Summary
MHC class I peptide complexes (pMHC) are routinely used to enumerate T cell populations, and are currently being evaluated as vaccines to tumors and specific pathogens. Herein we describe the structures of three generations of single-chain pMHC progressively designed for the optimal presentation of covalently associated epitopes. Our ultimate and novel design employs a versatile disulfide trap between an invariant MHC residue and a short C-terminal peptide extension. This general strategy is non-disruptive of native pMHC conformation and T cell receptor engagement. Indeed, cell surface expressed MHC complexes with disulfide-trapped epitopes are refractory to peptide exchange suggesting they will make safe and effective vaccines. Furthermore, we find that disulfide-trap stabilized, recombinant pMHC reagents reliably detect polyclonal CD8 T cell populations as proficiently as conventional reagents, and are thus well-suited to monitor or modulate immune responses during pathogenesis.
Introduction
Class I major histocompatibility complex (MHC) proteins serve a critical role in the adaptive immune response by binding short peptide fragments intracellularly and presenting them at the cell surface for surveillance by cytotoxic T lymphocytes (CTLs). Structural studies of human and murine MHC class I proteins have revealed that peptides of 8–10 residues in length are presented to TCRs in the context of a narrow groove formed by two antiparallel α-helices positioned above an eight-strand, antiparallel β-sheet [1–5]. This peptide-binding platform is comprised of the α1 and α2 domains of a polymorphic heavy chain (HC), which also contains an immunoglobulin-like α3 domain, a transmembrane segment, and a short cytoplasmic tail. Stable surface expression of MHC complexes requires the association of the peptide-HC assembly with β2-microglobulin (β2m), which contacts the α3 domain and supports the peptide-binding platform.
The generation of antigen-MHC complexes for priming and activating of specific CD8 T cell responses has been the goal of a number DNA vaccination strategies [6]. In some of these vaccination regimes the immunogen is encoded by a DNA delivery vector and is expressed in vivo by transfected dendritic cells (DC) upon vaccination [7]. The potency of such DNA vaccines is critically dependent on the presentation of processed antigenic epitopes by MHC class I on the surface of the transfected DCs. However, certain immunologically important tumor- or pathogen-derived antigens are not presented efficiently at the plasma membrane. This inefficiency results from the fact that the surface expression of pMHC is influenced by a variety of factors including efficiency of antigen processing [8, 9], specificity of peptide translocation into the ER [10], interference by viral pathogens [11], and the biogenesis and kinetic stability of the pMHC itself [12].
In addition to vaccine development, MHC proteins have been used extensively to construct soluble multimer reagents that have been successfully used for T cell enumeration and diagnostics [13–18] and more recently for the isolation of therapeutic antigen-specific T cells [19, 20]. A major challenge for the production and use of pMHC multimers is peptide dissociation. Indeed, it has been shown that naïve CD8 T cells can activate each other by cross presentation of dissociated peptide from soluble recombinant pMHC [21, 22]. This problem is especially relevant to potential clinical applications of pMHC reagents infused into patients for the manipulation of immune responses. These include, for example, antibody-mediated targeting of MHC class I complexes to the surface of tumor cells [23, 24] and direct in vivo modulation of T cells by recombinant pMHC [25, 26].
Therefore, to enhance antigenic peptide presentation and to prevent epitope dissociation from the MHC we engineered the pMHC as a covalent assembly. These engineered proteins, which we term single chain trimers (SCTs), consist of a single polypeptide comprised of an antigenic peptide followed by a flexible linker that connects the C terminus of the peptide to the N terminus of β2m and another flexible linker that connects the C terminus of β2m to the N terminus of the HC. In our initial studies we used the Kb-Ova complex as a platform to create the first SCT. These studies revealed that SCT transfected into cells assembled rapidly in the ER, exhibited extended cell surface half-life, and were more resistant to exogenous peptide exchange compared to Kb loaded with endogenous peptides [27]. This first generation SCT, with its high-affinity Ova constituent, has been particularly valuable in notable studies of the biology of MHC class I [28, 29].
Upon further characterization, we found that extremely high concentrations of competitor peptide could displace the Ova sequence more easily in the SCT than in native Kb-Ova [30]. This observation suggested that the peptide-β2m linker was disrupting optimal anchoring of the Ova C-terminus. If this were true, the extended cell surface half-life of SCTs relative to native Kb-Ova likely reflects their capacity to rebind the spacer-attached Ova rather than continuous groove occupancy. Therefore a second SCT generation was designed to open the C-terminal end of the MHC groove in order to better accommodate the C-terminal linker of the presented peptide [30]. Our goal was to optimize the SCT design for therapeutic and diagnostics applications, particularly when the incorporated class I ligand was not of high affinity. Therefore we crystallized and solved the structures of the first and second SCT generations in order to define the chemical basis of peptide binding and presentation by these constructs. Our structural data show that we have successfully re-engineered the SCT groove to accommodate the free extrusion of the C-terminal peptide linker.
Improving upon these original constructs, we engineered a third generation SCT by incorporating a disulfide trap in the region of the SCT groove involved in binding the C-terminus of the peptide known as the F pocket of the MHC. We reasoned that introduction of a disulfide trap would lock-in the linker-attached peptide and further stabilize the integrity of the SCT. Herein we present structural, chemical, and functional characterizations of this new construct and provide unequivocal evidence that the introduction of an F pocket disulfide improves C-terminal peptide anchoring, reliably precludes the binding of competitive peptides, and preserves TCR engagement. Altogether, these results suggested that an analogous disulfide bond could lock pMHC without the use of subunit linkers. Indeed, we demonstrate here the use of such disulfide to trap a synthetic peptide in the groove of a soluble, recombinant HC. We also show that tetramers constructed with disulfide-trapped pMHC (dt-pMHC) reliably detect pathogen-specific T cells. The diagnostic, vaccine and immune regulatory applications of these various pMHC designs are discussed.
Results
Crystal structure analysis of a single-chain pMHC trimer
The first generation SCT, designated here SCTWT, was designed by linking the C terminus of Ova (SIINFEKL, Ovalbumin, residues 257–264) using a (Gly4-Ser)3 linker to the N terminus of β2m and linking the C terminus of β2m to the N terminus of the Kb HC using a (Gly4-Ser)4 linker (Figure 1a). To probe the molecular means by which MHC proteins designed as single chain trimers bind and present peptides and to obtain a structural framework for their improvement we initiated a crystallographic investigation. Bacterially expressed and oxidatively refolded SCTWT protein crystallized in the primitive monoclinic space group P21 with two molecules per asymmetric unit (Table I). After initial refinement interpretable electron density was seen for the Ova peptide and linker residues immediately C-terminal to Ova. The final electron density map was of excellent quality with no ambiguities observed for main chain or side chain atoms of the Ova peptide. Interpretable density was also observed for the entire peptide-β2m linker. In contrast, no electron density was observed for the β2m-HC linker or for residues 278–280 at the C terminus of the HC. The final SCTWT model refined to 2.0 Å resolution has an R factor of 21.5% (Rfree 25.3%) with excellent geometry.
Figure 1. Single chain trimer designs and structural features.
(a) Schematic representations of the SCTs are shown. Ova, β2m, and HC, are represented as yellow, magenta, and cyan boxes, respectively. The linkers are represented as thin orange boxes. Linker sequences and residue numbering for Ova, β2m, and HC are shown above their respective representations. Disulfide bonds bridging the indicated cysteine residues are represented as green brackets. (b) Ribbon diagram of SCTWT. Ova and PBL are rendered as ball and stick models and colored as follows: Ova and PBL carbon atoms, yellow and orange, respectively; nitrogen atoms, blue; oxygen atoms, red. The protein is oriented with the Ova N terminus on the left and the membraneproximal α3 domain at the bottom. A possible conformation for BHL is represented and rendered as small orange balls. (c) Stereo representation of the experimental electron density for the disulfide bond between CysPBL2 and Cys84 in the dt-SCT and the surrounding region. A simulated-annealing, omit map (CNS) calculated at 1.80 Å is shown in cyan at a contour of 2.0σ. The FO-FC map was constructed from model phases calculated without the coordinates for Ova, PBL, and residues 76 through 87 and 137 through 148 of one of the molecules in the asymmetric unit. A ball and stick representation of the omitted region of dt-SCT is shown and superposed onto the electron density and colored as in (a) with sulfur atoms colored green.
Table 1.
Data collection, refinement, and structure comparisons statistics
| SCTWT | SCTY84A | SCTY84C-PBL2C | |
|---|---|---|---|
| Data collection | |||
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 66.2, 89.6, 88.5 | 66.1, 89.3, 88.5 | 66.6, 89.1, 88.6 |
| α=γ=90, β (°) | 111.0 | 111.0 | 110.9 |
| No. of Molecules Per ASU | 2 | 2 | 2 |
| Resolution (Å) | 20.0-2.00(2.07-2.00)b | 20-2.00(2.07-2.0) | 20-1.80(1.86-1.80) |
| Rsym a | 7.5(47.3) | 9.3(53.5) | 5.8(53.3) |
| I/σI | 23.4(3.80) | 17.7(3.07) | 30.3(3.39) |
| Completeness (%) | 98.4(97.2) | 99.9(99.7) | 99.9(100) |
| Redundancy | 6.8 | 6.3 | 6.8 |
| Refinement | |||
| Resolution (Å) | 20.0-2.00(2.07-2.00) | 20.0-2.00(2.07-2.00) | 20.0-1.80(1.86-1.80) |
| No. reflections | 64087(5963) | 64727(6111) | 89351(8411) |
| Rwork/Rfreeb | 21.5(34.1) / 25.3(35.1) | 21.0(30.8)/25.2(34.7) | 20.8(33.3)/23.9(34.2) |
| No. atoms | |||
| Protein | 6396 | 6382 | 6388 |
| Water | 639 | 700 | 755 |
| B-factors (Å2) | |||
| Protein | 35.8 | 30.2 | 34.9 |
| Water | 44.4 | 39.8 | 45.2 |
| R.m.s.d. from ideal values | |||
| Bond lengths (Å) | 0.0088 | 0.0086 | 0.0093 |
| Bond Angles (°) | 1.56 | 1.55 | 1.678 |
| Structural Comparisonsd | |||
| R.m.s.d. (Å) from 1VAC(c2) | |||
| Kb-Ova / HC / α1α2 / β2m / Ova | 1.96/1.15/0.61 /0.98/0.52 | 2.15/1.34/0.78/0.83/0.48 | 2.15/1.37/0.81 /0.97/0.51 |
| R.m.s.d. (Å) from 1T0M(p21) | |||
| Kb/HC/α1α2/β2m | 0.79/0.72/0.55/0.85 | 0.64/0.68/0.46/0.45 | 0.72/0.66/0.55/0.82 |
Statistics as defined in SCALEPACK
Values in parentheses are for data in the highest resolution shell
Statistics as defined in CNS
R.m.s.d. values calculated based on an alignment of main-chain atoms of the indicated complexes or domains using Lsqkab in CCP4
Our structural data reveals that the Ova portion of the single chain is bound in the MHC groove in canonical fashion (Figure 1b). The anchoring residues IleP2, PheP5 and LeuP8 are buried in the complex and point toward the β-sheet platform in manner similar to that observed in the native complex (Figure 2a and 2b) [31]. The peptide-β2m linker extrudes from the C-terminal end of the peptide-binding groove, traverses over the C-terminal, TCR-proximal surface of the α1 helix, and joins the N terminus of β2m on the membrane-proximal side of the peptide-binding platform (Figure 1b, see also Figures 2b and 3b). This linker does not adopt any regular secondary structure and is seen in different conformations in each complex of the asymmetric unit consistent with its solvent exposure (Figure 1b).
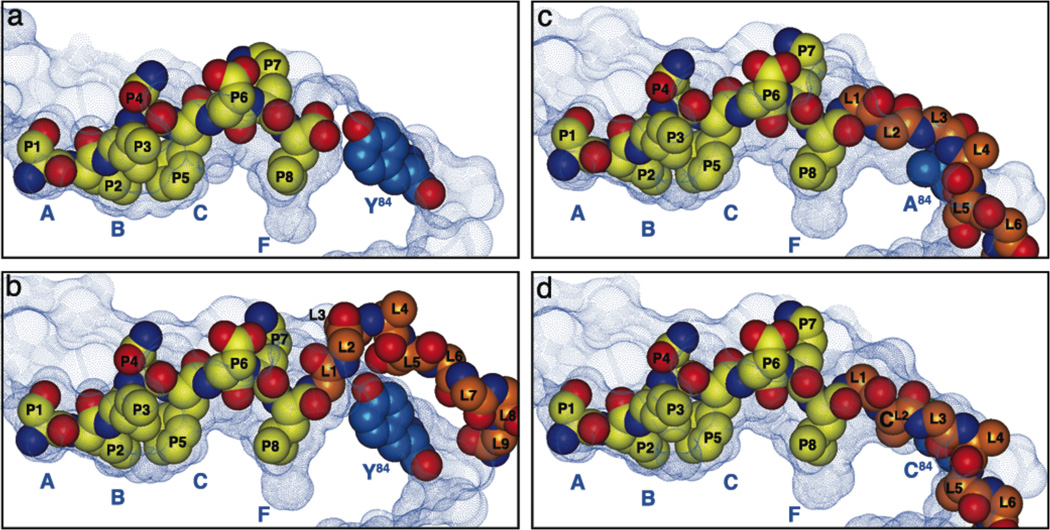
Figure 2. Linker accommodation in three generations SCT.
The solvent-accessible surfaces of the peptide-binding grooves of Kb (a), SCTWT (b), SCTY84A (c), and dt-SCT (d) are displayed as blue dotted surfaces. Ova, as well as the Ova portion and the first nine PBL residues in SCTWT, or the first six PBL residues in SCTY84A and the dt-SCT are represented as CPK models and colored as in Figure 1b. Tyr84, as well as Ala84, and Cys84 are also depicted as CPK models and colored in light blue. Kb pocket locations are indicated underneath each surface. For clarity PBL residues are labeled L1 through L9 for SCTWT, and L1 through L6 for SCTY84A and dt-SCT.
Structure-based design of novel SCTs
A closer inspection of the SCTWT F pocket (Figure 2b) reveals that Tyr84 precludes optimal linker positioning by forcing a near 90° turn in the protein main chain at the first linker residue. Moreover the closed nature of the SCT groove constrained the linker extrusion over a conserved TCR antigen recognition region [32, 33]. In order to improve linker accommodation and to potentially enhance peptide-MHC association, we re-engineered the peptide-binding groove of the SCTWT protein. Our design was based on examination of MHC class I and class II structures, since class II molecules allow binding of peptides with C-terminal extensions [31, 34]. These comparisons revealed that the invariant Tyr84 in class I proteins is prominent in closing the MHC groove around the peptide C terminus.
Accordingly we mutated Tyr84 to an Ala to create SCTY84A (Figure 1a). Initial studies of immune recognition of this SCT were highly suggestive that this mutation allows relaxation of the peptide-β2m linker [35]. Next, we introduced a stabilizing disulfide trap between the peptide and the MHC by engineering a disulfide bond between a cysteine introduced in place of Tyr84 and a cysteine introduced at the second position in the peptide-β2m linker (Figure 1a). We hypothesized that a Cys at position 84 would maintain an opening in the MHC groove and that a disulfide bond at this position would re-establish C-terminal anchoring. This SCT construct was designated dt-SCT (disulfide-trapped SCT).
We crystallized and solved the structures of SCTY84A and dt-SCT. These proteins crystallized under similar crystallization conditions with the same space group and similar unit cell dimensions as the SCTWT (Table I). Diffraction data to 2.00 Å (SCTY84A), and 1.80 Å (dt-SCT) were used for refinement of the final atomic models, which have R factors of 21.0% (Rfree 25.2%) for SCTY84A, and 20.8% (Rfree 23.9%) for dt-SCT. The electron density maps for each of the SCT molecules were of excellent quality (Figure 1c). Interpretable density was seen for residues 1 through 7 and 11 through 15 in the first linker of SCTY84A, and residues 1 through 7 and 12 through 15 in the first linker of the dt-SCT. Furthermore, the electron density in the F pocket of dt-SCT verifies the formation of the engineered disulfide bond, which we designed to enable durable peptide anchoring (Figure 1c). No interpretable electron density was seen for the β2m-HC linker and for residues 278 through 280 at the C terminus of the HC in any of the SCT structures.
To see how the peptide-β2m linker is accommodated in the new SCT constructs and to assess the structural changes in each MHC groove we calculated the solvent accessible surface [36] of the α1α2 domain in each SCT using a probe radius of 1.4 Å (Figure 2). Comparison with the solvent accessible surface of native Kb reveals the way in which Tyr84 closes the C-terminal end of the Kb groove (Figure 2a). In contrast to SCTWT (Figure 2b), the structures of SCTY84A and dt-SCT (Figure 2c and 2d, respectively) have a novel channel that accommodates the extrusion of the peptide-β2m linker away from the TCR-proximal surface of the α1 helix. Thus, the groove opening by the Y84A and Y84C mutations not only optimized peptide-MHC interactions, but also minimized the possibility that linker residues would hinder immune recognition of antigen.
Structural and functional equivalence between SCT and native pMHC
Structural alignment of all of the single chain platform domains to native Kb bound to Ova yielded pair-wise all-atom r.m.s.d. values of 1.14 Å for SCTWT, 1.32 Å for SCTY84A, and 1.32 Å for dt-SCT indicating that only minimal structural perturbations of the antigen presenting platform are created in each SCT (Figure 3a). Most of the main chain differences were found in solvent-exposed, flexible loops and the C-terminal regions of the α1α2 domains. Importantly wild-type Kb-Ova crystallized with different lattice contacts from the SCTs and comparisons of our structures with Kb and Kbm8 complexes with similar lattice interactions indicated even smaller differences (Table I).
Figure 3. Functional and structural equivalence of the SCT constructs and native H2-Kb-Ova.
(a) The peptide-binding platforms of SCTWT, gray; SCTY84A, yellow; dt-SCT, magenta are superposed onto the platform of native Kb-Ova, cyan. The superposition is based on the overlap of the main chain atoms of HC residues 1 through 182. The superposed structures are depicted as ribbon tubes. Side chains for Ova residues in each SCT and in the native complex are rendered as ball and stick models and colored correspondingly. (b) Response of OT-1 T cells to LM1.8 cells transfected with Kb and fed 10 µM Ova (panel 1, cyan); or transfected with SCTWT (panel 2, gray), SCTY84A (panel 3, yellow), and dt-SCT (panel 4, magenta) is compared to the OT-1 response to control LM1.8 transfectants (H2-Kb, no peptide, shown in red in each panel) in a 51Cr release assay. (c) and (d) The Ova atomic positions in each SCT are compared to the native peptide. A close-up view of the superposed Ova peptides is shown at a near 90° rotation from the orientation in (a). Aligned underneath the peptides (d) are quantified positional differences, which were graphed to denote per residue main chain (top) and side chain (bottom) differences in reference to native Ova.
These initial comparisons led us to believe that the engineered perturbations in each SCT would have negligible effects on T cell recognition. To test this hypothesis the SCT constructs were introduced into the LM1.8 cell line and tested as targets in a chromium-release assay against Kb-Ova reactive OT-1 T cells (Figure 3b). As we anticipated, the OT-1 T cell clone recognized each SCT efficiently in contrast to control LM1.8 Kb-transfected cells. This indicated that each SCT construct folded properly in the ER, was expressed at the cell surface, and retained the native antigen recognition elements involved in TCR activation.
We also compared the conformations of Ova in each of the SCT constructs to that of Ova in the native complex. Alignment of the antigen-binding platforms revealed only minor differences in the main chain and side chain atomic positions for the anchoring Ova residues in each SCT compared to Kb-Ova (Figure 3c and 3d). Likewise, the conformations of solvent exposed Ova residues are also very similar with the most significant differences seen for the side chain of LysP7 (Figure 3c and 3d). Thus our SCT proteins can capably present peptides in a manner very similar to their native counterparts.
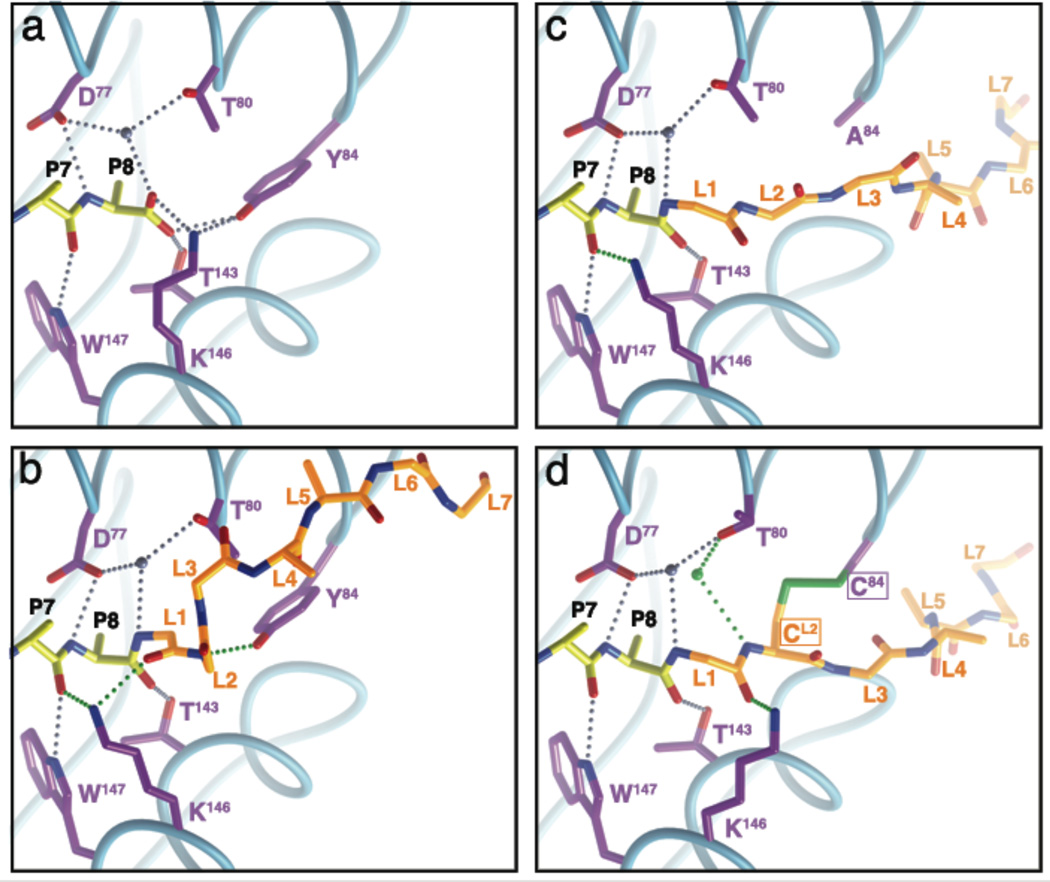
Reconfiguration of F-pocket peptide anchoring
Since the SCT format could impact anchoring of the C-terminus of the peptide, we analyzed the hydrogen bonding established in the F pockets of each SCT (Figure 4) and compared them to that in native Kb-Ova (Figure 4a). In each structure, the amide nitrogen atom of GlyPBL1 occupies nearly the same position as one of the terminal oxygen atoms of native Ova. A structurally conserved water molecule, which mediates a number of important hydrogen bonds between the peptide C terminus and conserved MHC residues, is present in all SCT structures and is coordinated similarly as in the native structure. On the other hand Lys146, which normally hydrogen bonds to Tyr84 and the other terminal oxygen atom of Ova, adopts one of two conformations in the different SCT structures. In the SCTWT (Figure 4b) and SCTY84A (Figure 4c) structures, Lys146 forms a hydrogen bond with the carbonyl oxygen atom of LysP7, while in dt-SCT (Figure 4d) it hydrogen bonds to the carbonyl oxygen of GlyPBL1. A second water molecule is also located in the F pocket of dt-SCT (Figure 4d), which is coordinated by the amide nitrogen atom of CysPBL2 and the Oδ1 oxygen atom of Thr80. Thus the SCTY84A and the dt-SCT seem to establish unique F pocket hydrogen bonding networks compared to the native Kb-Ova complex; however, as noted above, there are no major conformational differences between each SCT generation and the native complex associated with the MHC groove re-engineering.
Figure 4. Re-engineering of F pocket peptide anchoring.
HC residues involved in hydrogen bonding to the main chain of LysP7 and LeuP8 in Kb-Ova (a); LysP7, LeuP8, GlyPBL1 and GlyPBL2 in SCTWT (b) and SCTY84A (c); and LysP7, LeuP8, GlyPBL1 and CysPBL2 in dt-SCT (d) are shown and colored in purple. Peptide and linker are represented as in Figure 1(b) with side chains beyond Cβ atom omitted. Preserved hydrogen bonds are shown as small silver balls, while novel ones are depicted in green. Preserved water molecules are colored in gray, while the additional water molecule in dt-SCT is colored in green. The main chain traces of the peptide binding platforms are shown as thin cyan tubes. The complex is oriented as in Figure 2b. For clarity PBL residues are labeled L1 through L7.
Stability of dt-SCT over SCTWT and SCTY84A
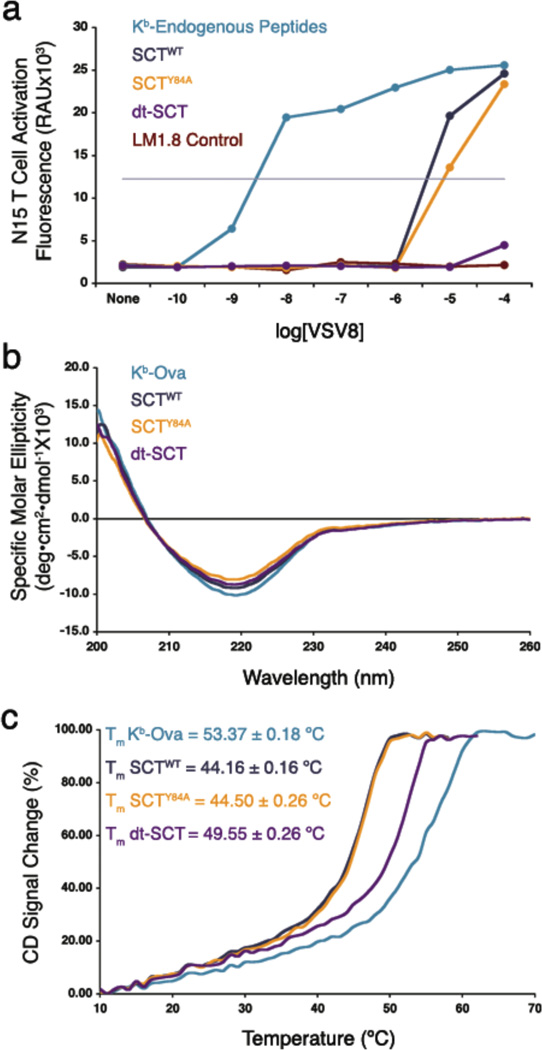
An important goal of expressing class I pMHC as SCT was to achieve a kinetically stable assembly that retains and presents a single peptide determinant. To test the ability of the disulfide trap to secure the Ova sequence in the dt-SCT protein, we used a highly sensitive T cell hybridoma expressing the N15 TCR to detect exogenous VSV8 peptide binding to Kb SCT constructs expressed on LM1.8 cells (Figure 5a). We compared the relative accessibility of exogenous peptide binding to the various Kb constructs by determining the concentration of VSV8 peptide required to achieve half-maximal activation of the N15 hybridoma (Figure 5a). Our finding show that SCTWT and SCTY84A proteins were greater that 1000-fold more resistant to exogenous peptide binding compared with native Kb, confirming published findings [30]. Most impressively, the novel disulfide trap makes the SCT at least 100-fold more refractory that the previous SCT constructs and greater that 105 times more resistant to exogenous peptide binding than native Kb.
Figure 5. SCT protein stability and prevention of peptide exchange.
(a) N15 hybridoma activation to detect VSV8 binding to native Kb or SCT proteins. LM1.8 cells expressing the indicated constructs were incubated for 24 hours with increasing concentration of exogenous VSV8 peptide. IL-2 production was measured via proliferation of CTL L-2 cells, which was in turn detected by Alamar blue fluorescence at 590 nm. Horizontal gray line shows the level of half-maximal hybridoma activation. (b) Far UV CD spectra of SCTWT, SCTY84A, dt-SCT, and Kb-Ova. The measured CD signal is given as [Molar Ellipticity]r, the molar ellipticity per residue. (c) Thermostability as measured by CD. Increase in the CD signal at 220 nm as a function of temperature was normalized to a scale of 0 to 100. The Tm (see Experimental Procedures) for each protein is denoted in the legend.
We also tested the thermostability of purified, recombinant SCTs by following their denaturation profiles as a function of temperature using CD spectroscopy. As a control we looked at the far UV spectra of the SCT proteins at the beginning of each temperature denaturation (10°C), which show that each construct adopts similar secondary structure in the solution conditions used in these experiments (Figure 5b). Reproducibly higher Tm values were observed for the dt-SCT protein (49.55 °C) when compared with those of SCTWT and SCTY84A (44.16 °C and 44.50 °C, respectively). This Tm increase is comparable to that observed when a better anchoring residue was introduced in a peptide bound to Kbm8 [37]. These thermostability data (Figure 5c) along with the peptide competition analysis (Figure 5a) demonstrate that the dt-SCT forms a stable assembly in which the disulfide trap effectively prevents peptide dissociation. Furthermore, we have also found that when relatively low affinity peptide complexes with a different MHC allele (Ld) were compared, the disulfide trap improved peptide occupancy relative to the native complex (data not shown).
Characterization of SCT-based staining reagents
We and others have recently shown that tetramers can be constructed using SCTs and they reliably stain clonal T cell populations [30, 38, 39]. To determine whether the SCT engage its cognate TCR in a manner indistinguishable from the native pMHC we performed competitive staining of the same T cell sample with tetramers made of SCT or native Kb-Ova but conjugated to different fluorochromes. When tested individually, the tetramers constructed with SCTWT or SCTY84A stained OT-1 cells comparably to each other and to conventional tetramers (Figure 6a and data not shown). More importantly, the staining of SCT tetramers displayed a striking correlation to the staining of conventional tetramers when tested in combination demonstrating that SCT tetramers retained specificity for the OT-1 TCR (Figure 6a).
Figure 6. Application of disulfide-trap technology for soluble reagent generation.
Flow cytometry was used to detect staining of CD8+ lymphocytes by conventional and engineered pMHC tetramers. All plots are gated on CD8+ cells, and percentages of CD8+ cells are given for relevant quadrants. (a) OT-1 transgenic splenocytes were stained individually (first row) with conventional Kb-Ova tetramers conjugated to APC (left panel), SCTY84A tetramers conjugated to PE (middle panel), or Kb-Ova based dt-pMHC tetramers conjugated to PE (right panel). These cells were then double stained (second row) with conventional Kb-Ova and SCTY84A tetramers (left panel) or Kb-Ova and dt-pMHC tetramers (right panel). (b) H2-Kb-Ova-specific polyclonal T cells were generated by infecting B6 mice intravenously with 5×103 cfu of L.m.-Ova. After 7 days, splenocytes were harvested and stained as described in (a).
We next determined whether polyclonal T cells responding to the native class I complex recognize equivalently SCT-based tetramers. Six B6 mice were infected with a strain of Listeria monocytogenes that expresses ovalbumin. To determine the extent of overlap in CD8 T cell populations that recognize each type of pMHC, tetramers of both conventional and SCT configurations were used to stain splenocytes from the infected mice (Figure 6b). These studies revealed that similar numbers of CD8 T cells were detected by each tetramer alone (Figure 6b). More significantly, when conventional and SCT tetramers linked to different fluorochromes were used to simultaneously stain the B6 splenocytes, the same polyclonal subset of antigen-specific T cells bound equivalently to both tetramers (Figure 6b and Supplemental Table I). Similar results were obtained after infection of B6 mice with gammaherpesvirus 68 expressing ovalbumin (γHV68-Ova, data not shown). Neither conventional nor SCT tetramers stained cells from uninfected B6 mice (Supplemental Figure 1a); negative control H2-Kb tetramers with an irrelevant peptide (SIYRYYGL) did not stain OT-1, uninfected B6, or pathogen-infected B6 splenocytes (Supplemental Figure 1b). Thus these findings demonstrate that SCT proteins have a native structure capable of specifically engaging polyclonal, pathogen-infected CD8 T cells. This finding further validates the use of SCT-based vaccines to elicit physiologic immunity.
Generation of disulfide-trapped pMHC reagents
We next considered the possibility of generating pMHC tetramers in which only the peptide is permanently attached to the class I HC. We reasoned that covalent attachment of synthetic peptide through a disulfide bond would preclude the need for stabilizing linkers involving β2m. The result would be a minimally modified class I pMHC with a synthetic peptide disulfide-trapped to the peptide-binding groove. We designated these recombinant molecules disulfide-trap pMHC, or dt-pMHC.
To generate these complexes, soluble Kb HC carrying the Y84C mutation and C-terminal biotinylation sequence [40] was refolded with β2m and a modified Ova peptide. This peptide was C-terminally extended to include a Gly-Cys sequence for disulfide bond formation analogous to the one observed in the dt-SCT structure. To demonstrate conclusively that tetramers of the dt-pMHC format are valid reagents that mimic exactly the native MHC class I determinant, we used the system described above to perform dual tetramer staining. OT-1 staining by both tetramers either alone or in combination clearly verified that this TCR is detected equivalently by disulfide-trapped and conventional tetramers (Figure 6a). Likewise, each tetramer stained similar numbers of CD8 T cells from six individual L.m.-Ova infected mice, and when staining simultaneously with both tetramers, the antigen-specific CD8 T cells were bound by both tetramers (Figure 6b and Supplemental Table I). Furthermore, tetramer staining of splenocytes from B6 mice infected with γHV68-Ova yielded very similar patterns (data not shown). These results demonstrate that disulfide-trap technology can be used to covalently attach a synthetic peptide antigen to a recombinant MHC HC. Our data furthermore demonstrate that this novel approach will make stain reagents with the specificity and sensitivity to enumerate polyclonal CD8 T cells. As discussed below, construction of disulfide-trap tetramers should be universally applicable and offers a promising, alternative approach to make straining reagents to pathogen-derived epitopes in situations where conventional tetramers are unstable or short-lived.
Discussion
In order to develop novel MHC I-based reagents for vaccines and probes for pathogen surveillance, we engineered three consecutive SCT designs that are characterized here biophysically and functionally. Our studies show that the progressive SCT generations manifest dramatic improvements in linker accommodation, C-terminal peptide anchoring, and improved resistance to exogenous peptide exchange. Although the data presented here is based on the well-defined Kb-Ova model system, our approaches are completely general and will readily extend to other mouse and human class I alleles. To date we and others have reported first generation SCT constructs with human HLA-A2, -B27, and -E as well as murine H2-Kb, -Ld, and -Db each bound by different peptide ligands [38, 39, 41]. Furthermore, we expect that the novel disulfide trap technology characterized here will also extend to yet untested class I complexes due to the tight integration of our engineering with the highly conserved chemical basis of class I C-terminal peptide anchoring. In particular, since Tyr84 is invariant in all mouse and human class I alleles we anticipate that our disulfide-trap positioning can be readily employed to generate analogous trapped pMHC from other class I alleles as well. In strong support of the general applicability, we have incorporated an analogous trap in two complexes known to contain relative poor binding peptides – human HLA-A2 bound by the G280 peptide involved in melanoma detection as well as mouse H2-Ld bound by the QL9 peptide involved in allorecognition, and human HLA-A2 bound by the G280 peptide involved in melanoma detection (data not shown).
Our structural data shows that the Y84C mutation restructures the Kb groove to open a channel allowing the linker to freely extrude from the peptide-binding platform. In addition, experimental electron density maps for the dt-SCT protein unambiguously show the presence of a disulfide bond between CysPBL2 and Cys84. We also present experimental evidence that the presence of this disulfide bridge increases the thermal stability of the SCT format and effectively prevents binding of competitor peptides. Consequently, incorporation of disulfide traps will have great potential for broadening the application of SCTs to include physiologically important antigenic peptides that are not tight binders.
Recently, three studies on the use of first generation SCT in DNA vaccinations for cancer prevention have been reported. In one of these studies a DNA construct encoding a breast cancer epitope derived from mammoglobin-A presented in the context of HLA-A2 as a SCT was used successfully in vaccination of mice to induce the specific expansion of epitope-positive CTLs [42]. In the second study, mice vaccinated with SCT proteins presenting an immunodominant epitope derived from HPV-16 E6 protein in the context of H2-Kb exhibited significantly increased E6 peptide-specific responses compared to mice immunized with DNA encoding E6 protein. Moreover, 100% of the mice vaccinated with the SCT encoding DNA were protected against lethal challenge with E6-expressing tumors [43]. Using a similar approach, SCT-based vaccines were very recently found to be efficacious in the mouse model of human ovarian cancer [44]. Based on the findings reported here, it is highly likely that the SCTs incorporating disulfide traps could offer further advantages due to their improved stability and resistance to exogenous peptide exchange.
Our findings with the SCT disulfide trap and its ability to improve F pocket anchoring suggested that the linkers involving β2m might be eliminated to make tetramers with only a disulfide-trapped peptide. More specifically we tested whether an E. coli produced, soluble HC with a Y84C mutation could be refolded in the presence of a synthetic peptide with a GC extension. As reported here, this approach worked beautifully to produce disulfide-trap Kb-Ova tetramers and these dt-pMHC tetramers clearly stained pathogen-induced polyclonal α-Kb-Ova T cells with the same specificity as conventional wild-type tetramers.
Recombinant dt-pMHC molecules feature a significant advantage over their single-chain counterparts. Bacterial production of SCT is dependent on successful N-formyl methionine (fMet) cleavage to reveal the N-terminus for the correct SCT incorporated peptide sequence. However, this cleavage event depends on the amino acid size following the initiator fMet [45, 46]. Therefore, bacterial expression of SCT proteins is limited to those that have antigenic peptide sequences, such as Ova, that allow for efficient fMet cleavage. Due to these sequence limitations it is clear that the most efficient way to introduce the engineered disulfide bond into recombinant pMHC is the dt-pMHC approach introduced and validated in this study.
Significance
Herein we present the structural and functional characterization of three generations of MHC I proteins designed as single chain trimers. We connected peptide, β2m, and HC with flexible linkers into a single polypeptide. This initial design was improved by reengineering the MHC groove to accommodate the peptide-β2m linker, and by disulfide trapping to enhance peptide-MHC association. Importantly, these improvements were made without any structural or functional compromises, as assessed by crystallography, highly sensitive T cells activation assays, and flow cytometry of pathogen-specific monoclonal and polyclonal T cells. Due to their preassembled nature SCT proteins bypass conventional antigen processing, express efficiently at the cell surface [27, 30], and potently stimulate CD8 T cells in vivo [47]. These properties afford us a unique opportunity to broaden the production of antigen-specific DNA vaccines to include the induction of subdominant immune responses that in many cases are critically important in producing a broadly effective immunity against tumors [48, 49] and pathogens [50–52]. Likewise, the novel coupling of peptide and class I HC using our method of covalent disulfide bond trapping represents a versatile approach for generating soluble covalently linked pMHC reagents. In the case of MHC complexes that involve weak-binding peptides such reagents may have dramatically improved shelf life and quality of staining. Thus our disulfide-trap SCT and soluble pMHC reagents have exciting application for the diagnosis and modulation of T cell responses to pathogens and tumors.
Experimental Procedures
Protein Expression and Purification
Each SCT (Figure 1a) was expressed in the bacterial strain BL21CodonPlus®(DE3)RIL (Stratagene, La Jolla CA) and insoluble inclusion bodies were prepared as previously described [53]. To form the SCT proteins inclusion bodies were solubilized in 6 M Gdn•HCl, 10 mM Tris pH 8.0, and 10 mM β-mercaptoethanol and refolded under oxidative conditions at 4 °C using a rapid dilution method as described previously [53]. Briefly, each SCT was injected in five separate batches spaced over an 8-hour period into 500 ml of refolding buffer. After an overnight incubation the refolding reaction was concentrated to 4 ml and the formed SCT proteins were purified using size exclusion and anion exchange chromatographies as previously described [53]. Prior to crystallization the purified SCT proteins were exchanged in buffer containing 20 mM HEPES pH 7.5, and 20 mM NaCl. Electrospray mass spectral analysis of each SCT confirmed the presence of abundant peaks at 47,003 Da, 46,909 Da, and 46,985 Da respectively corresponding to the predicted molecular weights of SCTWT, SCTY84A, and properly oxidized dt-SCT.
Crystallization and Data Collection
Crystals of each SCT were obtained by hanging-drop, vapor-diffusion method. Protein at 6 to 8 mg/ml was equilibrated at 20 °C against 13% (w:v) polyethylene glycol (PEG) 10,000 and 100 mM MES pH 6.2 for SCTWT; 14% PEG 6,000 and 100 mM MES pH 6.4 for SCTY84A; 13% PEG 6,000 and 100 mM MES pH 6.3 for dt-SCT. Small crystals obtained in these drops were used to microseed protein hanging drops equilibrated against similar conditions with marginally lower concentrations of PEG. Prior to data collection, crystals were soaked in crystallization buffer to which ethylene glycol was added to a final concentration of 20% (v:v) as cryoprotectant. X-ray diffraction data were collected on the ID-19 beamline at APS. 360 frames were collected for each crystal; each frame representing a 1.0° oscillation range. These data were indexed and integrated using DENZO (HKL Suite, HKL Research, Inc., Charlottesville VA), and scaled and merged using SCALEPACK (HKL Suite, HKL Research, Inc., Charlottesville VA) (Table I). Wilson scaling was applied to the final output structure factor amplitudes [Collaborative Computing Project 4 (CCP4), Daresbury Laboratory, Warrington UK [54]].
Structure Determination and Refinement
Initial phases for each structure were obtained by rigid-body refinement of the coordinates of H2-Kbm8 (PDB 1RJY) against each of the data sets. Extensive model building was performed with the modeling program O (O version 6.22, Uppsala Software Factory, Sweden) using 2FO-FC, FO-FC, and 2FO-FC composite omit maps [CNS, Yale University, New Haven CT [55]]. Atomic refinement was done employing simulated annealing, energy minimization, and restrained B-factor refinement protocols as implemented in CNS and CCP4.
Computational Analysis
Graphical representations were primarily created using Ribbons [56]. Molecular surfaces of the peptide-binding grooves were generated using InsightII (Biosym Technologies, San Diego CA). Superpositions and r.m.s.d. calculations were obtained using Lsqkab (CCP4). R.m.s.d. values between the aligned peptides were calculated using CNS [55]. HBPLUS [57] was used to catalogue contacting atoms and putative hydrogen bonds.
T Cell Assays
OT-1 T cells were assayed in a 4-hour 51Cr release assays as previously described [27]. Target cells for these assays were LM1.8-Kb cells [30] incubated with or without 10 µM exogenous peptide; or LM1.8 cells transduced with SCT constructs using a pMSCV-based retroviral vector (Clontech, Mountain View CA) and the Vpack retroviral expression system (Stratagene). Kb-VSV8 reactive N15 hybridoma cells [58] were a gift of Dr. Hsiu-Ching, Dana-Farber Cancer Institute, Harvard Medical School. For peptide competition assays, LM1.8 cells (2×104/200µl/well) expressing native Kb or the SCT constructs were incubated for 24 hours in a 96-well plate with N15 hybridoma cells (2×105/200 µl/well) in the continuous presence of the indicated concentrations of VSV8 peptide. Relative amounts of IL2 secretion were measured by proliferation of CTLL-2 cells as quantified using Almar blue (BioSource Int., Camarillo CA).
Thermal Denaturation Studies
Purified SCT proteins and Kb-Ova were dialyzed against buffer containing 10 mM K2HPO4/KHPO4 pH = 7.5, and 150 mM NaCl. The proteins were diluted to 475 nM in buffer containing 20 mM K2HPO4/KHPO4 pH = 7.5, and 150 mM NaCl. CD spectra at 10° C were collected between 260 and 200 nm at 0.5 nm increments on Jasco-810 instrument (Jasco Inc., Easton MD) equipped with Peltier temperature controller. The thermal denaturation profiles were monitored by the change in CD signal at 220 nm as a function of temperature. Thermal scan data were collected at a 1.0 °C interval from 10 to 70 °C with a temperature ramp rate of 50 °C/hour. Thermal denaturation curves were scaled from 0% to 100% to provide plots of the percent of signal change versus temperature. The Tm is the temperature at which the CD signal change is half of the total signal change. All measurements were made at least four times and averaged; standard deviations of each measurement were also calculated and presented.
Tetramer Construction
Peptides were obtained commercially or synthesized on an Applied Biosystems Model 432A peptide synthesizer. H2-Kb-Ova (SIINFEKL) and negative control H2-Kb-SIYR (SIYRYYGL) tetramers were constructed according to conventional methods [59]. SCTY84A tetramers were prepared in a similar manner, as described previously[30]. The construction of disulfide-trapped (dt-pMHC) tetramers was accomplished by refolding synthetic SIINFEKLGC peptide with β2m and H2-Kb HC with the mutation Y84C. The dt-pMHC tetramers and controls included the mutation C121S to remove the unpaired cysteine from Kb. Molecular weights of each recombinant MHC were confirmed by mass spectrometry. For tetramerization, phycoerythrin-(PE-) or allophycocyanin-(APC-)streptavidin (BD Biosciences, San Jose CA) was added at a molar ratio of 1 fluorochrome-conjugated streptavidin to 4 biotinylated MHC. The stock concentration of all tetramer preparations was 4.3 ± 0.2 µM MHC monomer, and tetramer staining was calibrated on OT-1 transgenic T cells to obtain matching fluorescence intensity for tetramers with different fluorochromes.
Listeria Infections and Tetramer Staining
B6 mice were infected by tail vein injection with 5×103 cfu of Listeria monocytogenes expressing ovalbumin (L.m.-Ova, a kind gift of Hao Shen, University of Pennsylvania) in 200 µl saline. After 7 days, splenocytes were stained with tetramers and FITC-conjugated anti-CD8 (BD Biosciences). Uninfected OT-1 splenocytes were stained in the same manner. All plots are gated on CD8 cells, and dead cells were excluded by propidium iodide staining. A FACSCalibur instrument and CELLQuest software (BD Biosciences) were used for data acquisition and analysis. Greater than 15,000 CD8+ events were collected per staining in order to reliably determine the frequency of tetramer-positive CD8 T cells. Percentages of tetramer-positive CD8 T cells for six L.m.-Ova infected B6 mice are listed in Supplemental Table 1. Negative control tetramers stained less than 0.8% of CD8 T cells (Supplemental Figure 1). Representative data are shown for one of the six B6 mice.
Supplementary Material
Flow cytometry was used to detect staining of CD8+ lymphocytes as in Figure 6. All plots are gated on CD8+ cells, and percentages of CD8+ cells are given for relevant quadrants. (a) Kb-Ova, SCTY84A, and Kb-Ova based dt-pMHC tetramers were used to stain splenocytes from uninfected B6 mice. Minimal background staining was detected. (b) Kb-SIYR tetramers were constructed with either APC or PE fluorochromes and verified by staining 2C T cells (not shown). These control tetramers did not stain OT-1 transgenic splenocytes (left), uninfected B6 splenocytes (middle), or splenocytes from mice infected with L.m.-Ova (right) as described for Figure 6.
Acknowledgements
This work was supported in part by NIH grants AI055849 (THH) and AI27568 (JMC). The authors would like to thank Dr. Emil Unanue and lab for help with Listeria infections, Dr. Skip Virgin and lab for help with γHV68 infections, and Ms. Tina Primeau for help with T cell assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones EY. MHC class I and class II structures. Curr Opin Immunol. 1997;9:75–79. doi: 10.1016/s0952-7915(97)80162-8. [DOI] [PubMed] [Google Scholar]
- 2.Madden DR. The three-dimensional structure of peptide-MHC complexes. Annu Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 3.Wilson IA, Fremont DH. Structural analysis of MHC class I molecules with bound peptide antigens. Semin Immunol. 1993;5:75–80. doi: 10.1006/smim.1993.1011. [DOI] [PubMed] [Google Scholar]
- 4.Natarajan K, Li H, Mariuzza RA, Margulies DH. MHC class I molecules, structure and function. Rev Immunogenet. 1999;1:32–46. [PubMed] [Google Scholar]
- 5.Young AC, Nathenson SG, Sacchettini JC. Structural studies of class I major histocompatibility complex proteins: insights into antigen presentation. Faseb J. 1995;9:26–36. doi: 10.1096/fasebj.9.1.7821756. [DOI] [PubMed] [Google Scholar]
- 6.Boyd D, Hung CF, Wu TC. DNA vaccines for cancer. IDrugs. 2003;6:1155–1164. [PubMed] [Google Scholar]
- 7.Porgador A, Irvine KR, Iwasaki A, Barber BH, Restifo NP, Germain RN. Predominant role for directly transfected dendritic cells in antigen presentation to CD8+ T cells after gene gun immunization. J Exp Med. 1998;188:1075–1082. doi: 10.1084/jem.188.6.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenlohr LC, Yewdell JW, Bennink JR. Flanking sequences influence the presentation of an endogenously synthesized peptide to cytotoxic T lymphocytes. J Exp Med. 1992;175:481–487. doi: 10.1084/jem.175.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Del Val M, Schlicht HJ, Ruppert T, Reddehase MJ, Koszinowski UH. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell. 1991;66:1145–1153. doi: 10.1016/0092-8674(91)90037-y. [DOI] [PubMed] [Google Scholar]
- 10.Heemels MT, Ploegh H. Generation, translocation, and presentation of MHC class I-restricted peptides. Annu Rev Biochem. 1995;64:463–491. doi: 10.1146/annurev.bi.64.070195.002335. [DOI] [PubMed] [Google Scholar]
- 11.Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 12.Deng Y, Yewdell JW, Eisenlohr LC, Bennink JR. MHC affinity, peptide liberation, T cell repertoire, and immunodominance all contribute to the paucity of MHC class I-restricted peptides recognized by antiviral CTL. J Immunol. 1997;158:1507–1515. [PubMed] [Google Scholar]
- 13.Romero P, Dunbar PR, Valmori D, Pittet M, Ogg GS, Rimoldi D, Chen JL, Lienard D, Cerottini JC, Cerundolo V. Ex vivo staining of metastatic lymph nodes by class I major histocompatibility complex tetramers reveals high numbers of antigen-experienced tumor-specific cytolytic T lymphocytes. J Exp Med. 1998;188:1641–1650. doi: 10.1084/jem.188.9.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greten TF, Slansky JE, Kubota R, Soldan SS, Jaffee EM, Leist TP, Pardoll DM, Jacobson S, Schneck JP. Direct visualization of antigen-specific T cells: HTLV-1 Tax11-19-specific CD8(+) T cells are activated in peripheral blood and accumulate in cerebrospinal fluid from HAM/TSP patients. Proc Natl Acad Sci U S A. 1998;95:7568–7573. doi: 10.1073/pnas.95.13.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker EB, Haley D, Miller W, Floyd K, Wisner KP, Sanjuan N, Maecker H, Romero P, Hu HM, Alvord WG, Smith JW, 2nd, Fox BA, Urba WJ. gp100(209-2M) peptide immunization of human lymphocyte antigen-A2+ stage I-III melanoma patients induces significant increase in antigen-specific effector and long-term memory CD8+ T cells. Clin Cancer Res. 2004;10:668–680. doi: 10.1158/1078-0432.ccr-0095-03. [DOI] [PubMed] [Google Scholar]
- 16.Monsurro V, Nagorsen D, Wang E, Provenzano M, Dudley ME, Rosenberg SA, Marincola FM. Functional heterogeneity of vaccine-induced CD8(+) T cells. J Immunol. 2002;168:5933–5942. doi: 10.4049/jimmunol.168.11.5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Labrador FX, He XS, Berenguer M, Cheung RC, Wright TL, Greenberg HB. The use of class-I HLA tetramers for the detection of hepatitis C virus NS3-specific CD8(+) T cells in patients with chronic infection. J Immunol Methods. 2004;287:91–99. doi: 10.1016/j.jim.2004.01.023. [DOI] [PubMed] [Google Scholar]
- 18.Gratama JW, van Esser JW, Lamers CH, Tournay C, Lowenberg B, Bolhuis RL, Cornelissen JJ. Tetramer-based quantification of cytomegalovirus (CMV)-specific CD8+ T lymphocytes in T-cell-depleted stem cell grafts and after transplantation may identify patients at risk for progressive CMV infection. Blood. 2001;98:1358–1364. doi: 10.1182/blood.v98.5.1358. [DOI] [PubMed] [Google Scholar]
- 19.Yee C, Savage PA, Lee PP, Davis MM, Greenberg PD. Isolation of high avidity melanoma-reactive CTL from heterogeneous populations using peptide-MHC tetramers. J Immunol. 1999;162:2227–2234. [PubMed] [Google Scholar]
- 20.Cobbold M, Khan N, Pourgheysari B, Tauro S, McDonald D, Osman H, Assenmacher M, Billingham L, Steward C, Crawley C, Olavarria E, Goldman J, Chakraverty R, Mahendra P, Craddock C, Moss PA. Adoptive transfer of cytomegalovirus-specific CTL to stem cell transplant patients after selection by HLA-peptide tetramers. J Exp Med. 2005;202:379–386. doi: 10.1084/jem.20040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ge Q, Stone JD, Thompson MT, Cochran JR, Rushe M, Eisen HN, Chen J, Stern LJ. Soluble peptide-MHC monomers cause activation of CD8+ T cells through transfer of the peptide to T cell MHC molecules. Proc Natl Acad Sci U S A. 2002;99:13729–13734. doi: 10.1073/pnas.212515299. Epub 12002 Oct 13728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schott E, Bertho N, Ge Q, Maurice MM, Ploegh HL. Class I negative CD8 T cells reveal the confounding role of peptide-transfer onto CD8 T cells stimulated with soluble H2-Kb molecules. Proc Natl Acad Sci U S A. 2002;99:13735–13740. doi: 10.1073/pnas.212515399. Epub 12002 Oct 13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savage P, Cowburn P, Clayton A, Man S, Lawson T, Ogg G, Lemoine N, McMichael A, Epenetos A. Anti-viral cytotoxic T cells inhibit the growth of cancer cells with antibody targeted HLA class I/peptide complexes in SCID mice. Int J Cancer. 2002;98:561–566. doi: 10.1002/ijc.10219. [DOI] [PubMed] [Google Scholar]
- 24.Lev A, Noy R, Oved K, Novak H, Segal D, Walden P, Zehn D, Reiter Y. Tumor-specific Ab-mediated targeting of MHC-peptide complexes induces regression of human tumor xenografts in vivo. Proc Natl Acad Sci U S A. 2004;101:9051–9056. doi: 10.1073/pnas.0403222101. Epub 2004 Jun 9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masteller EL, Warner MR, Ferlin W, Judkowski V, Wilson D, Glaichenhaus N, Bluestone JA. Peptide-MHC class II dimers as therapeutics to modulate antigen-specific T cell responses in autoimmune diabetes. J Immunol. 2003;171:5587–5595. doi: 10.4049/jimmunol.171.10.5587. [DOI] [PubMed] [Google Scholar]
- 26.Maile R, Wang B, Schooler W, Meyer A, Collins EJ, Frelinger JA. Antigen-specific modulation of an immune response by in vivo administration of soluble MHC class I tetramers. J Immunol. 2001;167:3708–3714. doi: 10.4049/jimmunol.167.7.3708. [DOI] [PubMed] [Google Scholar]
- 27.Yu YY, Netuschil N, Lybarger L, Connolly JM, Hansen TH. Cutting edge: single-chain trimers of MHC class I molecules form stable structures that potently stimulate antigen-specific T cells and B cells. J Immunol. 2002;168:3145–3149. doi: 10.4049/jimmunol.168.7.3145. [DOI] [PubMed] [Google Scholar]
- 28.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, French AR, Sunwoo JB, Lemieux S, Hansen TH, Yokoyama WM. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 29.Choudhuri K, Wiseman D, Brown MH, Gould K, van der Merwe PA. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature. 2005;436:578–582. doi: 10.1038/nature03843. [DOI] [PubMed] [Google Scholar]
- 30.Lybarger L, Yu YY, Miley MJ, Fremont DH, Myers N, Primeau T, Truscott SM, Connolly JM, Hansen TH. Enhanced immune presentation of a single-chain major histocompatibility complex class I molecule engineered to optimize linkage of a C-terminally extended peptide. J Biol Chem. 2003;278:27105–27111. doi: 10.1074/jbc.M303716200. Epub 22003 May 27105. [DOI] [PubMed] [Google Scholar]
- 31.Fremont DH, Stura EA, Matsumura M, Peterson PA, Wilson IA. Crystal structure of an H-2Kb-ovalbumin peptide complex reveals the interplay of primary and secondary anchor positions in the major histocompatibility complex binding groove. Proc Natl Acad Sci U S A. 1995;92:2479–2483. doi: 10.1073/pnas.92.7.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia KC, Degano M, Speir JA, Wilson IA. Emerging principles for T cell receptor recognition of antigen in cellular immunity. Rev Immunogenet. 1999;1:75–90. [PubMed] [Google Scholar]
- 33.Garcia KC, Teyton L, Wilson IA. Structural basis of T cell recognition. Annu Rev Immunol. 1999;17:369–397. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 34.Kersh GJ, Miley MJ, Nelson CA, Grakoui A, Horvath S, Donermeyer DL, Kappler J, Allen PM, Fremont DH. Structural and functional consequences of altering a peptide MHC anchor residue. J Immunol. 2001;166:3345–3354. doi: 10.4049/jimmunol.166.5.3345. [DOI] [PubMed] [Google Scholar]
- 35.Lybarger L, Wang X, Harris MR, Virgin HWt, Hansen TH. Virus subversion of the MHC class I peptide-loading complex. Immunity. 2003;18:121–130. doi: 10.1016/s1074-7613(02)00509-5. [DOI] [PubMed] [Google Scholar]
- 36.Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 37.Miley MJ, Messaoudi I, Metzner BM, Wu Y, Nikolich-Zugich J, Fremont DH. Structural basis for the restoration of TCR recognition of an MHC allelic variant by peptide secondary anchor substitution. J Exp Med. 2004;200:1445–1454. doi: 10.1084/jem.20040217. Epub 2004 Nov 1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oved K, Lev A, Noy R, Segal D, Reiter Y. Antibody-mediated targeting of human single-chain class I MHC with covalently linked peptides induces efficient killing of tumor cells by tumor or viral-specific cytotoxic T lymphocytes. Cancer Immunol Immunother. 2005;54:867–879. doi: 10.1007/s00262-005-0666-5. Epub 2005 May 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greten TF, Korangy F, Neumann G, Wedemeyer H, Schlote K, Heller A, Scheffer S, Pardoll DM, Garbe AI, Schneck JP, Manns MP. Peptide-beta2-microglobulin-MHC fusion molecules bind antigen-specific T cells and can be used for multivalent MHC-Ig complexes. J Immunol Methods. 2002;271:125–135. doi: 10.1016/s0022-1759(02)00346-0. [DOI] [PubMed] [Google Scholar]
- 40.Chen I, Howarth M, Lin W, Ting AY. Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat Methods. 2005;2:99–104. doi: 10.1038/nmeth735. Epub 2005 Jan 2021. [DOI] [PubMed] [Google Scholar]
- 41.Crew MD, Cannon MJ, Phanavanh B, Garcia-Borges CN. An HLA-E single chain trimer inhibits human NK cell reactivity towards porcine cells. Mol Immunol. 2005;42:1205–1214. doi: 10.1016/j.molimm.2004.11.013. Epub 2005 Jan 1208. [DOI] [PubMed] [Google Scholar]
- 42.Jaramillo A, Narayanan K, Campbell LG, Benshoff ND, Lybarger L, Hansen TH, Fleming TP, Dietz JR, Mohanakumar T. Recognition of HLA- A2-restricted mammaglobin-A-derived epitopes by CD8+ cytotoxic T lymphocytes from breast cancer patients. Breast Cancer Res Treat. 2004;88:29–41. doi: 10.1007/s10549-004-8918-1. [DOI] [PubMed] [Google Scholar]
- 43.Huang CH, Peng S, He L, Tsai YC, Boyd DA, Hansen TH, Wu TC, Hung CF. Cancer immunotherapy using a DNA vaccine encoding a singlechain trimer of MHC class I linked to an HPV-16 E6 immunodominant CTL epitope. Gene Ther. 2005;12:1180–1186. doi: 10.1038/sj.gt.3302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hung CF, Calizo R, Tsai YC, He L, Wu TC. A DNA vaccine encoding a single-chain trimer of HLA-A2 linked to human mesothelin peptide generates anti-tumor effects against human mesothelin-expressing tumors. Vaccine. 2006;7:7. doi: 10.1016/j.vaccine.2006.06.087. [DOI] [PubMed] [Google Scholar]
- 45.Hirel PH, Schmitter MJ, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc Natl Acad Sci U S A. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lathrop BK, Burack WR, Biltonen RL, Rule GS. Expression of a group II phospholipase A2 from the venom of Agkistrodon piscivorus piscivorus in Escherichia coli: recovery and renaturation from bacterial inclusion bodies. Protein Expr Purif. 1992;3:512–517. doi: 10.1016/1046-5928(92)90069-9. [DOI] [PubMed] [Google Scholar]
- 47.Primeau T, Myers NB, Yu YY, Lybarger L, Wang X, Truscott SM, Hansen TH, Connolly JM. Applications of major histocompatibility complex class I molecules expressed as single chains. Immunol Res. 2005;32:109–122. doi: 10.1385/ir:32:1-3:109. [DOI] [PubMed] [Google Scholar]
- 48.Kawakami Y, Eliyahu S, Jennings C, Sakaguchi K, Kang X, Southwood S, Robbins PF, Sette A, Appella E, Rosenberg SA. Recognition of multiple epitopes in the human melanoma antigen gp100 by tumor-infiltrating T lymphocytes associated with in vivo tumor regression. J Immunol. 1995;154:3961–3968. [PubMed] [Google Scholar]
- 49.Tsai V, Southwood S, Sidney J, Sakaguchi K, Kawakami Y, Appella E, Sette A, Celis E. Identification of subdominant CTL epitopes of the GP100 melanoma-associated tumor antigen by primary in vitro immunization with peptide-pulsed dendritic cells. J Immunol. 1997;158:1796–1802. [PubMed] [Google Scholar]
- 50.van der Most RG, Murali-Krishna K, Whitton JL, Oseroff C, Alexander J, Southwood S, Sidney J, Chesnut RW, Sette A, Ahmed R. Identification of Db- and Kb-restricted subdominant cytotoxic T-cell responses in lymphocytic choriomeningitis virus-infected mice. Virology. 1998;240:158–167. doi: 10.1006/viro.1997.8934. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y, Webster RG, Woodland DL. Induction of CD8+ T cell responses to dominant and subdominant epitopes and protective immunity to Sendai virus infection by DNA vaccination. J Immunol. 1998;160:2425–2432. [PubMed] [Google Scholar]
- 52.Fu TM, Friedman A, Ulmer JB, Liu MA, Donnelly JJ. Protective cellular immunity: cytotoxic T-lymphocyte responses against dominant and recessive epitopes of influenza virus nucleoprotein induced by DNA immunization. J Virol. 1997;71:2715–2721. doi: 10.1128/jvi.71.4.2715-2721.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitaksov V, Fremont DH. Structural definition of the H-2Kd peptidebinding motif. J Biol Chem. 2006;281:10618–10625. doi: 10.1074/jbc.M510511200. Epub 12006 Feb 10610. [DOI] [PubMed] [Google Scholar]
- 54.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 55.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse- Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 56.Carson M. Ribbon Models of Macromolecules. J Mol Graphics. 1987;5:103–106. [Google Scholar]
- 57.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 58.Chang HC, Smolyar A, Spoerl R, Witte T, Yao Y, Goyarts EC, Nathenson SG, Reinherz EL. Topology of T cell receptor-peptide/class I MHC interaction defined by charge reversal complementation and functional analysis. J Mol Biol. 1997;271:278–293. doi: 10.1006/jmbi.1997.1169. [DOI] [PubMed] [Google Scholar]
- 59.Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigenspecific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow cytometry was used to detect staining of CD8+ lymphocytes as in Figure 6. All plots are gated on CD8+ cells, and percentages of CD8+ cells are given for relevant quadrants. (a) Kb-Ova, SCTY84A, and Kb-Ova based dt-pMHC tetramers were used to stain splenocytes from uninfected B6 mice. Minimal background staining was detected. (b) Kb-SIYR tetramers were constructed with either APC or PE fluorochromes and verified by staining 2C T cells (not shown). These control tetramers did not stain OT-1 transgenic splenocytes (left), uninfected B6 splenocytes (middle), or splenocytes from mice infected with L.m.-Ova (right) as described for Figure 6.