Abstract
One strategy for controlling transmission of insect-borne disease involves replacing the native insect population with transgenic animals unable to transmit disease. Population replacement requires a drive mechanism to ensure the rapid spread of linked transgenes, the presence of which may result in a fitness cost to carriers. Medea selfish genetic elements have the feature that when present in a female, only offspring that inherit the element survive, a behavior that can lead to spread. Here we derive equations that describe the conditions under which Medea elements with a fitness cost will spread, and the equilibrium allele frequencies achieved. Of particular importance, we show that whenever Medea spreads, the non-Medea genotype is driven out of the population, and we estimate the number of generations required to achieve this goal for Medea elements with different fitness costs and male-only introduction frequencies. Finally, we characterize two contexts in which Medea elements with fitness costs drive the non-Medea allele from the population: an autosomal element in which not all Medea-bearing progeny of a Medea-bearing mother survive, and an X-linked element in species in which X/Y individuals are male. Our results suggest that Medea elements can drive population replacement under a wide range of conditions.
Keywords: population replacement, mosquito, maternal effect, introgression, dengue, malaria
Introduction
Mosquitoes are vectors for a number of important human diseases, including malaria and dengue fever. Replacement of insect disease vectors with modified counterparts refractory to pathogen transmission is a long-established concept for disease prevention (reviewed in Braig and Yan 2001; Gould and Schliekelman 2004; Sinkins and Gould 2006), and genes that inhibit the mosquito's ability to transmit Plasmodium or dengue have been identified (de Lara Capurro et al. 2000; Ito et al. 2002; Moreira et al. 2002; Franz et al. 2006; Corby-Harris, et al. 2010). However, the expression of these genes is not expected to result in a fitness benefit to carriers (Schmid-Hempel 2005; Tripet et al. 2008), and a large percentage of the wild population will need to be refractory in order to achieve substantial levels of disease control (Boete and Koella 2002). Therefore, effective population replacement is generally thought to require that genes conferring disease refractoriness be coupled with a mechanism, such as linkage with a selfish genetic element, for driving them through the wild population (Braig and Yan 2001; Gould and Schliekelman 2004; Sinkins and Gould 2006).
Maternal-effect lethal selfish genetic elements were first described in the flour beetle Tribolium castaneum and are known by the acronym Medea (maternal-effect dominant embryonic arrest). Tribolium Medea, which sits at a fixed chromosomal position, has the feature that when present in females, only progeny that inherit the element-containing chromosome survive (Beeman et al. 1992). In contrast, heterozygous Medea-bearing males give rise to wildtype and Medea-bearing progeny with equal frequency when mated to wildtype females. Therefore, Medea enhances its transmission relative to competing non-Medea-bearing homologous chromosomes (hereafter referred to as the non-Medea allele) by causing the death of progeny that do not carry a copy of Medea found in the mother. Synthetic Medea elements have been generated that drive population replacement in Drosophila (Chen et al. 2007).
Medea's ability to spread relies on the elimination from the population of non-Medea alleles in the offspring of heterozygous females mated with non-Medea or heterozygous males. Selection against the non-Medea allele is weak at low and high Medea allele frequencies, when such crosses are rare. Therefore, if there is selection against the Medea allele (carriers experience a cost), this process produces a threshold frequency (an unstable equilibrium), below which Medea will be lost, and above which Medea will spread to a stable equilibrium. Wade and Beeman (1994) showed that if the presence of Medea does not result in a fitness (fecundity) cost to carriers, Medea spreads to fixation for all degrees of maternal effect lethality, though the rate of Medea increase is initially very slow. They also showed that if the presence of Medea results in a decrease in fecundity independent of maternal-effect killing, the frequency of the Medea allele could still increase to a stable internal equilibrium provided that fitness costs were recessive, or if dominant, small. These authors, and Smith (Smith 1998), showed that Medea's ability to spread in the face of fitness costs could be enhanced if progeny of a Medea-bearing mother compete with each other for resources. In this context, known as family-level, or soft selection (Wade, 1985; Kelly 1992), the death of non-Medea offspring within the family of a Medea-bearing mother frees limited resources for sibling Medea-bearing progeny. In the work below we assume no family-level selection because this assumption provides a more conservative estimate of Medea's potential as a population replacement drive mechanism. That said, some mosquitoes, such as Aedes aegypti, an important vector of dengue, breed in small containers that may often be resource-limited for larval growth (Clements 1999), suggesting that family-level selection could be important in some contexts, a topic that should be further explored. Population genetic models of Hastings (1994), Smith (1998) and Chen et al. (2007) show that, in the absence of family-level selection, Medea elements with significant dominant fitness costs can still spread, provided they are introduced above a critical introduction frequency. Previous work has focused on the fate of the Medea allele. However, it is the fate of Medea-bearing genotypes that is important for population replacement.Chen et al. (2007) showed that, at least under some conditions, when Medea elements with fitness costs are introduced at frequencies that result in spread to an internal equilibrium allele frequency, non-Medea individuals are nonetheless rapidly eliminated from the population. However, it is not clear to what extent this conclusion can be generalized.
Here we characterize the dynamics of Medea elements in large, unstructured populations, focusing particular attention on the behavior of Medea elements with fitness costs that show some degree of dominance, located on autosomes or sex chromosomes. We show that whenever Medea spreads, it eliminates the non-Medea genotype. We provide equations describing the conditions under which elimination of the non-Medea genotype constitutes a stable equilibrium, and the equilibrium allele frequencies attained. We also calculate the introduction frequencies and number of generations required for Medea elements with specific fitness costs to bring about population replacement. Finally, we identify several scenarios - incomplete zygotic rescue by a single copy of Medea in the progeny of Medea-bearing mothers, or X linkage in a X/Y male species - in which the spread of Medea results in elimination of the non-Medea chromosome from the population, and we provide equations describing the conditions under which loss of the non-Medea chromosome constitutes a stable equilibrium. Our results suggest there are a wide variety of conditions under which Medea can drive population replacement in a relatively short time (∼1-2 years), using male-only release sizes achievable in the mid-twentieth century. These observations, coupled with the fact that the synthetic form of Medea is the only gene drive mechanism that is both well understood at the molecular level and that has been demonstrated to drive population replacement, suggest Medea as a target for further development.
The model
We use a deterministic model to examine the invasion of synthetic Medea elements into populations. Terms are defined in Table 1. Table 2 presents the frequencies of parental genotypes in the population, and offspring frequencies and genotypes produced by each mating. The deterministic model assumes an infinite population with random mating and discrete, non-overlapping generations. Expression of the toxin/antidote genes that make up Medea, and/or the cargo genes linked to Medea (genes conferring resistance to pathogen transmission), may result in a fitness cost to carriers. Fitness costs may also arise through tight linkage between Medea and a nearby deleterious allele or insertion-dependent effects on the expression of nearby genes. These are fitness costs not associated with the Medea-killing itself. We consider three types of fitness cost: an embryonic fitness cost (cE,Het and cE,Homo), a maternal fecundity loss (cD,Het and cD,Homo), and a paternal fertility loss (cS,Het and cS,Homo). cD,Het, cS,Het, cD,Homo, and cS,Homo act on the genotypes of the parents, resulting in a fecundity loss in females and a fertility loss in males. For example, if wildtypes have a fertility/fecundity of 1, then in heterozygous females this cost can be interpreted as meaning heterozygous females lay only (1-cD,Het) fertile eggs, while heterozygous males only successfully fertilize (1-cS,Het) eggs. An embryonic cost refers to the fraction of Medea-bearing embryos dying as juveniles. Since costs are likely to be borne by both parents (e.g. insertion site-dependent effects; consequences of toxin/antidote expression) or by the female alone (e.g. costs associated with expression of a maternal toxin or a transgene mediating disease resistance), we do not consider paternal fitness costs in isolation. In some of what follows, it is more convenient to frame the discussion in terms of fitness (Vx) rather than the fitness costs induced by the construct. For example, the fitness of a homozygous female is VD,Homo=1-cD,Homo.
Table 1.
A list of abbreviations. All abbreviations in the text are listed here
| SMM | Fraction of the male population homozygous for Medea |
| SM+ | Fraction of the male population heterozygous for Medea |
| S++ | Fraction of the male population homozygous for the non-Medea allele |
| DMM | Fraction of the female population homozygous for Medea |
| DM+ | Fraction of the female population heterozygous for Medea |
| D++ | Fraction of the female population homozygous for the non-Medea allele |
| GMM | Fraction of the population homozygous for Medea |
| Gm+ | Fraction of the population heterozygous for Medea |
| G++ | Fraction of the population homozygous for the non-Medea allele |
| ′ | A prime in all above refers to the next generation |
| cS,genotype | Male fertility loss at a given genotype |
| cD,genotype | Female fecundity loss at a given genotype |
| cE,genotype | Rate of embryonic death at a given genotype, independent of the maternal effect Medea killing |
| p | Allele frequency of the Medea element |
| q | Allele frequency of the WT element |
| W | Mean fitness |
| VS,genotype | Male fertility retained at a given genotype(1-cS,genotype). When homozygous fitness is equal to the |
| square of heterozygous fitness, the fitness cost is said to be multiplicative. | |
| VD,genotype | Female fecundity retained at a given genotype(1-cD,genotype). When homozygous fitness is equal to the |
| square of heterozygous fitness, the fitness cost is said to be multiplicative. | |
| VE,genotype | Embryonic viability retained at a given genotype(1-cE,genotype). When homozygous fitness is equal to |
| the square of heterozygous fitness, the fitness cost is said to be multiplicative. | |
| t0 | Fraction of WT offspring of a Medea-bearing mother that die. |
| t1 | Fraction of heterozygous offspring of a homozygous Medea mother that live. |
Table 2.
Parental genotype frequency, fitness effects, and offspring frequency.Mating frequencies are the product of the genotype frequencies of the male and female parents. A reduction of fertility, fecundity or embryonic viability leads to a reduction in the frequency of viable offspring of some genotypes. When μ0=1, all non-Medea offspring of Medea heterozygous mothers will live. When μ1=1, all heterozygous Medea offspring of Medea homozygous mothers will live. By summing all the families and dividing by the mean fitness of the population, we find the genotype frequencies of the offspring. This is explicitly done in the text.
| Parental Genotype Frequency | Fitness/Fecundity/Fertility | Offspring Frequency | ||||||
|---|---|---|---|---|---|---|---|---|
| Family | Male | Female | Mating | Male | Female | Homo | Het | WT |
| 1 | SMM | DMM | SMM*DMM | VS, Homo | VD, Homo | VE, Homo | ||
| 2 | SM+ | DMM | SM+*DMM | VS, Het | VD, Homo | ½ VE, Homo | ½VE,Het μ1 | |
| 3 | S++ | DMM | S++ *DMM | 1 | VD, Homo | VE, Het μ1 | ||
| 4 | SMM | DM+ | SMM*DM+ | VS, Homo | VD, Het | ½ VE, Homo | ½ VE, Het | |
| 5 | SM+ | DM+ | SM+*DM+ | VS, Het | VD. Het | ¼ VE,Homo | ½ VE. Het | ¼ μ0 |
| 6 | S++ | DM+ | S++*DM+ | 1 | VD, Het | ½ VE, Het | ½ μ0 | |
| 7 | SMM | D++ | SMM*D++ | VS, Homo | 1 | VE. Het | ||
| 8 | SM+ | D++ | SM+*D++ | VS. Het | 1 | ½ VE, Het | ½ | |
| 9 | S++ | D++ | S++* D++ | 1 | 1 | 1 | ||
We consider two types of Medea-dependent lethality. The term t0 refers to the fraction of non-Medea progeny from heterozygous Medea mothers that die. Typically, we will consider t0=1, meaning that embryos from Medea-bearing mothers that fail to inherit Medea always die. We also consider situations in which heterozygous offspring of homozygous Medea mothers have a probability of dying. This death, also considered by Smith (Smith 1998), and observed by Beeman et al. (Beeman et al. 1992) in Tribolium, and by Chen et al. (Chen et al. 2007) in Drosophila, may represent incomplete zygotic rescue of maternal-effect lethality associated with two maternal copies of the toxin gene and one copy of the zygotic antidote. The fraction that die by this mechanism is modeled as t1, with t1=1 meaning that all heterozygous progeny die. Except where noted specifically, t1=0. It is sometimes useful to consider the fraction of progeny that live (μ): μ0=1-t0 and μ1=1-t1.
Given the assumptions above, the equations for the genotype frequencies in generation n + 1 from those in generation n are
where the mean fitness, W, equals the numerator of the right sides of these equations
In all generations after the introduction, male and female genotype distributions are the same.
Throughout the text we follow allele fitness as a way of understanding the fate of Medea in populations. By allele fitness we mean the probability that a given allele in a zygote that has survived possible Medea-dependent killing will be passed in the next generation to a zygote that also survives Medea-dependent killing, given a specific set of population genotype frequencies. This term incorporates fitness losses associated with Medea-dependent maternal-effect killing, as well as killing-independent fitness costs associated with Medea. In order to understand the dynamics of Medea spread, and how this depends on allele frequency and fitness, we need to be able to describe Medea allele and genotype frequencies, and allele and genotype fitnesses, over generations. In a Medea-bearing population the fate of an individual depends on the genotype of its mother as well as its own genotype. Thus, knowledge of one genotype frequency after a single round of random mating is not sufficient to characterize the population. However, after a single round of mating, the range of possible genotype frequencies for a given allele frequency is constrained. Subsequent generations of mating further constrain the possible genotype frequencies for a given allele frequency. In the supplementary materials we use these observations as the basis for a method by which genotype frequencies and fitness values can be calculated with respect to Medea allele frequency.
When Medea spreads, non-Medea individuals are eliminated from the population
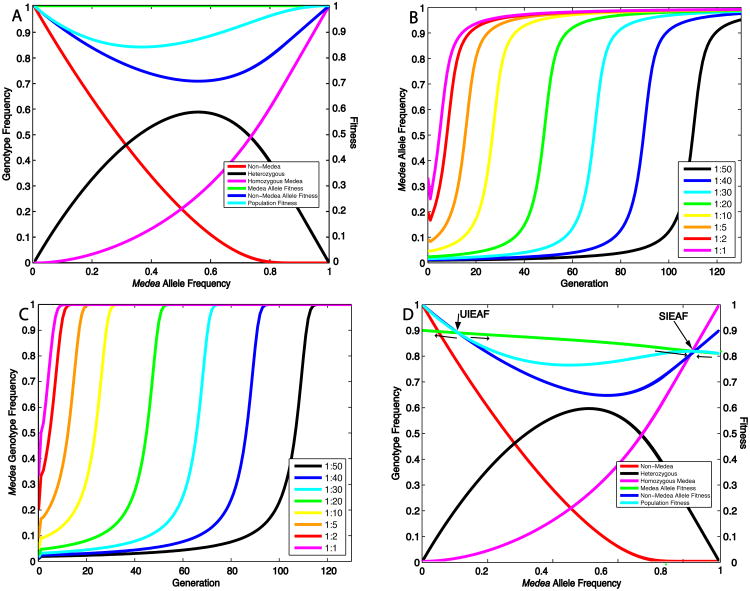
Medea increases in frequency by killing alternative non-Medea alleles, thereby causing a relative increase in the population frequency of the Medea allele. Medea-bearing individuals and alleles experience no direct benefit from this killing, but non-Medea alleles experience Medea-dependent death (a fitness loss) in each generation that is dependent on the Medea allele frequency. The relationship between genotype and Medea allele frequency, and between allele and population fitness and Medea allele frequency, for an autosomal Medea with no fitness cost, is illustrated in Figure 1A. The Medea allele spreads to fixation because its fitness is always greater than that of the non-Medea allele. This result agrees with that of earlier works (Hastings 1994; Wade and Beeman 1994; Smith 1998). The rate of Medea spread depends dramatically on the introduction frequency. If Medea is released into a population at low frequencies there is a long lag phase during which the frequency of Medea alleles and individuals increases only slowly because the frequency of Medea-dependent killing is low (Fig. 1B, C) (Wade and Beeman 1994). This lag phase is followed by roughly 15 generations that accounts for a dramatic loss of non-Medea alleles and individuals (Fig. 1B, C). If Medea is released at higher frequencies the lag phase is shortened, but in other respects the population trajectories are very similar (Fig. 1B, C).
Figure 1.
Characteristics of Medea allele and genotype spread as a function of introduction frequency, and of allele and population fitness as a function of Medea allele frequency. (A) The frequency of individuals lacking Medea (Non-Medea), heterozygotes (Heterozygous), and homozygotes for Medea (Homozygous Medea), are plotted with respect to Medea allele frequency. The fitness of the Medea allele, the non-Medea allele, and the population is also shown. (B) Medea allele frequency is plotted as a function of the number of generations for different introduction ratios of homozygous Medea/non-Medea males into a population of non-Medea females. (C) The population frequency of individuals with Medea (Medea Genotype Frequency) is plotted as a function of generations for different introduction ratios of homozygous Medea/non-Medea males into a population of non-Medea females. (D) Plot of allele and population fitness, and genotype frequency, as a function of Medea allele frequency, for a Medea that carries a 10% embryonic fitness cost. The UIEAF and SIEAF are indicated. Thin arrows indicate the directions in which the Medea allele frequency moves on either side of the UIEAF and SIEAF.
We now consider the fate of autosomal Medea alleles that have a fitness cost not associated with Medea-dependent killing. As an example, we begin by considering the fate of a Medea carrying a 10% multiplicative embryonic fitness cost (Fig. 1D) (homozygotes carry a 19% fitness cost). The fitness curves for the Medea and non-Medea allele cross at two positions. These define the unstable internal equilibrium allele frequency (UIEAF) and the stable internal equilibrium allele frequency (SIEAF) (the stable internal equilibrium of Wade and Beeman (1994), and Smith (1998)), the point to which all gamete frequencies converge when Medea spreads (See also discussion of Supp. Fig 1A). At these two points Medea-dependent killing of non-Medea alleles is balanced by natural selection-dependent loss of Medea alleles. If Medea is present at a frequency below the UIEAF, it is driven out of the population. In contrast, if the Medea allele frequency is above the UIAEF but below the SIAEF, the frequency of Medea increases over the generations towards the SIEAF, even though this is associated with a decrease in overall population fitness (Fig. 1D). The SIEAF represents a stable upper limit on the Medea allele frequency since when the Medea allele frequency is higher than the SIEAF, non-Medea alleles have, on average, a higher relative fitness; they lack the fitness costs associated with being in Medea homozygotes (they are in heterozygotes), and because Medea is now common they only rarely suffer the cost of death due to maternal-effect lethality in non-Medea progeny. In consequence, the SIEAF also represents a local population fitness maximum (Fig. 1D).
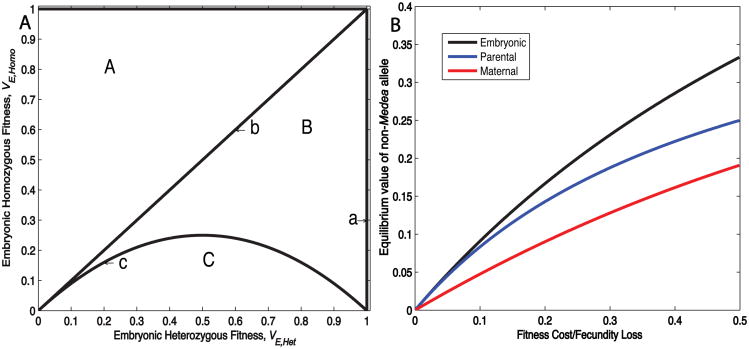
In order to characterize more generally the fitness conditions under which Medea spreads, and the genotype and allele equilibria achieved when spread occurs, we solved for the equilibrium values of this system, setting and Recalling that genotype frequencies must sum to one, GMM + GM+ + G++ = 1, and assuming that all fitness values are greater than 0, we find 4 biologically possible equilibria: populations consisting of only non-Medea individuals, all three genotypes (the UIEAF), only Medea-bearing individuals (the SIEAF), or only homozygous Medea individuals. In order to analyze the stability of these equilibria, we use the standard linear stability analysis for difference equations (see Supplemental Materials for details). To simplify the model we limit our analysis to cases where there are 2 independent variables (homozygous and heterozygous fitness). These variables are plotted against each other to create a parameter space diagram in which regions of feasibility and stability or instability are indicated (Fig. 2A). We consider a Medea with an embryonic fitness cost, and t1=0 and t0=1. A similar analysis and plots for parental or maternal fitness costs are presented in the supplemental materials. We are particularly interested in the case in which heterozygotes experience a fitness cost, and this cost is less than that experienced by homozygotes, because we believe this is the most likely scenario to be encountered with real, engineered Medea elements carrying genes that mediate disease refractoriness.
Figure 2.
Equilibrium characteristics of autosomal Medea elements with fitness costs. (A) Diagram partitioning (VHet, VHomo) fitness parameter space into regions in which linear stability analysis indicate qualitatively similar behaviors are observed. This diagram is identical for a Medea with parental fitness cost. Qualitative behavior changes as each curve is crossed, with the occurrence of a bifurcation. Transcritical bifurcation occurs as Equilibrium 3 moves through Equilibrium 4 (i.e. the two collide), with the two equilibria exchanging stability. Curve c separates regions B and C. On this curve, Equilibrium 2 and 3 are coincident. Transcritical bifurcation occurs as the two equilibria collide, with the two equilibria exchanging stability. (B) Stable internal equilibrium values of the non-Medea allele are plotted as a function of fitness cost/fecundity loss for embryonic, sex-independent parental, or maternal costs.
(1) All non-Medea individuals
G++ = 1, GM+ = GMM= 0;
This equilibrium is always stable unless VE,Het=1 (Fig. 2A, line a), at which value the linear stability analysis is inconclusive. In other words, if the presence of Medea results in a fitness cost to heterozygotes, very low-level introductions of Medea will result in loss of the Medea allele. Numerical results indicate that when fitness costs are purely recessive (VE,Het = 1; VE,Het ≤ 1), this equilibrium is unstable, implying that low-frequency introductions of such a Medea can result in spread, even if the fitness of homozygotes is close to zero.
(2) All three genotypes present in population
Equilibrium 2 is only biologically feasible if VE,Homo≥VE,Het (1-VE,Het) (Fig. 2A, regions A and B). It is unstable when VE,Het<1 and VE,Homo>VE,Het(1-VE,Het) (Fig. 2A, regions A and B). At VE,Het=1 (Fig. 2A, line a), this equilibrium is coincident with equilibrium 1 (G++=1, GM+=0 and GMM=0) and unstable as determined through numerical simulations. This analysis implies that in broad regions of fitness space, populations containing all three genotypes will not persist because the equilibria they are associated with are unstable, and all real populations are subject to perturbations. The equilibrium values described by the above equations in regions A and B represent the UIEAF.
(3) No non-Medea individuals in the population
This equilibrium is biologically feasible if VE,Homo≤VE,Het (Fig. 2A, regions B and C). It is unstable for VE,Homo<VE,Het(1-VE,Het) (Fig. 2A, region C), and stable for VE,Homo>VE,Het(1-VE,Het) (Fig. 2A, region B). When VE,Homo=VE,Het (Fig. 2A, line b) this equilibrium is coincident with equilibrium 4. When VE,Homo=VE,Het(1-VE,Het) (Fig. 2A, line c), this equilibrium is coincident with equilibrium 2. This analysis implies that within a biologically important region of fitness space (VE,Homo≤VE,Het and VE,Homo>VE,Het(1-VE,Het) (Fig. 2A, region B)), Medea elements will, if released at frequencies greater than the UIEAF, spread such that eventually all individuals carry Medea. The stable equilibrium values described by the above equations (region B) represent the SIEAF.
(4) All Medea homozygous individuals in the population
G++ = 0, GM+ = 0, GMM= 1;
By linear analysis, this equilibrium is stable if VE,Homo>VE,Het (Fig. 2A, region A), and unstable if VE,Homo<VE,Het (Fig. 2A, regions B and C). When VE,Homo=VE,Het, this equilibrium is coincident with equilibrium 3 (Fig. 2A, line b) and stable. This analysis implies that when a Medea carries a fitness cost, and homozygotes are less fit than heterozygotes (and t1=0), unless a population begins with no non-Medea alleles, the population will always contain non-Medea alleles.
Calculations using maternal and paternal fitness costs, also lead to 4 equilibria (details can be found in Supplementary Materials). Several simple conclusions emerge from this analysis for each type of fitness cost, provided that VHet<1 and VHomo<VHet. First, if such a Medea is introduced at low frequency it will be lost from the population. Second, populations with all three genotypes are unstable and will ultimately either lose Medea or lose wildtype individuals. Third, there are large regions of fitness space (region B in Figure 2A, Fig. S2) within which - if a Medea is present at a frequency greater than the UIEAF - it will spread to a stable equilibrium (the SIEAF). At this equilibrium non-Medea alleles remain (Fig. 2B, Fig. S1) (see supplement for derivation of equations), but non-Medea individuals are absent. This last feature of Medea, that if spread occurs non-Medea individuals are driven from the population, is important because the epidemiology of insect-borne diseases such as malaria indicates that disease prevention through population replacement will require that most insects be refractory to disease transmission (Boete and Koella 2002).
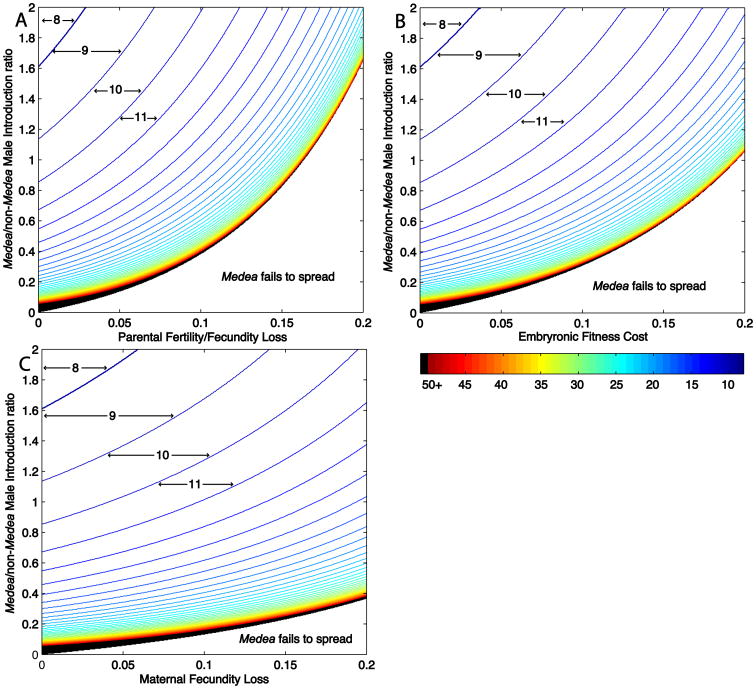
The rate at which Medea spreads and eliminates non-Medea individuals is a function of its introduction frequency and the nature and size of the fitness cost
In order for population replacement to be useful in the real world, the drive mechanism must be able to spread genes quickly through the wild population. The relationship between fitness cost, introduction frequency (expressed as the ratio of homozygous Medea/non-Medea males present in the population) and number of generations required to bring non-Medea individuals below a specific frequency (1%), is illustrated in Fig. 3A, 3B and 3C, for different kinds of fitness cost (all assume multiplicative fitness costs, t0=1, t1=0). The lower bound on these graphs (the black border) defines for any given fitness cost, the critical homozygous Medea/non-Medea male introduction ratio (CMIR), below which Medea will not spread. This number is of practical interest because the release of Medea-bearing males is most appropriate for population replacement in mosquito populations since it is technically feasible to release only males (Knipling et al. 1968; Catteruccia et al. 2005; Smith et al. 2007), and females bite and transmit disease while males do not. In addition, the release of homozygous males only is likely to maximize spread since it forces all Medea-bearing individuals to mate with non-Medea-bearing individuals, maximizing drive in the next, heterozygous, generation. A sex-independent parental fitness cost (Fig. 3A) requires the highest introduction frequency because in the first generation two copies of the fitness cost are born by homozygous Medea fathers used to initiate population replacement, leading to a decrease in their reproductive output and the effective introduction frequency. Medeas carrying an embryonic fitness cost (Fig. 3B) require a somewhat lower introduction frequency because there is no cost in the first, parental generation. A female-specific fitness cost (Fig. 3C) requires the lowest introduction frequency because no fitness cost is incurred in the first, parental generation, while in subsequent generations the costs are limited to females.
Figure 3.
When an autosomal Medea with a fitness cost and t1=0 spreads, it drives the elimination of non-Medea individuals, but not non-Medea alleles, from the population. (A) Plot describing the number of generations required for Medea to be present in 99% of individuals, for a Medea element with a parental fertility/fecundity cost. Homozygous Medea male:non-Medea male introduction ratios are indicated on the Y-axis, and parental fertility/fecundity cost on the X axis. Area between lines indicates regions of parameter space within which a specific number of generations (indicated by numbers and arrows) are required for the frequency of Medea individuals to reach 99% or greater. Line color, shown in the heat map in the lower right, provides a measure of how many generations are required. Black lines (50+) indicate that fifty or more generations are required. The border between the black-lined region and the lower unlined region defines the critical male introduction ratio (CMIR). (B) Plot describing the number of generations required for Medea to be present in 99% of individuals, for a Medea element with an embryonic fitness cost. (C) Plot describing the number of generations required for Medea to be present in 99% of individuals, for a Medea element with a maternal fecundity cost.
If offspring heterozygous for an autosomal Medea experience a fitness cost avoided by homozygotes, the non-Medea chromosome may be eliminated from the population
In the above discussion of autosomal Medea elements we have assumed that heterozygous Medea offspring of homozygous Medea mothers always survive. We now relax this assumption. Such a situation could easily arise if progeny of homozygous Medea-bearing mothers (which inherit two dosages of the toxin) cannot, or can only imperfectly be rescued from death by a single copy of Medea (a single copy of the antidote) in the zygote (0<t1<1) (see also Fig. S3 in (Chen et al. 2007)). Smith makes a related point, though details of his model differ from ours with respect to the fate of homozygous Medea progeny of homozygous Medea mothers (Smith 1998): we assume that homozygous progeny of homozygous Medea mothers show 100% survival, as observed in (Chen et al. 2007), while he does not. To understand the consequences of this novel cost for the fate of Medea we calculate the equilibrium values for one particular case, a multiplicative embryonic fitness cost (VE,Homo=VE,Het2) and t0=1. Again there are four possible equilibria. The feasibility and stability of equilibria within t1, embryonic fitness parameter space are plotted in Fig. 4A.
Figure 4.
When Medea is located on an autosome, and heterozygous Medea offspring of homozygous Medea mothers do not always survive, the non-Medea allele experiences a cost that can result in its elimination from the population. (A) Diagram partitioning (t1, VHet) parameter space into regions in which linear stability analysis indicates qualitatively similar behaviors are observed. Qualitative behavior changes as we cross each of these curves, with the occurrence of a bifurcation, as described in the legend to Figure 2. (B) Plot of allele and population fitness, and genotype frequency, as a function of Medea allele frequency, for a Medea that carries a 10% embryonic fitness cost and has t1=0.5. Compare with the Medea shown in Fig. 1D, in which t1=0. (C) Medea-bearing genotype frequency is plotted as a function of the number of generations for Medea elements with zero fitness cost and different levels of heterozygous offspring lethality (t1), introduced into a population of non-Medea females using a fixed 1:1 ratio of Medea:non-Medea males. (D) Plot of Medea allele frequency as a function of the number of generations for the zero fitness cost Medea elements in (C). (E) Plot as in (C) for Medea elements with a 10% embryonic fitness cost. (F) Plot of Medea allele frequency as a function of the number of generations for Medea elements with a 10% embryonic fitness cost.
(1) No Medea-bearing individuals in the population
G++=1, GMM=0, GM+=0.
This equilibrium is stable (Fig. 4A, regions A, B, and C) except when VE,Het=1 (Fig. 4A, line a), where the stability analysis is inconclusive. Simulations indicate that at this point the equilibrium is unstable. This equilibrium implies that if there is a fitness cost, regardless of t1, with very low introduction frequencies Medea will be lost.
(2) All three genotypes in the population
This equilibrium is not feasible for low fitness values (Figure 4A, region C), and is biologically feasible, but unstable when t1 is relatively high, (Fig. 4A, regions A and B) (See supplemental materials for algebraic expressions of equilibrium frequencies and the stability boundary). In short, populations containing all three genotypes will not persist; they will either lose Medea-bearing genotypes (equilibrium 1) or wildtypes (equilibrium 3 and equilibrium 4).
(3) No non-Medea individuals in the population
This equilibrium population is only feasible when t1≤2 (1-VE,Het), and is stable in Fig. 4A region A but unstable in Fig. 4A region C. This equilibrium indicates that there are biologically relevant regions (region A, primarily low t1 values) in which wildtype individuals are eliminated but non-Medea alleles remain the population.
(4) All Medea homozygous individuals in the population
G++=0, GM+=0, and GMM=1.
This equilibrium is stable when t1>2(1-VE,Het) (Figure 4A, region B) and unstable when t1<2(1-VE,Het) (Figure 4A, regions A and C). At the equality, the analysis is inconclusive but the equilibrium is coincident with equilibrium 3. This equilibrium indicates that there are biologically relevant situations (region B; high Medea allele fitness and high t1) in which the spread of Medea results in the non-Medea allele being driven from the population.
The behavior of Medea elements with t1>0 is illustrated in Fig. 4. Figs. 4C, D show the behavior of Medea elements with no fitness cost, in which heterozygous progeny of homozygous mothers do not die (t1=0), sometimes die, (t1=0.25; t1=0.5; t1=0.75), or always die (t1=1.0). Figs. 4E, F show the behavior of Medea elements that carry a 10% embryonic fitness cost, with t1 values as above. In both cases non-Medea individuals are eliminated from the population, though values of t1>0 result in modest delays. When a Medea carries a fitness cost, values of t1>0 result in a decrease in the non-Medea equilibrium allele frequency, which can go to 0 for high values of t1 (Fig. 4F).
The mechanism by which values of t1>0 can lead to loss of the non-Medea allele from the population can be understood by considering the changing fitness of the non-Medea allele for t1=0 and t1>0, as the frequency of Medea increases. Fig. 6B provides an example, for a Medea with a 10% embryonic fitness cost and t1= 0.5. When such a Medea allele is present at frequencies just above the UIAEF, most Medea alleles are in heterozygotes, and non-Medea alleles experience a Medea-dependent fitness cost similar to that for a Medea with a 10% embryonic fitness cost and t1=0 (Fig. 4B, compare with Fig. 1D). However, in contrast to the case of an element with t1=0, in which the fitness of the non-Medea allele recovers as the frequency of males carrying Medea increases (Fig. 1D), for a Medea with a 10% fitness cost and t1=0.5, the fitness of the non-Medea allele continues to remain low, never becoming greater than that of the Medea allele (Fig. 4B). This occurs because non-Medea alleles from heterozygous males face a new, 50% risk of death in heterozygous progeny when female parents are homozygous for Medea. As Medea spreads, the frequency of heterozygotes becomes rare with respect to the frequency of Medea-bearing individuals (which are mostly homozygotes), while heterozygotes make up the majority of individuals carrying the non-Medea allele. Therefore, the fitness costs from this form of death are born primarily by the non-Medea allele. This results in the non-Medea allele being eliminated from the population, an outcome also reflected in population fitness, which - following an initial decrease resulting from high levels of Medea-dependent killing - increases continuously as Medea spreads to fixation (Fig. 4B).
When a Medea on the X chromosome in a X/Y male species spreads, the non-Medea allele experiences a novel cost that drives it to extinction
The non-Medea allele also experiences a novel cost when Medea is located on the X in a X/Y male heterogametic species. This cost arises because in each generation X-linked non-Medea alleles present in heterozygous Medea female parents have a 50% probability of ending up in a male progeny, which are doomed to death because they cannot be rescued by a paternally derived Medea allele. Therefore, the non-Medea X allele experiences a minimum 50% probability of death in the subsequent generation each time it finds itself in a heterozygous Medea female, and the probability of finding itself in a heterozygous Medea female (as opposed to a non-Medea female) increases as the frequency of Medea increases. The consequences of this novel cost are outlined below.
Sx represents the fraction of the total population that is male of genotype x and Dx the fraction of the total population that is female of genotype x. For a Medea on the X, the standard iterative Medea equations are modified as follows:
where the mean fitness, W, equals
Recalling S+Y+SMY+D+++DM++DMM =1, and setting S′+Y=S+Y, D′MM=DMM, D′M+=DM+, and D′++=D++ we find 4 equilibria, only 2 of which, populations consisting of all wildtypes or all Medea, are feasible and stable for biologically relevant fitness values (VE>.54). Details of the other two equilibria, populations consisting of all genotypes, or a mixture of heterozygous and homozygous Medea genotypes are left to Supplemental Materials.
| (1) |
Equilibrium (1) is stable for all values of VE,Het except VE,Het=1. At VE,Het=1 the linear stability analysis is inconclusive, but simulations indicate that the equilibrium is unstable. In other words, if the presence of Medea results in a fitness cost to carriers, low-level introductions of Medea will result in loss of the Medea allele.
| (2) |
This equilibrium is stable for VE,Het>1/2, unstable for VE,Het<1/2, and the analysis is inconclusive at VE,Het=1/2. This equilibrium is particularly interesting because it suggests that if Medea becomes established, there is a broad range of physiologically relevant conditions (VE,Het>1/2) under which, if the Medea is introduced above the UIEAF, the non-Medea allele is eliminated from the population.
These features of an X-linked Medea are illustrated in Fig. 5A, which plots allele fitness versus Medea allele frequency for a X-linked Medea with a 10% embryonic fitness cost. The fitness costs associated with the non-Medea allele remain higher than those associated with the Medea allele for all Medea allele frequencies above the UIEAF. This forces the non-Medea allele out of the population, and is also reflected in changes in population fitness as Medea spreads. Population fitness initially decreases as a result of Medea-dependent killing of non-Medea alleles; it then increases continuously as Medea spreads and the killing of non-Medea male progeny declines. The dynamics of Medea spread for representative elements located on the X carrying an embryonic fitness cost are illustrated in Figs. 5B-D. Non-Medea individuals are rapidly eliminated from the population, though the times required are somewhat longer, and CMIRs somewhat higher than those required for an autosomal element with a similar fitness cost (compare with Fig. 3A-C), since introduced males carry only one copy of Medea.
Figure 5.
When Medea located on the X chromosome in a male heterogametic species spreads, the non-Medea allele is eliminated from the population. (A) Plot of allele and population fitness as a function of Medea allele frequency, for a Medea that carries a 10% embryonic fitness cost, located on the X chromosome. Lines and labels and other conditions are as in Fig. 1D. (B) Medea genotype frequency is plotted as a function of the number of generations for Medea elements on the X with different levels of an embryonic fitness cost, introduced into a population of non-Medea females using a fixed 1:1 ratio of Medea:non-Medea males. (C) Plot describing the number of generations required for Medea to be present in 99% of individuals, for a Medea element on the X with a 10% embryonic fitness cost. Compare with Fig. 3B. (D) Medea allele frequency is plotted as a function of the number of generations for the elements shown in (B).
Discussion
Here we use a deterministic model to show that Medea selfish genetic elements can drive rapid population replacement under a wide range of conditions, provided that they are introduced above a critical introduction frequency (or critical male introduction ratio), determined by the fitness costs associated with Medea. Stochastic effects (drift, founder effects) will soften this transition such that Medea will sometimes spread when introduced below the CMIR, and sometimes fail to spread when introduced above it, as recently modeled for the case of the Wolbachia drive system (Jansen et al. 2008). A detailed analysis of Medea behavior in finite populations remains to be carried out.
A critical feature of Medea's potential as a drive mechanism, highlighted throughout this work, is that under all conditions in which spread occurs, even when Medea carries a fitness cost and non-Medea alleles remain in the population, loss of non-Medea individuals from the population constitutes a stable equilibrium, implying that spread of Medea results in elimination of these individuals from the population. In some cases, when autosomal Medea elements have a t1>0, or when Medea is located on the X, loss of the non-Medea allele also constitutes a stable equilibrium, implying the non-Medea allele is eliminated from the population. The rate of Medea spread is a function of introduction ratio, fitness costs, and number of elements. Low fitness costs allow rapid spread at relatively low Medea/non-Medea male introduction ratios, while high fitness costs require higher introduction ratios in order for spread to occur quickly, or at all.
Practical population replacement requires that transgenic individuals be refractory to disease transmission for many generations. Medea elements that are autosomally linked with t1>0, or that are X-linked in a male heterogametic species (such as the malaria vector, Anopheles gambiae) could be useful in this regard since having no non-Medea alleles in the population serves to maximize the number of genes for disease refractoriness in individual females in the population, thereby delaying the appearance of insects permissive for disease transmission as these genes (which are presumed not to confer an overall fitness benefit to carriers) mutate to inactivity.
The ability to eliminate a specific allele from the population also provides a method by which first-generation elements can be removed from the population in favor of second-generation elements. This is likely to be important in several contexts. First, as noted above, genes conferring disease refractoriness will eventually mutate to inactivity. In addition, pre-existing diversity and mutation within the pathogen population may select for pathogen populations resistant to first-generation transgenes. Finally, it possible, though unlikely, that the presence of specific transgenes will facilitate the appearance of novel pathogens or unwanted ecosystem effects. As discussed in Chen et al. (2007), second generation Medea elements can be generated that will spread at the expense of first generation elements, when both elements are located at the same chromosomal position. If second generation autosomal elements having t1>0 are used, or the elements are X-linked, first generation elements can be eliminated from the population during this process. The use of such second generation elements carries a price in that somewhat higher introduction frequencies are needed than with an autosomal Medea element having t1=0 (Chen et al. 2007, Fig. S3; and compare Fig. 3B with Fig. 5C). But given the importance that control over the fate of released transgenes is likely to have for the acceptance of population replacement as a strategy for disease prevention, this may be a small price to pay. This strategy does not restore the population to its pre-transgenic state, but it does provide a method for removing specific transgenes from the population.
What are the contexts in which area-wide population replacement with Medea can realistically be carried out? Our results suggest that in order for Medea to drive rapid population replacement within 10-20 generations (roughly 1-2 years), Medea/non-Medea male introduction ratios of between 1:10 and 1:1 are needed, depending on the nature and size of fitness costs. These numbers represent optimistic estimates because they assume that Medea males are competitive with wild males. Wild populations of Aedes aegypti and some Anopheles species have been estimated to range from 10,000-20,000 adults per village (Scott et al. 2000; Taylor et al. 2001). These sizes are small compared with those associated with classical sterile male release in other insects; 68,000 per week in the case of the screw worm fly, and ∼109 in the case of Mediterranean fruit flies (Dyck et al. 2005). With respect to mosquitoes, weekly factory production of 1,000,000 Aedes aegypti could be achieved routinely in the 1960s. Large numbers of Anopheles males have also been produced in factory environments using mid-twentieth century technologies (Knipling et al. 1968). In some contexts it may also be possible to take advantage of naturally-occurring changes in mosquito population size to provide an environment in which Medea can more easily gain a foothold within a population. For example, while wet season populations of Anopheles adults per village in Mali can reach ∼15,000, in the dry season these populations consist of only 1,000-3,000 adults (Taylor et al. 2001). These encouraging points notwithstanding, it is important to emphasize that area-wide population replacement remains a daunting task. Disease-endemic regions can be very large (thousands of square miles), and consist of many villages, requiring that the number of Medea males to be released be scaled accordingly. Finally, we note that the models examined here make a number of assumptions: infinite population size, non-overlapping generations, no age structure within the population, random mating, and no migration. This kind of model is often used to gain basic insights into population genetic processes. However, it provides only a qualitative snapshot of the conditions under which Medea can succeed in driving population replacement. It will be important to carry out more detailed modeling that takes account of the biology of specific pest species, as well as other variables that can influence rate of spread and functional lifetime in the wild.
Supplementary Material
Acknowledgments
Y.H., A.L.L. and F.G. were supported by NIH grant R01-AI54954-0IA2 and a grant to the Regents of the University of California from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health initiative. B.A.H, C.M.W. and J.T.S were supported by grants to B.A.H from NIH (R01 GM072879; R01 GM070956; DP1 OD003878), and the Weston Havens Foundation. Support was also provided by a grant to the Regents of the University of California from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health initiative. We thank M. Legros for making helpful suggestions on model analysis.
Literature Cited
- Beeman RW, Friesen KS, Denell RE. Maternal-effect selfish genes in flour beetles. Science. 1992;256:89–92. doi: 10.1126/science.1566060. [DOI] [PubMed] [Google Scholar]
- Boete C, Koella JC. A theoretical to predicting the success of genetic manipulation of malaria mosquitoes in malaria approach control. Malar J. 2002;1:3. doi: 10.1186/1475-2875-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig HR, Yan G. The spread of genetic constructs in natural insect populations. In: Letourneau DK, B BE, editors. Genetically Engineered Organisms: Assessing Environmental and Human Health Effects. CRC Press; 2001. pp. 251–314. [Google Scholar]
- Burt A, Trivers R. Genes in conflict. Belknap Press; Cambridge, MA: 2006. [Google Scholar]
- Catteruccia F, Benton JP, Crisanti A. An Anopheles transgenic sexing strain for vector control. Nat Biotechnol. 2005;23:1414–1417. doi: 10.1038/nbt1152. [DOI] [PubMed] [Google Scholar]
- Chen CH, Huang H, Ward CM, Su JT, Schaeffer LV, Guo M, Hay BA. A synthetic maternal-effect selfish genetic element drives population replacement in Drosophila. Science. 2007;316:597–600. doi: 10.1126/science.1138595. [DOI] [PubMed] [Google Scholar]
- Clements AN. The Biology of Mosquitoes: Sensory Reception and Behavior. CABI; Oxfordshire, UK: 1999. [Google Scholar]
- Corby-Harris V, Drexler A, Watkins de Jong L, Antonova Y, Pakpour N, Ziegler R, Ramberg F, Lewis EE, Brown JM, Luckhart S, Riehle MA. Activation of Akt signaling reduces the prevalance and intensity of malaria parasite infection and lifespan in Anopheles stephensi mosquitoes. PloS Pathog. 2010;6:e1001003. doi: 10.1371/journal.ppat.1001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lara Capurro M, Coleman J, Beerntsen BT, Myles KM, Olson KE, Rocha E, Krettli AU, James AA. Virus-expressed, recombinant single-chain antibody blocks sporozoite infection of salivary glands in Plasmodium gallinaceum-infected Aedes aegypti. Am J Trop Med Hyg. 2000;62:427–433. doi: 10.4269/ajtmh.2000.62.427. [DOI] [PubMed] [Google Scholar]
- Dyck VA, Hendrichs J, Robinson AS, editors. Sterile Insect Technique: Principles and Practice in Area-Wide Integrated Pest Management. Springer; Dordrecht, The Netherlands: 2005. [Google Scholar]
- Franz AW, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, James AA, Olson KE. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc Natl Acad Sci U S A. 2006;103:4198–4203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould F, Schliekelman P. Population genetics of autocidal control and strain replacement. Annu Rev Entomol. 2004;49:193–217. doi: 10.1146/annurev.ento.49.061802.123344. [DOI] [PubMed] [Google Scholar]
- Hastings IM. Selfish DNA as a method of pest control. Philos Trans R Soc Lond B Biol Sci. 1994;344:313–324. doi: 10.1098/rstb.1994.0069. [DOI] [PubMed] [Google Scholar]
- Hay BA, Chen CH, Ward CM, Huang H, Su JT, Guo M. Engineering the genomes of wild insect populations: Challenges, and opportunities provided by synthetic Medea selfish genetic elements. J Insect Physiol. doi: 10.1016/j.jinsphys.2010.05.022. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Ghosh A, Moreira LA, Wimmer EA, Jacobs-Lorena M. Transgenic anopheline mosquitoes impaired in transmission of a malaria parasite. Nature. 2002;417:452–455. doi: 10.1038/417452a. [DOI] [PubMed] [Google Scholar]
- Jansen VA, Turelli M, Godfray HC. Stochastic spread of Wolbachia. Proc Biol Sci. 2008;275:2769–2776. doi: 10.1098/rspb.2008.0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK. Kin selection in density regulated populations. J Theor Biol. 1992;157:447–461. doi: 10.1016/s0022-5193(05)80663-0. [DOI] [PubMed] [Google Scholar]
- Knipling EF, Laven H, Craig GB, Pal R, Kitzmiller JB, Smith CN, Brown AW. Genetic control of insects of public health importance. Bull World Health Organ. 1968;38:421–438. [PMC free article] [PubMed] [Google Scholar]
- Moreira LA, Ito J, Ghosh A, Devenport M, Zieler H, Abraham EG, Crisanti A, Nolan T, Catteruccia F, Jacobs-Lorena M. Bee venom phospholipase inhibits malaria parasite development in transgenic mosquitoes. J Biol Chem. 2002;277:40839–40843. doi: 10.1074/jbc.M206647200. [DOI] [PubMed] [Google Scholar]
- Schmid-Hempel P. Evolutionary ecology of insect immune defenses. Annu Rev Entomol. 2005;50:529–551. doi: 10.1146/annurev.ento.50.071803.130420. [DOI] [PubMed] [Google Scholar]
- Scott TW, Morrison AC, Lorenz LH, Clark GG, Strickman D, Kittayapong P, Zhou H, Edman JD. Longitudinal studies of Aedes aegypti (Diptera: Culicidae) in Thailand and Puerto Rico: population dynamics. J Med Entomol. 2000;37:77–88. doi: 10.1603/0022-2585-37.1.77. [DOI] [PubMed] [Google Scholar]
- Sinkins SP, Gould F. Gene drive systems for insect disease vectors. Nat Rev Genet. 2006;7:427–435. doi: 10.1038/nrg1870. [DOI] [PubMed] [Google Scholar]
- Smith NG. The dynamics of maternal-effect selfish genetic elements. J Theor Biol. 1998;191:173–180. doi: 10.1006/jtbi.1997.0579. [DOI] [PubMed] [Google Scholar]
- Smith RC, Walter MF, Hice RH, O'Brochta DA, Atkinson PW. Testis-specific expression of the beta2 tubulin promoter of Aedes aegypti and its application as a genetic sex-separation marker. Insect Mol Biol. 2007;16:61–71. doi: 10.1111/j.1365-2583.2006.00701.x. [DOI] [PubMed] [Google Scholar]
- Taylor C, Toure YT, Carnahan J, Norris DE, Dolo G, Traore SF, Edillo FE, Lanzaro GC. Gene flow among populations of the malaria vector, Anopheles gambiae, in Mali, West Africa. Genetics. 2001;157:743–750. doi: 10.1093/genetics/157.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet F, Aboagye-Antwi F, Hurd H. Ecological immunology of mosquito-malaria interactions. Trends Parasitol. 2008;24:219–227. doi: 10.1016/j.pt.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ. Hard selection, soft selection, kin selection, and group selection. Am Nat. 1985;125:61–73. [Google Scholar]
- Wade MJ, Beeman RW. The population dynamics of maternal-effect selfish genes. Genetics. 1994;138:1309–1314. doi: 10.1093/genetics/138.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.