Abstract
Cholesterol is an essential structural component of cellular membranes and serves as a precursor for several classes of signaling molecules. Cholesterol exerts its effects and is, itself, regulated in large part by engaging in specific interactions with proteins. The full complement of sterol-binding proteins that exist in mammalian cells, however, remains unknown. Here, we describe a chemoproteomic strategy that uses clickable, photoreactive sterol probes in combination with quantitative mass spectrometry to globally map cholesterol-protein interactions directly in living cells. We identified over 250 cholesterol-binding proteins, including many established and previously unreported interactions with receptors, channels, and enzymes. Prominent among the newly identified interactions were enzymes that regulate sugars, glycerolipids, and cholesterol itself, as well as those involved in vesicular transport and protein glycosylation and degradation, pointing to key nodes in biochemical pathways that may couple sterol concentrations to the control of other metabolites and protein localization and modification.
The membrane bilayer serves as a physical barrier that defines the outer boundary of cells and segregates their interior into distinct compartments that perform specialized functions. Among the many lipid constituents of mammalian cell membranes, cholesterol is special in that it is a major regulator of membrane fluidity and contributes to the formation of specific membrane structures such as caveolae and other lipid microdomains1. Beyond its role in membrane structure, cholesterol also serves as a metabolic precursor for a diverse array of signaling molecules, including oxysterols2, steroids3, and bile acids4. Deregulation of cholesterol uptake and metabolism is the basis for a range of human diseases that include cardiovascular disorders5 skin6, and developmental7,8 defects, as well as lysosomal storage syndromes9,10 and neurodegeneration10,11.
Cholesterol's central role in mammalian physiology mandates that the concentrations of this lipid are tightly regulated in cells. Meeting this objective poses an unusual challenge for cells, since the vast majority of cholesterol is embedded within the lipid bilayer. Multiple sterol-sensing pathways have been identified that communicate alterations in the membrane concentrations of cholesterol to the transcriptional and post-transcriptional control of sterol biosynthetic, uptake, and transport pathways12–14. Beyond these primary control points, cholesterol has also been found to interact with a number of other proteins by both covalent15 and non-covalent mechanisms16,17. These cholesterol-protein interactions are thought to regulate protein stability, localization, and activity.
Despite the tremendous progress that has been made in characterizing specific cholesterol-protein interactions, our understanding of the full spectrum of proteins that regulate, and are regulated by, cholesterol remains incomplete, in large part due to a lack of global methods for mapping proteins that physically interact with sterols in living cells. The biochemical assessment of cholesterol-protein interactions has, to date, been restricted to a limited canon of assays, that include direct in vitro binding using radiolabeled cholesterol and purified proteins18 and modification of proteins with radiolabeled photoreactive cholesterol analogues19,20. While these methods have been used to successfully characterize individual protein-cholesterol interactions, the approaches also lack key features, most notably a means for affinity enrichment, that have precluded their incorporation into chemoproteomic platforms for the global discovery of sterol-binding proteins in mammalian cells.
Here, we expand the repertoire of available methods for mapping sterol-binding proteins by introducing a chemoproteomic approach to label, enrich, and identify cholesterol-interacting proteins from living cells. We apply this approach to identify more than 250 cholesterol-binding proteins in HeLa cells, including several proteins that are known to biosynthesize, transport, and regulate cholesterol, as well as a large suite of proteins for which no prior interaction with cholesterol has been described.
RESULTS
Design of clickable, photoreactive sterol probes
We reasoned that chemoproteomic probes for mapping cholesterol-binding proteins in living cells would need to possess three general features: 1) a photoreactive group for ultraviolet (UV) light-induced crosslinking to probe-interacting proteins, 2) a latent affinity handle, such as an alkyne group for conjugation to azide-reporter tags by copper-catalyzed azide-alkyne cycloaddition (CuAAC or click) chemistry21, which enables detection, enrichment, and identification of probe-interacting proteins22, and 3) an intact cholesterol scaffold, such that after integration of photocrosslinking and `clickable' groups, the resulting probe could still interact with most cholesterol-binding proteins. With these considerations in mind, we designed and synthesized a set of sterol probes (Fig. 1a) each of which contains a photoactivatable diazirine group at the 6-position of the steroid core (using standard numbering), which is a modification that has been previously shown to minimally perturb the biophysical properties of cholesterol23. The probes also possess an alkyne incorporated via an ester linkage into the alkyl side-chain of cholesterol. While this ester linkage, which was chosen for ease of synthesis, would be expected to increase the polarity of the alkyl side-chain, we favored this modification over further perturbations to the steroid core, which we believed should serve as the major basis for recognition for a large fraction of cholesterol-binding proteins. The probes differ in the diastereomeric relationship between the C3-alcohol and C5-hydrogen groups appended to the cholesterol core (termed cis, trans, and epi hereafter) and were synthesized from a common precursor - the bile acid hyodeoxycholic acid (Supplementary Note).
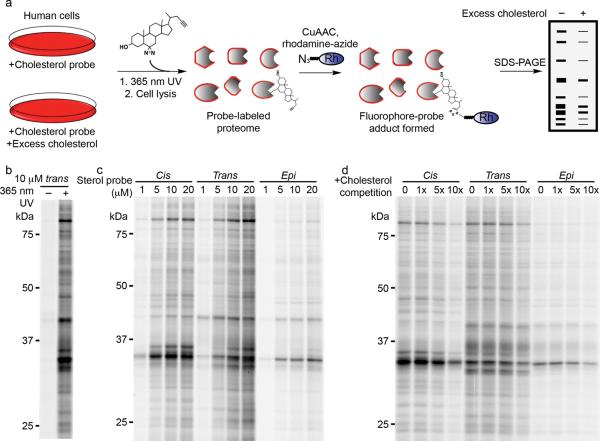
Figure 1. Clickable photoreactive sterol probes.
(a) Structures of cholesterol and three diastereomeric sterol chemoproteomic probes. (b) Three-dimensional structures of cholesterol and sterol probes as determined by x-ray crystallography; cholesterol structure derived from PDBID: 3GKI24. In a and b, from top to bottom; cholesterol (pink), trans-sterol probe (green), epi-sterol probe (cyan), cis-sterol probe (purple).
We obtained X-ray structures of the keto-acid intermediates corresponding to each cholesterol probe to verify their relative and absolute stereochemistry (Fig. 1b). As compared to the three-dimensional structure of cholesterol (extracted from a structure in complex with NPC124, Fig 1b), the trans probe exhibits the most similarity to cholesterol in terms of stereochemistry and molecular topology. Due to its 3α-OH stereochemistry, the epi probe instead most resembles epicholesterol. The cis probe, while appearing bent in the crystal structure (Fig. 1b), is probably the most flexible of the probes due to its structurally distinct cis-decalin-type A–B ring fusion25. This ring fusion stereochemistry, along with its 3α-OH stereochemistry, likely allow for the cis probe to adopt bent cholesterol-like conformations in solution that retain the equatorial orientation of the C3 hydroxyl group.
Gel profiling of sterol-binding proteins in human cells
We first assessed sterol probe labeling of cellular proteins using SDS-PAGE analysis (Fig. 2a; see Online Methods for more details). Probe labeling for virtually all detected proteins was found to be UV-irradiation dependent (Fig 2b), indicating that these interactions reflect non-covalent binding events (versus the post-translational modification of proteins by cholesterol, as has been reported in select instances15). Concentration-dependent increases in protein labeling were observed for all three probes (1–20 μM probe), with the cis- and trans-sterol probes showing stronger overall protein-labeling profiles than the epi probe (Fig. 2c). We chose conditions that produce substantial, but sub-maximal labeling (10 μM probe) for further evaluation.
Figure 2. Gel-based profiling of sterol-binding proteins in HeLa cells.
(a) Scheme for treatment of live cells with sterol probes and competitive treatments. (b) HeLa cells treated with 10 μM trans-sterol probe, with and without 365 UV irradiation before click chemistry and SDS-PAGE analysis. (c) Concentration-dependent labeling of live HeLa cells with each probe (cis, epi, trans) at 1, 5, 10, and 20 μM. (d) Competition of sterol probe labeling profiles (10 μM probes) with increasing cholesterol from 0, 10 (1×), 50 (5×), and 100 μM (10×). Fluorescence gel images shown in grayscale.
We found that the protein labeling profiles for all three sterol probes were competed to variable degrees by excess cholesterol (1×, 5× or 10× cholesterol supplemented as a mβCD complex), with the cis-sterol probe showing the greatest sensitivity (Fig. 2d). Steroids or sterols with structures similar to cholesterol also blocked many of the protein labeling events observed for the trans-sterol probe, while structurally less-related steroids, exhibited little or no competition (Supplementary Fig. 1)..
We next set out to develop a mass spectrometry (MS) method to enrich, identify, and quantify sterol-binding proteins in human cells.
MS profiling of sterol-binding proteins in human cells
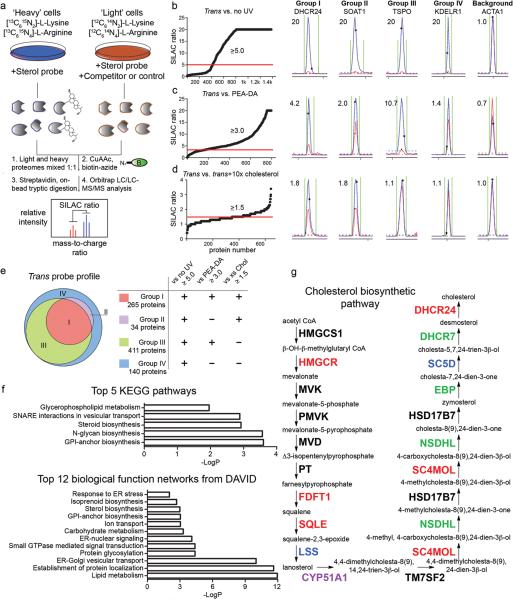
We aimed to characterize sterol-binding proteins using biotin-streptavidin methods coupled with SILAC (Stable isotope labeling by amino acids in cell culture) MS26, so that we could distinguish with precision proteins that specifically interact with sterol probes and assess the sensitivity of these interactions to competition by excess cholesterol (Fig. 3a). For our initial analyses, isotopically `heavy' HeLa cells were treated with the trans-sterol probe (20 μM) for 30 min and irradiated with UV light (5 min), while `light' cells received either no probe treatment prior to UV light exposure (vehicle control), or received the trans-sterol probe (20 μM), but were not irradiated (no-UV control). Cells were then harvested, their heavy and light proteomes mixed 1:1, and probe-labeled proteins conjugated to an azide-biotin tag by click chemistry and enriched using streptavidin chromatography. Enriched proteins were then digested on-bead with trypsin and the resulting tryptic peptide mixture analyzed by LC-MS methods as described in the Online Methods. Proteins that exhibited heavy:light SILAC ratios of ≥ 5 for trans-sterol probe versus both vehicle and no-UV light control reactions were designated as sterol-interacting proteins. About 850 proteins met these criteria (Fig. 3b and Supplementary Tables 1 and 2).
Figure 3. MS-based profiling of sterol-binding proteins in HeLa cells.
(a) Scheme for enrichment and analysis of sterol probe labeling profiles in mammalian cells by biotin-streptavidin methods and SILAC MS analysis. (b–d) Heavy/light ratio plots for total proteins identified in experiments that compared the labeling profiles of the trans-sterol probe versus no-UV light control (b; 20 μM trans probe / 20 μM trans probe with no UV), the PEA-DA probe (c; 20 μM trans probe / 20 μM PEA-DA probe), and 10× cholesterol competition (d; 10 μM trans probe / 10 μM trans probe + 100 μM cholesterol). Representative MS1 traces with calculated ratios for proteins that fall into Groups I-IV, as well as the MS1 traces for a non-specific background protein, are shown to the right of the global ratio plots. Ratios of > 20 are listed as 20. (e) Venn diagram showing the distribution of Group I-IV proteins for the trans-sterol probe labeling profile. (f) Top-five pathways determined by searching Group I proteins on the KEGG database, and top-12 biological function networks determined by searching Group I proteins on the DAVID gene ontology server. (g) Trans-sterol probe labeling profile for the cholesterol biosynthetic pathway, with colors reflecting each enzyme's Group designation (black: not detected).
We next compared the proteome-labeling profiles of the trans-sterol probe with the cis and epi probes by SILAC. The mean labeling intensity ratio for probe-enriched proteins in the trans-versus-cis comparison was 0.9 (trans/cis; standard deviation = 0.57) (Supplementary Fig. 2), indicating that the trans and cis probes display similar protein-interaction profiles in cells. In contrast, the mean ratio, as well as the standard deviation, of the trans-versus-epi comparison were much higher (2.1 and 1.2 for trans/epi, respectively; Supplementary Fig. 2), suggesting that the stereochemistry of the sterol hydroxyl group impacts probe-protein interactions in a variable manner.
Nearly 700 of the identified sterol-binding proteins showed strong selectivity (≥ 3-fold higher signals) for the trans-sterol probe over a non-steroidal neutral lipid probe containing a diazirine and alkyne group embedded within an N-palmitoylethanolamine structure [N-palmitoylethanolamine-diazirine-alkyne (PEA-DA; Supplementary Note) (Fig. 3c and Supplementary Table 1). Only 18 proteins from the sterol-binding group showed the opposite profile, exhibiting greater labeling with the PEA-DA probe (heavy/light signal ratio < 1.0) (Fig. 3c).
We next performed competitive profiling experiments where the trans-sterol probe was evaluated for protein labeling in cells treated with excess cholesterol. Light and heavy-labeled HeLa cells were treated with the trans-sterol probe (10 μM) in the presence or absence of 10× (100 μM) cholesterol, respectively. Approximately 300 proteins showed at least a 50% decrease in trans-sterol probe labeling intensity in cells treated with excess cholesterol (Fig. 3d and Supplementary Table 3). Over 250 of these cholesterol-sensitive targets also showed selective labeling by the trans-sterol probe compared to the PEA-DA probe (Supplementary Table 1), and the majority (> 60%) of the competed proteins also showed evidence of cholesterol competition in experiments performed with the cis-sterol probe (Supplementary Fig. 3). We also performed parallel DNA microarray experiments and found that cholesterol-sensitive proteins, in general, showed no evidence of gene expression changes at the 30 min time point of competitive analysis (Supplementary Fig. 4, Supplementary Table 4).
Finally, we controlled for the level of isotopically labeled amino acid incorporation in our SILAC experiments by treating heavy and light HeLa cells each with 20 uM trans-sterol probe, and combining and processing their proteomes as described above. The overall median and mean ratio for proteins detected in this experiment was 1.0 (Supplementary Table 3), indicative of > 95% heavy amino acid incorporation, and therefore ratios from other SILAC studies are presented without any correction factors.
Together, these quantitative MS experiments enabled us to distribute the proteins that interacted with the trans-sterol probe into four groups: 1) Group I, which were both sensitive to cholesterol competition and selective for the trans-sterol probe over the PEA-probe (265 total proteins), 2) Group II, which were sensitive, but not selective (34 total proteins), 3) Group III, which were selective, but not sensitive (411 total proteins), and 4) Group IV, which were neither sensitive nor selective (140 total proteins) (Fig. 3e and Supplementary Table 1). Representative MS1 traces for sterol-interacting proteins from each group are shown in Fig. 3b–d. We next examined in more detail the 265 proteins found in Group I.
Analysis of Group I cholesterol-binding proteins
Our further analysis of Group I proteins included both computational and experimental inquiries. We first noted that Group I contains many proteins that are known to bind to cholesterol. These include Scap12; caveolin (CAV1)16; tetraspanin CD8220; the sterol transport protein ARV127; and the sterol biosynthetic enzymes, HMG-CoA reductase (HMGCR)17, which is known to be regulated by cholesterol through allosteric binding17,18 (Supplementary Table 5). We found that the trans-sterol probe interacted with many other enzymes in the sterol biosynthetic pathway (Fig. 3f and g). Interestingly, FDFT1 and SQLE, two upstream enzymes in the sterol biosynthetic pathway that do not directly handle sterols as substrates or products were identified in Group I (Fig. 3g), indicating that they may, like HMGCR, interact with cholesterol through an allosteric mechanism. Group II–IV also contained additional proteins known to interact with cholesterol, including, for instance, the lysosomal sterol transporter NPC124 (Supplementary Table 5).
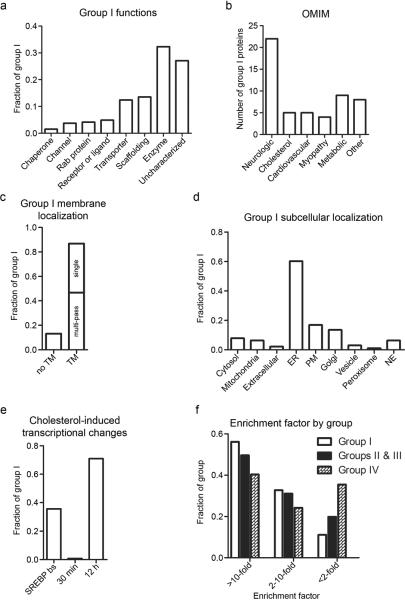
A broader survey of Group I revealed that it contains representatives from virtually all major classes of proteins, including G-protein coupled receptors, a class of receptors that have been proposed to bind cholesterol to stabilize certain functional conformations28, ion channels, transporters, and enzymes (Fig. 4a). Automated KEGG pathway and DAVID analysis tools29,30 identified several types of proteins that were enriched in Group I beyond the aforementioned sterol biosynthetic enzymes, including glycerophospholipid metabolic enzymes, protein glycosylation and degradation pathways, and protein networks that regulate membrane structure and dynamics (Fig. 3f). When considered in relation to human disease by analysis with the OMIM database, Group I was most highly enriched in proteins linked to neurological disorders, as well as cardiovascular and metabolic diseases (Fig. 4b). Group I also contained a substantial number of proteins of uncharacterized function (Fig. 4a), including several that cluster into sequence-related sub-families (such as YIF1A/YIF1B, FNDC3A/FNDC3B, UNC84A/UNC84B, DPY19L1/DPY19L4, FAM114A1/FAM114A2, FAM134A/FAM134B/FAM134C; Supplementary Table 1).
Figure 4. Analysis of Group I proteins.
(a) Breakdown of Group I proteins by biochemical functions. (b) Group I proteins known to be genetically associated with human disease based on the OMIM database; the `cholesterol' group represents diseases from all other groups that are known to manifest via aberrant cholesterol homeostasis. (c) Fraction of Group I proteins that possess known or predicted transmembrane (TM) domains. TM proteins are further divided into single- versus multi-pass TM proteins. (d) Known or predicted subcellular localization of Group I proteins. Subcellular localization predictions were made by examining protein sequences by the PSORT II algorithm (http://psort.hgc.jp/form2.html). (e) Cholesterol regulation of Group I proteins at the mRNA level. `SREBP bs' denotes the fraction of Group I proteins with SREBP transcription factor binding sites in the gene/promoter regions based on the Qiagen SABiosciences transcription factor database (http://www.sabiosciences.com/chipqpcrsearch.php?app=TFBS). `30 min' and `12 h' denote the fraction of Group I proteins with substantial (≥ two-fold) changes in mRNA levels after 100 μM cholesterol treatment for the indicated time. (f) Levels of enrichment (≥ 2-fold, 2–10 fold, ≥ 10-fold) of Groups I, II & III, and IV proteins in trans-sterol probe data sets compared to their abundance in unenriched membranes.
Evaluation of the subcellular distribution of Group I proteins revealed, perhaps not surprisingly, that the vast majority (87%) are known or predicted integral membrane proteins (Fig. 4c). These proteins were near-equally distributed between single- and multi-pass transmembrane proteins (Fig. 4c) and were dispersed across all subcellular membrane compartments with a notable enrichment in known or predicted ER proteins (Fig. 4d). Even though only a limited number of soluble proteins (13%) were found in Group I, the composition of this subset of proteins is intriguing, including three kinases (HK1, HK2, and ADPGK) that convert glucose to glucose-6-phosphate, a key metabolic substrate for glycolysis and the pentose phospate pathway. Both HK1 and HK2 are known to associate with cholesterol-rich regions of the mitochondria, although historically this interaction has been suggested to occur indirectly through binding VDAC channels31. Our chemoproteomic data indicate that HK1 and HK2 may also interact directly with cholesterol to facilitate their binding to mitochondria.
Cholesterol is known to regulate the expression of a large and diverse set of genes primarily through the Scap/SREBP system12,14. As noted above, very few of the Group I proteins showed evidence of gene expression changes at the 30 min time point of our chemoproteomic analyses. Interestingly, however ~35% of the Group I proteins possess predicted SREBP transcription factor-binding sites in their promoter regions, and we determined that ~70% of these proteins exhibit greater than two-fold changes (up or down) in their corresponding mRNA levels at 12 h following treatment of HeLa cells with cholesterol (100 μM) (Fig 4e, Supplementary Fig. 4, and Supplementary Tables 1 and 4). These data suggest that many proteins may be regulated by cholesterol at both the transcriptional (gene expression) and post-transcriptional (direct binding) levels.
We finally evaluated the degree to which our chemoproteomic method facilitated the enrichment and detection of sterol-binding proteins compared to analyses performed in unenriched proteomes. Group I proteins showed an average of 11 and 80 spectral counts in unenriched and probe-enriched proteomes, respectively, equating to an average enrichment factor of 7.3 (Supplementary Table 6). Additionally, more than 50% of Group I proteins were enriched > 10-fold (and conversely fewer than 10% of these proteins were enriched < two-fold) in the trans-probe data sets (Fig. 4f). We further observed no positive correlation between a protein's absolute spectral count values in the unenriched versus trans probe-enriched samples (Supplementary Fig. 5), indicating that Group I proteins were not biased toward abundant integral membrane proteins.
Experimentally validating cholesterol-protein interactions
To validate seven representative Group I proteins, including six novel sterol-binding proteins, we recombinantly expressed the proteins in HeLa cells and treated these cells with the trans-sterol probe, followed by CuAAC coupling to an azide-rhodamine tag, separation of proteins by SDS-PAGE, and detection of probe-labeled proteins by in-gel fluorescence scanning (Supplementary Fig. 6). We also confirmed that these sterol-protein interactions were competitively inhibited by excess cholesterol without alterations in the expression level of the recombinant proteins (Supplementary Fig. 6–7).
DISCUSSION
Our strategy to map sterol-binding proteins on a global scale operates in living cells, which is important, given that the transport of cholesterol to discrete subcellular compartments and membrane microdomains is controlled by complex transport machinery1. This work thus builds and significantly extends upon previous efforts that used radioactive, photocrosslinking probes19,20 by providing a means to not only detect, but also affinity-enrich and identify sterol-binding proteins from mammalian cells.
We should also mention some potential limitations of our approach and ways to address these shortcomings in the future. First, our initial set of probes may fail to detect certain sterol-protein interactions that are impaired by the diazirine and/or esterified alkyne modifications to the structure of cholesterol. The ester linkage might also be susceptible to cleavage by endogenous esterases, which could reduce the sensitivity of our proteomic profiles. These problems could be addressed by creating second-generation probes where the diazirine is moved to other locations on the sterol backbone and the esterified alkyne is replaced by distinct linkage chemistries. It might also be valuable to create probes that are tailored to target proteins that bind to specific subsets of sterols (oxysterols2, bile acids4, steroids, etc), as well as to perform competitive profiling where sterol analogues or other small-molecules are tested for their ability to block specific sterol-protein interactions. We furthermore do not yet understand how, in most instances, sterol probes bind to their target proteins. Here, developing a method to map the precise sites of sterol crosslinking to proteins would be valuable, and we note that the ester modification in our probe structures could provide a means to selectively release (through base hydrolysis) sterol-modified peptides from affinity-enrichment matrices. It would also be interesting, in future studies, to extend our analysis to additional cell types and conditions (such as, cells treated with statins or other cholesterol-lowering reagents), which could identify additional sterol-interacting proteins. Finally, we focused most of our attention in this study on Group I proteins because they exhibited evidence of both selective binding to sterols over other lipids and sensitivity to cholesterol competition; however, Groups II–IV also contain proteins known to interact with cholesterol. We do not know why cholesterol failed to compete trans probe labeling of known sterol-binding proteins like NPC1, but we should mention that competition was observed for NPC1 with the cis sterol probe (competition ratio = 2.1). Such probe-dependent variations in cholesterol competition could reflect differences in the local concentration of individual probes and/or cholesterol in specific organelles like the lysosome, or they may reflect differences in protein affinities for probes versus cholesterol. Regardless, that bona fide cholesterol-interacting proteins were found in Groups I, II, and III underscores the importance of considering the potential biological relevance for the entire 800+ sterol-protein interactions reported herein.
Online Methods
Chemical probe synthesis
See Supplementary Note.
Cell culture and live cell labeling
HeLa cells were grown at 37 °C under a humidified 5% CO2 atmostphere, in a culture medium consisting of high-glucose DMEM (HyClone) supplemented with 10% fetal bovine serum (FBS; Gemini) and penicillin, streptomycin, and glutamine (Cellgro; PSQ). For SILAC experiments, the culture medium was replaced with SILAC DMEM (Thermo) supplemented instead with 10% dialyzed FBS, PSQ, and 100 μg/ml [13C6,15N4] L-arginine-HCl and [13C6,15N2] L-lysine-HCl (Sigma-Aldrich). Cells were passaged at least six times in isotopically labeled media before being utilized for analysis by LC-MS/MS.
To facilitate delivery to cells, all sterols and steroids, including the sterol probes, were complexed in aqueous solution to mβCD (Sigma-Aldrich) for at least twelve hours before dilution in culture medium for labeling at working concentrations indicated in Results, adapted from literature procedures32. The desired amount of sterol or steroid was added to a saturated aqueous mβCD (38 mM) solution to generate a concentrated stock, and agitated at room temperature overnight; solutions were filtered prior to use the following day. Aqueous stock solutions of the trans probe were prepared at 2 mM; the cis and epi probes were prepared at 1.2 mM; cholesterol and other sterols for competition were prepared at 4 mM; steroids were prepared at 5 mM. Non-steroidal lipids, C17-MAGE, and di-C15-DAG, as well as the PEA-DA probe were suspended in DMSO (10 mM) and diluted to working concentrations directly in culture medium.
Prior to live-cell labeling, aqueous stock solutions of each sterol probe or competitor (or DMSO stock solution of lipids) were combined in opaque centrifuge tubes, and then diluted to final working concentrations in culture medium under dim ambient light. Unmodified culture medium was then removed from the cells, and replaced with probe-containing medium. Cells were then incubated at 37 °C for 30 minutes in the dark to load the cells with sterol probe and competitors. After this time, cells were washed quickly with cold PBS, and then irradiated for five minutes under 365 nm ultraviolet light in a FB-UVXL-1000 UV Crosslinker (Fisher) in cold PBS. Cells were then harvested by scraping, and the cell pellet frozen at −80 °C until processing for gel or LC-MS/MS analysis.
Sample processing for analysis by SDS-PAGE or LC-MS/MS
Frozen cell pellets were thawed on ice and lysed in PBS by sonication. Protein concentrations of cell lysates were determined using the BCA protein assay on a microplate reader. Click chemistry was then performed as previously described33 directly in whole-cell lysates in PBS. For analysis by gel, 50 μg of protein was used, adjusted to a protein concentration of 1 mg/ml (50 μl), and was mixed with 20 μM rhodamine-azide, 1 mM Tris(2-carboxyethyl)phosphine (TCEP, Sigma-Aldrich), 100 μM Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) (Sigma-Aldrich), and 1 mM CuSO4 in PBS at room temperature. After 1 hour, samples were mixed with SDS sample loading buffer and loaded without boiling on a 10% SDS-PAGE gel, separated, and imaged using a Hitachi FMBIO-II flatbed fluorescence scanner.
For proteomic analysis, 1 mg of both heavy and light lysates were mixed in a 1:1 ratio, and then combined with 500 μM biotin-azide, 100 μM TBTA, 1 mM TCEP, and 1 mM CuSO4 in 400 μl PBS for 1 hour. Water (100 μl), methanol (500 μl) and chloroform (125 μl) were then added directly to the reaction mixture and mixed vigorously by vortexing. The biphasic solution was then centrifuged at 4000 rpm for 20 minutes at 4 °C, and protein was pelleted at the phase interface as a solid disk. Liquid layers were discarded, and the protein was washed further by sonication into 1:1 methanol/chloroform (500 μl first wash, 250 μl second wash) followed by centrifugation at 13.3k rpm for 10 minutes at 4 °C to re-pellet. The protein pellets were air-dried briefly, and then resuspended by sonication into 500 μl water containing 25 mM ammonium bicarbonate and 6 M urea. To this solution, 5 μl of a 1 M DTT solution in water was added, followed by 140 μl 10% SDS in water, and the solution was heated at 65 °C for 15 minutes. The samples were cooled briefly on ice, and then 40 μl of a 0.5 M iodoacetamide (Sigma-Aldrich) solution in water was added, and the samples were incubated at room temperature for 30 minutes in the dark. The samples were then diluted to 6 ml with PBS, and enriched over streptavidin (Thermo) (100 μl slurry) for 2 hours at room temperature. The beads were washed once with 10 ml 1% SDS in PBS, then three more times with 10 ml PBS. The beads were then transferred to a 1.5 ml screw-cap tube in 200 μl 25 mM ammonium bicarbonate/2 M urea in water, with 1 mM calcium chloride and 2 μg sequencing grade porcine trypsin (Promega), and then digested at 37 °C overnight. The digest supernatant was then collected by filtration of the resin, which was washed additionally with 100 μl PBS. The combined filtrate and wash for each sample was then acidifed with 16 μl formic acid, and then pressure-loaded onto a biphasic (strong cation exchange/reverse phase) capillary column for analysis by two-dimensional liquid chromatography separation in combination with tandem mass spectrometry (2D-LC-MS/MS). Unenriched samples were processed in the same fashion, but the enrichment step was omitted, and instead, 200 μg of processed protein was committed to tryptic digestion and MudPIT analysis.
Mass spectrometry and data processing
Mass spectrometry was performed using a Thermo Orbitrap Velos mass spectrometer. Peptides were eluted using a 5-step multidimensional LC-MS (MudPIT34) protocol (using 0%, 25%, 50%, 80%, and 100% salt bumps of 500 mM aqueous ammonium acetate, followed by an increasing gradient of aqueous acetonitrile/0.1% formic acid in each step) and data were collected in data-dependent acquisition mode (2 MS1 microscans (400–1800 m/z) and 30 data-dependent MS2 scans) with dynamic exclusion enabled (repeat count of 1, exclusion duration of 20 s) with monoisotopic precursor selection enabled. All other parameters were left at default values. Unenriched membrane preparations were eluted using the same chromatographic steps and instrument settings. SEQUEST35 searches allowed for variable oxidation of methionine (+15.9949), static modification of cysteine residues (+57.0215; iodoacetamide alkylation), and no enzyme specificity. Each data set was independently searched with light and heavy parameters files; for the light search, all other amino acids were left at default masses; for the heavy search, static modifications on lysine (+8.0142) and arginine (+10.0082) were specified. The precursor ion mass tolerance was set to 50 ppm and the fragment ion mass tolerance was the default assignment of 0. The data was searched using a human reverse-concatenated non-redundant (gene-centric) FASTA database that combines IPI and Ensembl identifiers. The resulting matched MS2 spectra were assembled into protein identifications, then filtered using DTASelect (version 2.0.47), and only half- or fully-tryptic peptides were accepted for identification, and only fully-tryptic peptides were considered for quantification. Peptides were restricted to a specified false positive rate of 1%. Redundant peptide identifications common between multiple proteins were allowed, but the database was restricted to a single consensus splice variant. SILAC ratios were quantified using in-house software as described (CIMAGE36). Briefly, extracted MS1 ion chromatograms (+/− 10 ppm) from both `light' and `heavy' target peptide masses (m/z) are generated using a retention time window (+/− 10 minutes) centered on the time when the peptide ion was selected for MS/MS fragmentation, and subsequently identified. Next, the ratio of the peak areas under the light and heavy signals (signal-to-noise ratio S/N >2.5) are calculated. Computational filters used to ensure that the correct peak-pair is used for quantification include a co-elution correlation score filter (R2 ≥ 0.8), removing target peptides with bad co-elution profile, and an “envelope correlation score” filter (R2 > 0.8) that eliminates target peptides whose predicted pattern of the isotopic envelope distribution does not match the experimentally observed high-resolution MS1 spectrum. Also, peptides detected as singletons, where only the heavy or light isotopically labeled peptide is detected and sequenced, but which pass all other filtering parameters, were given a standard ratio of 20, which is the maximum SILAC ratio reported herein. All reported SILAC experiments were run in duplicate, except the cholesterol competition, which was run in quadruplicate.
After automated processing, data sets were further filtered and analyzed manually based on more stringent and experiment-specific criteria. Only proteins which showed at least 2 unique identified and quantified peptides between four control runs (trans probe versus vehicle and versus `no UV', each in duplicate), and that showed at least ratio of 5.0 when the heavy, trans probe-enriched signal was compared to the background signal, were considered for further analysis by comparison to the PEA-DA labeling profile, and to cholesterol competition. Only proteins that showed at least a ratio of 3.0 when trans probe-labeled cells were compared to PEA-DA-labeled cells were considered `selective' and could be considered Group III or higher, and only proteins that showed at least a ratio of 1.5 when competed by 10× cholesterol, and showed at least 2 quantified unique peptides across four competition runs were considered `sensitive' for further analysis and consideration as Group II or higher. All experimental SILAC ratios presented are the mean of the median ratios of all quantified peptides for each protein from each replicate for each experiment.
Meta-analyses in Figure 4 of the sensitive and selective group of trans probe targets were performed using several online resources and servers. Automated gene ontology and pathway enrichment analyses were performed by uploading the Group I protein list to the DAVID bioinformatics website and performing enrichment analyses. Protein annotations in Figure 4 were obtained from primary literature sources, GeneCards.com, the Qiagen SABiosciences transcription factor database, and the OMIM database. Transmembrane domains were predicted, as necessary, by PSORT II and TMHMM prediction servers.
Microarray analysis of cholesterol-induced transcriptional changes
To verify that the observed cholesterol competition is most likely due to direct physical competition of probe binding, and not due to cholesterol-mediated expression changes for each sensitive target, we incubated HeLa cells with or without 20 μM cis probe and 100 μM cholesterol for 30 minutes and 12 hours, then harvested total cellular mRNA from each treatment using the Qiagen RNEasy kit, and submitted 8 μg total mRNA (combination of three biological replicates) per sample the TSRI DNA Array core facility, for quantitative transcriptomic analysis by microarray, using the HU133 Set GeneChip (Affymetrix), and the Affymetrix GeneChip Expression 3' Amplification One-Cycle protocol.
Recombinant expression and validation cholesterol competition of novel targets
To validate protein-cholesterol interactions identified by the trans sterol probe for which there is no previous evidence of cholesterol binding, SQLE, FDFT1, PGRMC1, POR, ADPGK, CYP20A1, and JAGN1 cDNAs were obtained (OpenBiosystems), and were recombinantly over-expressed as the full-length, unmodified proteins (except JAGN1, which was expressed with a C-terminal myc-his epitope-tag) in HeLa cells via transfection with Fugene HD according to the manufacturer's protocol. 48 hours post-transfection, cells were labeled in situ with either 5 or 10 uM trans probe, with or without 10× (50 μM or 100 μM) cholesterol. Both the competed and non-competed transfected samples were compared to a control (`Ctrl', Fig. 5) transfected with a distinct protein, on a 10% SDS-PAGE gel, and equal expression of the protein of interest between competed and non-competed samples was verified via western blot of the same samples on a gradient (4–20%) gel using commercial antibodies specific to each protein. Antibodies used in this study were: monoclonal mouse α-myc (1:10,000; invitrogen), monoclonal rabbit α-POR (1:1000; Sigma-Aldrich), monoclonal mouse α-FDFT1 (1:1000; Sigma-Aldrich), monoclonal mouse α-ADPGK (1:1000; Sigma-Aldrich), monoclonal rabbit α-PGRMC1 (1:2000; Sigma-Aldrich), monoclonal mouse α-SQLE (1:1000; Sigma-Aldrich), monoclonal mouse α-actin (1:2000; Sigma-Aldrich), and polyclonal rabbit α-CYP20A1 (1:100; Thermo).
Supplementary Material
Acknowledgments
We would like to thank Dr. Phil Baran for helpful advice on the synthesis of sterol probes. This work was supported by the NIH (CA132630) and the Skaggs Institute for Chemical Biology.
Footnotes
Author contributions. J.J.H., S.E.T., M.J.N., and B.F.C. designed the experiments; J.J.H., A.B.C., and S.E.T performed the experiments; J.J.H. and B.F.C. analyzed data; J.J.H. and B.F.C. wrote the manuscript.
Competing financial interests. The authors declare no competing financial interests.
References
- 1.Schroeder F, et al. Caveolin, sterol carrier protein-2, membrane cholesterol-rich microdomains and intracellular cholesterol trafficking. Subcell Biochem. 2010;51:279–318. doi: 10.1007/978-90-481-8622-8_10. [DOI] [PubMed] [Google Scholar]
- 2.Russell DW. Oxysterol biosynthetic enzymes. Biochim Biophys Acta. 2000;1529:126–35. doi: 10.1016/s1388-1981(00)00142-6. [DOI] [PubMed] [Google Scholar]
- 3.McLean KJ, Hans M, Munro AW. Cholesterol, an essential molecule: diverse roles involving cytochrome P450 enzymes. Biochem Soc Trans. 2012;40:587–93. doi: 10.1042/BST20120077. [DOI] [PubMed] [Google Scholar]
- 4.Russell DW. Fifty years of advances in bile acid synthesis and metabolism. J Lipid Res. 2009;50(Suppl):S120–5. doi: 10.1194/jlr.R800026-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Badimon L, Vilahur G. LDL-cholesterol versus HDL-cholesterol in the atherosclerotic plaque: inflammatory resolution versus thrombotic chaos. Ann N Y Acad Sci. 2012;1254:18–32. doi: 10.1111/j.1749-6632.2012.06480.x. [DOI] [PubMed] [Google Scholar]
- 6.Mirza R, et al. DHCR24 gene knockout mice demonstrate lethal dermopathy with differentiation and maturation defects in the epidermis. J Invest Dermatol. 2006;126:638–47. doi: 10.1038/sj.jid.5700111. [DOI] [PubMed] [Google Scholar]
- 7.Porter FD. Human malformation syndromes due to inborn errors of cholesterol synthesis. Curr Opin Pediatr. 2003;15:607–13. doi: 10.1097/00008480-200312000-00011. [DOI] [PubMed] [Google Scholar]
- 8.Herman GE. Disorders of cholesterol biosynthesis: prototypic metabolic malformation syndromes. Hum Mol Genet. 2003;12(Spec No 1):R75–88. doi: 10.1093/hmg/ddg072. [DOI] [PubMed] [Google Scholar]
- 9.Cianciola NL, Carlin CR, Kelley TJ. Molecular pathways for intracellular cholesterol accumulation: common pathogenic mechanisms in Niemann-Pick disease Type C and cystic fibrosis. Arch Biochem Biophys. 2011;515:54–63. doi: 10.1016/j.abb.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbaum AI, Maxfield FR. Niemann-Pick type C disease: molecular mechanisms and potential therapeutic approaches. J Neurochem. 2011;116:789–95. doi: 10.1111/j.1471-4159.2010.06976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinhofer I, Forss-Petter S, Kunze M, Zigman M, Berger J. X-linked adrenoleukodystrophy mice demonstrate abnormalities in cholesterol metabolism. FEBS Lett. 2005;579:5512–6. doi: 10.1016/j.febslet.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–40. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 13.Gill S, Chow R, Brown AJ. Sterol regulators of cholesterol homeostasis and beyond: the oxysterol hypothesis revisited and revised. Prog Lipid Res. 2008;47:391–404. doi: 10.1016/j.plipres.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Jeon TI, Osborne TF. SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol Metab. 2012;23:65–72. doi: 10.1016/j.tem.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heal WP, J.B., Bessin S, Wright MH, Magee AI, Tate EW. Bioorthogonal chemical tagging of protein cholesterylation in living cells. Chem Commun (Camb) 2011;47:4081–3. doi: 10.1039/c0cc04710d. [DOI] [PubMed] [Google Scholar]
- 16.Hoop CL, Sivanandam VN, Kodali R, Srnec MN, van der Wel PC. Structural characterization of the caveolin scaffolding domain in association with cholesterol-rich membranes. Biochemistry. 2012;51:90–9. doi: 10.1021/bi201356v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theesfeld CL, Pourmand D, Davis T, Garza RM, Hampton RY. The sterol-sensing domain (SSD) directly mediates signal-regulated endoplasmic reticulum-associated degradation (ERAD) of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase isozyme Hmg2. J Biol Chem. 2011;286:26298–307. doi: 10.1074/jbc.M111.244798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Motamed M, et al. Identification of luminal Loop 1 of Scap protein as the sterol sensor that maintains cholesterol homeostasis. J Biol Chem. 2011;286:18002–12. doi: 10.1074/jbc.M111.238311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thiele C, Hannah MJ, Fahrenholz F, Huttner WB. Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat Cell Biol. 2000;2:42–9. doi: 10.1038/71366. [DOI] [PubMed] [Google Scholar]
- 20.Charrin S, et al. A physical and functional link between cholesterol and tetraspanins. Eur J Immunol. 2003;33:2479–89. doi: 10.1002/eji.200323884. [DOI] [PubMed] [Google Scholar]
- 21.Rostovtsev VV, Green JG, Fokin VV, Sharpless KB. A stepwise Huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 22.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 23.Mintzer EA, Waarts BL, Wilschut J, Bittman R. Behavior of a photoactivatable analog of cholesterol, 6-photocholesterol, in model membranes. FEBS Lett. 2002;510:181–4. doi: 10.1016/s0014-5793(01)03271-9. [DOI] [PubMed] [Google Scholar]
- 24.Kwon HJ, et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell. 2009;137:1213–24. doi: 10.1016/j.cell.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts JD, Caserio M. Basic Principles of Organic Chemistry. Addison-Wesley; 1977. pp. 445–487. [Google Scholar]
- 26.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–30. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 27.Tong F, et al. Decreased expression of ARV1 results in cholesterol retention in the endoplasmic reticulum and abnormal bile acid metabolism. J Biol Chem. 2010;285:33632–41. doi: 10.1074/jbc.M110.165761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson AA, et al. GPCR stabilization using the bicelle-like architecture of mixed sterol-detergent micelles. Methods. 2011;55:310–7. doi: 10.1016/j.ymeth.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pastorino JG, Hoek JB. Regulation of hexokinase binding to VDAC. J Bioenerg Biomembr. 2008;40:171–82. doi: 10.1007/s10863-008-9148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christian AE, Haynes MP, Phillips MC, Rothblat GH. Use of cyclodextrins for manipulating cellular cholesterol content. J Lipid Res. 1997;38:2264–72. [PubMed] [Google Scholar]
- 33.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2012;9:84–9. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Washburn MP, Wolters D, Yates JR., 3rd. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 35.Eng J, McCormack AL, Yates JR. An approach to correlate MS/MS data to amino acid sequences in a protein database. J. Amer. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 36.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–5. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christian AE, Haynes MP, Phillips MC, Rothblat GH. Use of cyclodextrins for manipulating cellular cholesterol content. J Lipid Res. 1997;38:2264–72. [PubMed] [Google Scholar]
- 33.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Global profiling of dynamic protein palmitoylation. Nat Methods. 2012;9:84–9. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Washburn MP, Wolters D, Yates JR., 3rd. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 35.Eng J, McCormack AL, Yates JR. An approach to correlate MS/MS data to amino acid sequences in a protein database. J. Amer. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 36.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–5. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.