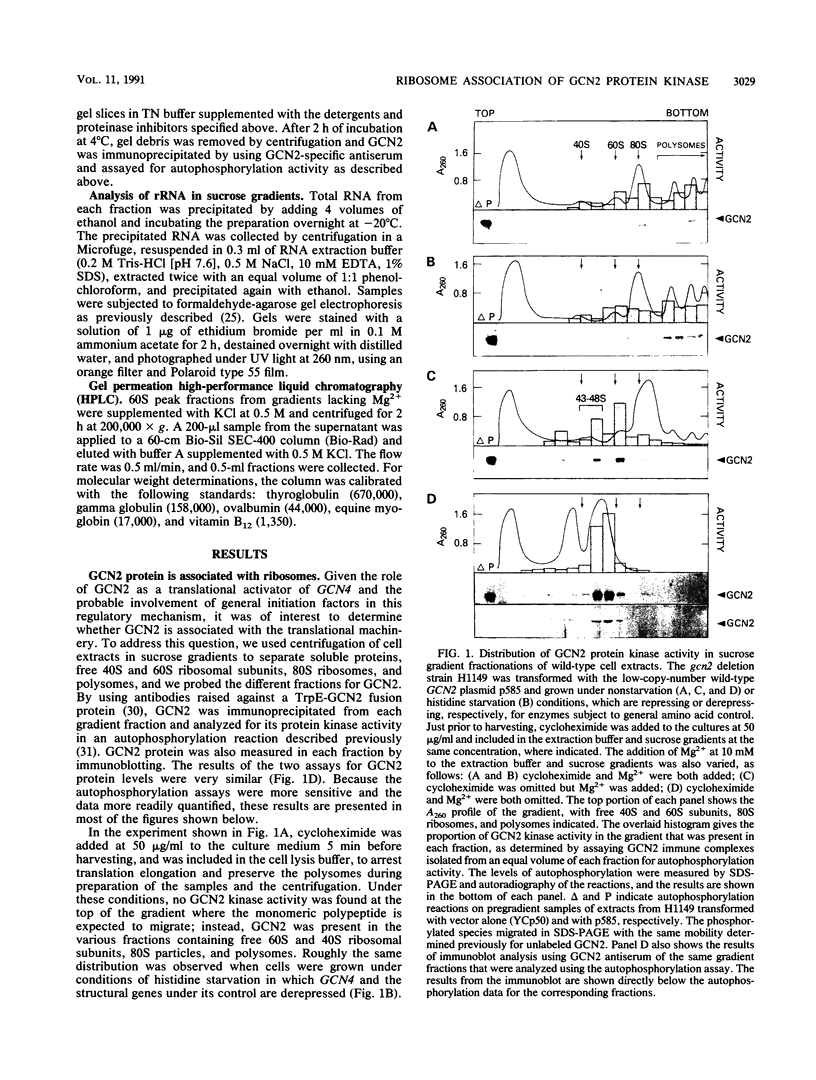
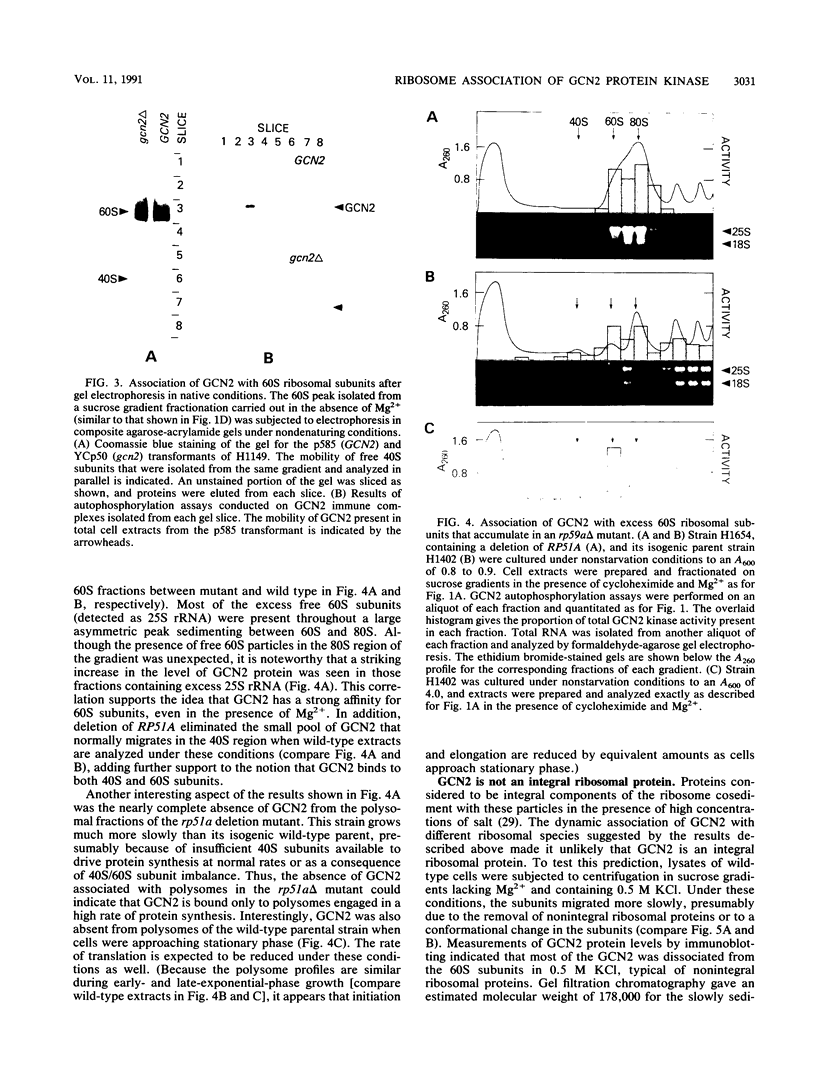
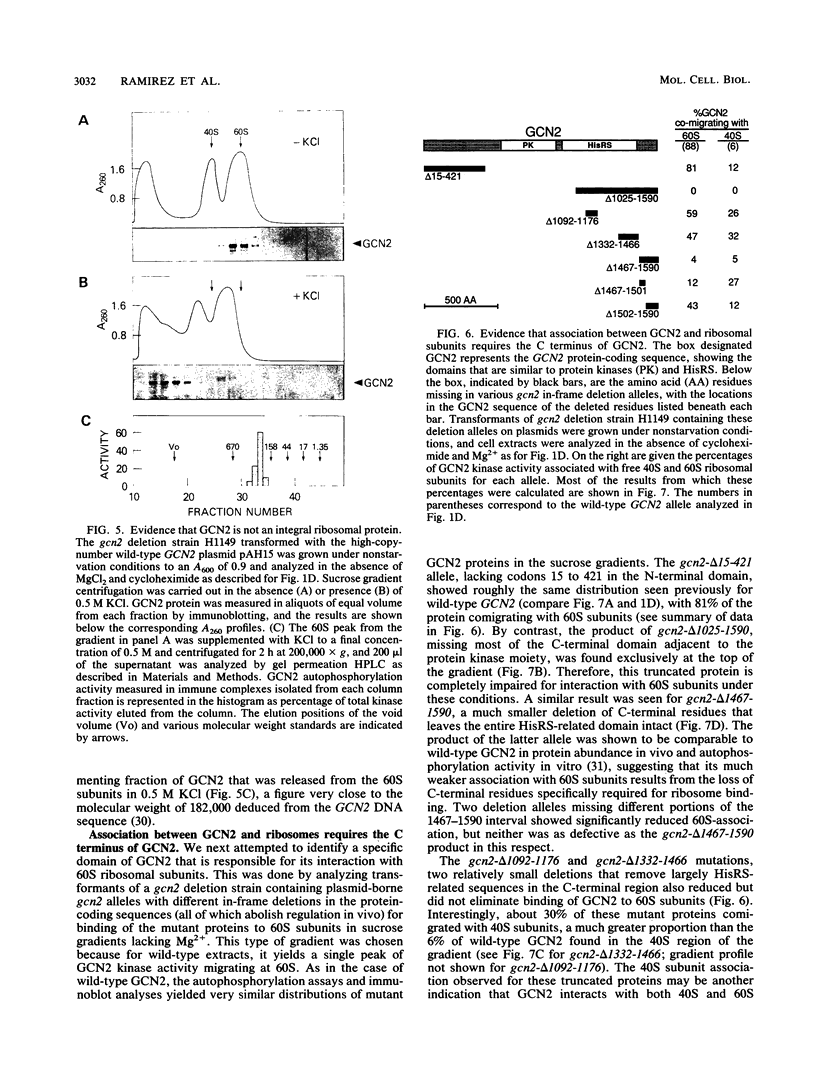
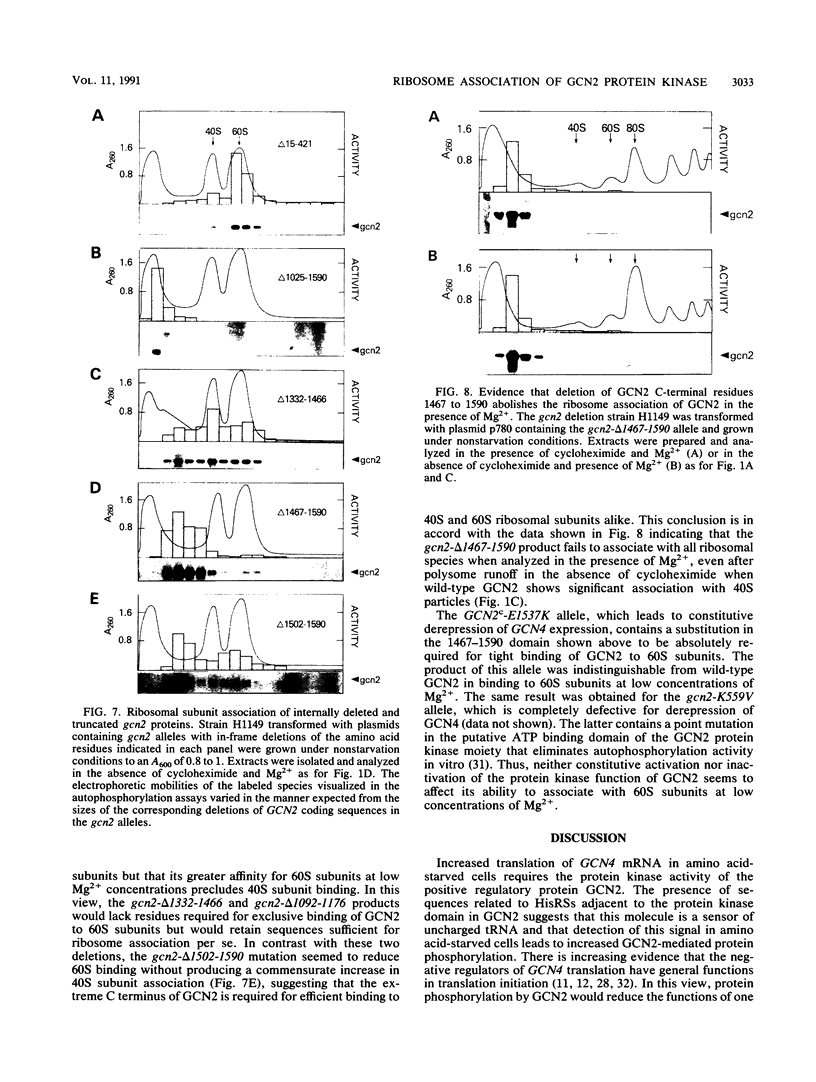
The GCN4 gene of the yeast Saccharomyces cerevisiae encodes a transcriptional activator of amino acid biosynthetic genes that is regulated at the translational level according to the availability of amino acids. GCN2 is a protein kinase required for increased translation of GCN4 mRNA in amino acid-starved cells. Centrifugation of cell extracts in sucrose gradients indicated that GCN2 comigrates with ribosomal subunits and polysomes. The fraction of GCN2 cosedimenting with polysomes was reduced under conditions in which polysomes were dissociated, suggesting that GCN2 is physically bound to these structures. When the association of 40S and 60S subunits was prevented by omitting Mg2+ from the gradient, almost all of the GCN2 comigrated with 60S ribosomal subunits, and it remained bound to these particles during gel electrophoresis under nondenaturing conditions. GCN2 could be dissociated from 60S subunits by 0.5 M KCl, suggesting that it is loosely associated with ribosomes rather than being an integral ribosomal protein. Accumulation of GCN2 on free 43S-48S particles and 60S subunits occurred during polysome runoff in vitro and under conditions of reduced growth rate in vivo. These observations, plus the fact that GCN2 shows preferential association with free ribosomal subunits during exponential growth, suggest that GCN2 interacts with ribosomes during the translation initiation cycle. The extreme carboxyl-terminal segment of GCN2 is essential for its interaction with ribosomes. These sequences are also required for the ability of GCN2 to stimulate GCN4 translation in vivo, leading us to propose that ribosome association by GCN2 is important for its access to substrates in the translational machinery or for detecting uncharged tRNA in amino acid-starved cells.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol. 1991 Jan;11(1):486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Foiani M., Hannig E. M., Hinnebusch A. G. Complex formation by positive and negative translational regulators of GCN4. Mol Cell Biol. 1991 Jun;11(6):3217–3228. doi: 10.1128/mcb.11.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Feng L., Donahue T. F. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Cigan A. M., Paddon C. J., Harashima S., Hinnebusch A. G. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M., Redman R., Kaplansky D. A. Evidence that the primary effect of phosphorylation of eukaryotic initiation factor 2(alpha) in rabbit reticulocyte lysate is inhibition of the release of eukaryotic initiation factor-2.GDP from 60 S ribosomal subunits. J Biol Chem. 1985 Aug 5;260(16):9491–9500. [PubMed] [Google Scholar]
- Gross M., Wing M., Rundquist C., Rubino M. S. Evidence that phosphorylation of eIF-2(alpha) prevents the eIF-2B-mediated dissociation of eIF-2 X GDP from the 60 S subunit of complete initiation complexes. J Biol Chem. 1987 May 15;262(14):6899–6907. [PubMed] [Google Scholar]
- Hanks S. K., Quinn A. M., Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988 Jul 1;241(4861):42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- Hannig E. M., Williams N. P., Wek R. C., Hinnebusch A. G. The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics. 1990 Nov;126(3):549–562. doi: 10.1093/genetics/126.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. E., Struhl K. Molecular characterization of GCD1, a yeast gene required for general control of amino acid biosynthesis and cell-cycle initiation. Nucleic Acids Res. 1988 Oct 11;16(19):9253–9265. doi: 10.1093/nar/16.19.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskas A., Cooper J. A. Autophosphorylation of the PDGF receptor in the kinase insert region regulates interactions with cell proteins. Cell. 1989 Sep 22;58(6):1121–1133. doi: 10.1016/0092-8674(89)90510-2. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levin D., London I. M. Regulation of protein synthesis: activation by double-stranded RNA of a protein kinase that phosphorylates eukaryotic initiation factor 2. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1121–1125. doi: 10.1073/pnas.75.3.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T. E. A simple general method to determine the proportion of active ribosomes in eukaryotic cells. Exp Cell Res. 1973 Aug;80(2):496–498. doi: 10.1016/0014-4827(73)90333-9. [DOI] [PubMed] [Google Scholar]
- Martin T. E., Hartwell L. H. Resistance of active yeast ribosomes to dissociation by KCl. J Biol Chem. 1970 Mar 25;245(6):1504–1506. [PubMed] [Google Scholar]
- Meurs E., Chong K., Galabru J., Thomas N. S., Kerr I. M., Williams B. R., Hovanessian A. G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990 Jul 27;62(2):379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989 May 19;57(4):585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Moldave K. Eukaryotic protein synthesis. Annu Rev Biochem. 1985;54:1109–1149. doi: 10.1146/annurev.bi.54.070185.005333. [DOI] [PubMed] [Google Scholar]
- Parent S. A., Fenimore C. M., Bostian K. A. Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast. 1985 Dec;1(2):83–138. doi: 10.1002/yea.320010202. [DOI] [PubMed] [Google Scholar]
- Roussou I., Thireos G., Hauge B. M. Transcriptional-translational regulatory circuit in Saccharomyces cerevisiae which involves the GCN4 transcriptional activator and the GCN2 protein kinase. Mol Cell Biol. 1988 May;8(5):2132–2139. doi: 10.1128/mcb.8.5.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. R., Söll D. Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu Rev Biochem. 1979;48:601–648. doi: 10.1146/annurev.bi.48.070179.003125. [DOI] [PubMed] [Google Scholar]
- Silverman S. J., Rose M., Botstein D., Fink G. R. Regulation of HIS4-lacZ fusions in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Oct;2(10):1212–1219. doi: 10.1128/mcb.2.10.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N. S., Matts R. L., Levin D. H., London I. M. The 60 S ribosomal subunit as a carrier of eukaryotic initiation factor 2 and the site of reversing factor activity during protein synthesis. J Biol Chem. 1985 Aug 15;260(17):9860–9866. [PubMed] [Google Scholar]
- Tzamarias D., Alexandraki D., Thireos G. Multiple cis-acting elements modulate the translational efficiency of GCN4 mRNA in yeast. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4849–4853. doi: 10.1073/pnas.83.13.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D., Roussou I., Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989 Jun 16;57(6):947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Warner J. R., Gorenstein C. The ribosomal proteins of Saccharomyces cerevisiae. Methods Cell Biol. 1978;20:45–60. doi: 10.1016/s0091-679x(08)62008-7. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Jackson B. M., Hinnebusch A. G. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Ramirez M., Jackson B. M., Hinnebusch A. G. Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol Cell Biol. 1990 Jun;10(6):2820–2831. doi: 10.1128/mcb.10.6.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N. P., Hinnebusch A. G., Donahue T. F. Mutations in the structural genes for eukaryotic initiation factors 2 alpha and 2 beta of Saccharomyces cerevisiae disrupt translational control of GCN4 mRNA. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7515–7519. doi: 10.1073/pnas.86.19.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinker S., Warner J. R. The ribosomal proteins of Saccharomyces cerevisiae. Phosphorylated and exchangeable proteins. J Biol Chem. 1976 Mar 25;251(6):1799–1807. [PubMed] [Google Scholar]