Abstract
Cationic polymers are commonly used to transfect mammalian cells, but their mechanisms of DNA delivery are unknown. This study seeks to decipher the mechanism by which plasmid DNA delivered by a class of cationic polymers traffics to and enters the nucleus. While studies have been performed to elucidate the mechanism of naked plasmid DNA (pDNA) import into the nuclei of mammalian cells, our objectives were to determine the effects of polymer complexation on pDNA nuclear import and the impact of polymer structure on that import. We have performed studies in whole cells and in isolated nuclei using flow cytometry and confocal microscopy to characterize how polymer-DNA complexes (polyplexes) are able to deliver their pDNA cargo to the nuclei of their target cells. The polymers tested herein include (i.) linear poly(ethylenimine) (JetPEI™), a polyamine, and (ii.) two poly(glycoamidoamine)s (PGAAs), polyamines that contain carbohydrate moieties (meso-galactarate, Glycofect™ (G4), and L-tartarate, T4) within their repeat units. Our results indicate that when complexed with the PGAAs, pDNA association with the nuclei was severely hampered in isolated nuclei compared to whole cells. When the pDNA was complexed with JetPEI™, there was slight inhibition of pDNA-nuclear interaction in isolated nuclei compared to whole cells. However, even in the case of PEI, the amount of pDNA imported into the nucleus increases in the presence of cytosolic extract, thus indicating that intracellular components also play a role in pDNA nuclear import for all polymers tested. Interestingly, PEI and G4 exhibit the highest reporter gene expression as well as inducing higher envelope permeability compared to T4, suggesting that the ability to directly permeabilize the nuclear envelope may play a role in increasing expression efficiency. In addition, both free T4 and G4 polymers are able to cross the nuclear membrane without their pDNA cargo in isolated nuclei, indicating the possibility of different modes of nuclear association for free polymers vs. polyplexes. These results yield insight to how the incorporation of carbohydrate moieties influences intracellular mechanisms and will prove useful in the rational design of safe and effective polymer-based gene delivery vehicles for clinical use.
Keywords: Gene delivery polymers, PEI, PGAAs, glycopolymer, nucleus, plasmid DNA, nuclear import
Introduction
Cationic polymer-based gene delivery systems are able to successfully compact plasmid DNA (pDNA) into polymer-pDNA complexes (polyplexes) and deliver their payload to cells, but they are less efficient than viruses and many exhibit high cytotoxicity.1–3 One well-studied and highly effective polycation vehicle is poly(ethylenimine) (PEI); this vehicle does exhibit toxicity, which has been a drawback for further clinical development.4–7 In response to the high cytotoxicity, several groups have developed cationic gene delivery polymers based on the structural characteristics of PEI8–12 to create a safe yet efficient gene delivery vehicle. For example, our group has created a library of poly(glycoamidoamine)s (PGAAs), which include PEI-like ethyleneamine moieties and a carbohydrate group within the polymer repeat unit to improve biocompatibility.13–16 These PGAA-based vehicles exhibit delivery efficacy, in most cases, comparable or better than PEI, but yield vastly higher cell viability profiles.13 Elucidating the mechanism of intracellular trafficking for these carbohydrate-containing polycation delivery vehicles is a long term goal, as understanding these mechanisms in detail will ultimately lead to a more rational design of safe and effective vehicles. Prior work in our group indicates that polyplexes formed with PGAAs enter the cell through different endocytic routes compared to polyplexes formed with PEI, illustrating the impact polymer structure has on plasmid DNA (pDNA) delivery.17 Once inside the cells, the intracellular trafficking mechanisms of PEI and the PGAAs vary greatly. Ultimately, the goal for polymer-based pDNA delivery vehicles is to transport their cargo to the nucleus. Herein, the effects of polymer structure on pDNA nuclear import are examined using the commercially-available vehicle JetPEI™ (Illkirch, France), poly(galactaramidopentaethylenetetramine) (G4, now being sold as Glycofect Transfection Reagent™) (Techulon, Blacksburg, VA), and poly(L-tartaramidopentaethylenetetramine) (T4).13
Previous research has shown that the sequence of the pDNA payload, particularly when it contains a human cytomegalovirus (CMV) early promoter sequence, can significantly impact its mechanism of nuclear entry.18 The CMV promoter has no known DNA nuclear targeting signal (DTS),19 and inclusion of a DTS in the sequence greatly enhances the transfection efficiency of CMV plasmids.19–21 For example, research on amphiphilic block copolymers by Yang et al. indicated that the structures activated an NF-κB-based pathway.22 That study also showed that pDNA transport to the nucleus was enhanced when there were NF-κB binding sites on the pDNA; interestingly, an increase in nuclear trafficking was not observed when the pDNA did not contain these binding sites.22 Similarly, Jeong et al. have studied poly(amido ethylenimine)-based vehicles for delivery of pDNA that incorporated NF-κB-binding motifs to enhance nuclear delivery.23 Their polymer vehicles are reducible once in the cytoplasm and showed enhanced pDNA delivery to the nucleus when compared to PEI, which did not degrade in the cytosol.23 However, plasmids containing a CMV promoter sequence complexed with polymers are also able to efficiently transfect cells and reach the nucleus, even without a DTS, indicating that polyplexes may go through different nuclear uptake pathways than naked plasmid DNA. It is unclear whether polymer-mediated pDNA delivery utilizes intracellular machinery such as classic nuclear localization signal (NLS) pathway, which employs the nuclear pore complexes (NPCs), or a different pathway, as is the case for glycosylated proteins that have been thought to utilize a sugar-dependent mechanism for nuclear import.24
Indeed, the structure of the polymer itself can greatly affect intracellular events during pDNA delivery. For example, prior studies have shown that pDNA complexed with PEI enhances nuclear trafficking compared to pDNA alone when microinjected into the cytosol.25 Based on these studies, the authors concluded that PEI’s ability to compact the pDNA into a nanoparticle size facilitated movement through the cytosol.25 Other studies have shown that PEI is capable of disrupting plasma membranes;7, 26 however, PEI-induced plasma membrane permeability did not lead to an increase in expression efficiency.27 Other studies have incorporated melittin into gene delivery peptides to improve their ability to permeabilize membranes, which in turn, increased pDNA delivery efficacy.28, 29 Our previous work has indicated that direct nuclear permeabilization is a possible route of nuclear import, and that the polymers that induced the highest nuclear envelope permeability also displayed the highest expression efficiency.30 Other studies have also shown that the addition of carbohydrate moieties to polymeric delivery vehicles can also affect nuclear trafficking and internalization when the carbohydrates are used as pendant groups on the polymer backbone.24, 31, 32 Further investigation into the role of the carbohydrates on these polymers indicate that they may rely on either association with nuclear shuttling proteins or an as yet unknown sugar-dependent mechanism.33
To investigate the effects of polymer structure on pDNA delivery, we studied how the presence or absence of a carbohydrate moiety in the repeated structure of the polymeric vehicle affects cytotoxicity and internalization of pDNA into the nucleus. To further determine if the presence of and type of carbohydrate moieties in polymeric delivery vehicles affect their mechanism(s) of cellular delivery, specifically, the import of pDNA into the nucleus, studies were performed using polyplexes formed with either JetPEI™ (PEI), Glycofect™ (G4), or T4 in cells and also isolated nuclei.
Materials and Methods
Materials
PEI (JetPEI™) and fluorescently labeled JetPEI-FluoF™ were purchased from Polyplus Transfection, Inc. (Illkirch, France). The polymer G4 (Glycofect™) was a generous gift from Techulon Inc. (Blacksburg, VA). The polymer T4 was synthesized as previously described13 and characterized using gel permeation chromatography (GPC) as previously described.30 The plasmid DNA used for the flow cytometry with labeled polymers and microscopy in whole cells was pCMV-luc from PlasmidFactory (Bielfield, Germany) and labeled with Cy5 using a LabelIT kit (Mirus, Madison, WI). The plasmid DNA used for experiments on isolated nuclei and for luciferase expression was gWiz-luc (Aldevron, Fargo, ND), also labeled with Cy5 with a LabelIT kit for flow cytometry and confocal microscopy studies. Both plasmids have similar sequences and contain a CMV promoter, and neither plasmid has a currently known DTS. Dithiothreitol (DTT), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), ethylene glycol tetraacetic acid (EGTA), potassium acetate, sucrose, magnesium acetate, protease inhibitor cocktail, digitonin, ATP, GTP, creatine phosphokinase, and creatine phosphate were from Sigma (St. Louis, MO). Coverslips used for all confocal microscopy experiments were 12 mm diameter No. 1 coverslips (Fisher Scientific, Pittsburgh, PA).
Cell culture
Cell culture media and supplements were purchased from Gibco (Carlsbad, CA). Human cervical adenocarcinoma (HeLa) cells were purchased from ATCC (Rockville, MD) and cultured according to the manufacturer’s specified conditions. Cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10 % fetal bovine serum (FBS), 100 µg/mL streptomycin, 0.25 µg/mL amphotericin, and 100 units/mL penicillin. Transfections were performed in serum-free medium (OptiMEM).
Expression efficiency
Luciferase activity was measured as previously described.13 Briefly, polyplexes were formed by adding 150 µL of polymer solution diluted to the appropriate N/P ratio to 150 µL of a 0.02 mg/mL solution of gWiz-Luc pDNA (Aldevron, Fargo, ND). Polyplexes were incubated for 1 hour at room temperature prior to transfection. Twenty-four hours after transfection, luminescence was measured using luciferase substrate (Promega, Madison, WI) on a plate reader (GENios Pro, Tecan, Research Triangle Park, NC). Cells were transfected in triplicate and the average luminescence was used to calculate transfection efficiency based on luciferase activity. Where indicated, cells were transfected in the presence of 0.006 mg/mL (20 µM) of nocodazole for the first 4 hours of transfection to disrupt microtubules or 10 µg/mL (30 µM) of aphidicolin for 24 hours during transfection to halt the cell cycle. Expression efficiency in the presence/absence of nocodazole and the presence/absence of aphidicolin was measured 24 hours after transfection.
Labeling polymers
In order to label T4 and G4, the polymers were dissolved in methanol at a concentration of 1 mM for T4 and 0.5 mM for G4. FITC-isothiocyanate (Sigma, St. Louis, MO) was dissolved in methanol and added dropwise to the polymer solution while stirring to reach a final molar ratio of 3:1 FITC: polymer. The polymer and FITC were reacted for 48 hours in the dark at room temperature with continuous stirring. Excess dye was separated from the polymer using dialysis with a 1 kDa MWCO membrane.
Isolation of HeLa nuclei for flow cytometry
HeLa cells were pelleted by centrifugation at 200 × g at 4 °C for 10 minutes and resuspended in 2 mLs of ice-cold phosphate-buffered saline (PBS) containing 2 mM DTT, 1 µg/mL protease inhibitor cocktail and 40 µg/mL digitonin. The cells were incubated on ice for 5 minutes as described previously34 to permeabilize the cells. Nuclei from the permeabilized cells were then pelleted at 2,000 × g at 4 °C for 10 minutes and resuspended in 2 mLs of ice-cold PBS containing 2 mM DTT and 1 µg/mL protease inhibitor cocktail and incubated on ice for 5 minutes. Nuclei were pelleted as described above and resuspended in 500 µL of PBS prior to flow cytometry analysis. Flow cytometry was performed on a BD FACSCanto II flow cytometer (BD Biosciences, San Jose, CA), and a minimum of 50,000 events per sample was collected.
Nuclear association in transfected cells
In order to determine nuclear association, cells were plated in 6 well plates at a density of 250,000 cells/well and incubated at 37 °C and 5 % CO2 for 24 hours. Polyplexes were formed by adding 250 µL of FITC-labeled polymer diluted to the appropriate N/P ratio to 250 µL of a 0.02 mg/mL Cy5-labeled pDNA solution. The 500 µL of polyplex solution was allowed to incubate for 1 hour at room temperature before being diluted with 1 mL of reduced-serum medium (OptiMEM) and added to cells. Cells were washed with 1 mL of PBS/well before being transfected, and cells were transfected with 5 µg pDNA/well. Four hours after transfection, the polyplex-containing OptiMEM was aspirated off, cells were washed with 1 mL of PBS/well, and 5 mLs of supplemented DMEM was added to each well. Twenty-four hours after transfection, nuclei were isolated using the procedure described in the previous section. Nuclei were pelleted and resuspended in 500 µL PBS before being analyzed for FITC and Cy5 fluorescence using flow cytometry. A minimum of 50,000 events was collected for each sample. To further study the nuclear interactions of the polyplexes, nuclei from the flow cytometry experiment were pelleted by centrifugation at 2,000 × g, 4 °C for 10 minutes and resuspended 1 mL of PBS containing 100 µg/mL of 2 MDa rhodamine-labeled dextran (Molecular Probes, Eugene, OR). The nuclei were incubated in the dextran-containing PBS for 15 minutes, and then pelleted by centrifugation at 2,000 × g in the conditions described above and resuspended in 1 mL of 4 % paraformaldehyde (PFA) solution. The nuclei were fixed in the PFA solution for 15 minutes at room temperature. After that, nuclei were pelleted, washed in PBS and deposited on poly-L-lysine (PLL)-coated coverslips. The nuclei incubated on the coverslips overnight at 4 °C before being mounted onto microscopy slides and analyzed via confocal microscopy. The degree of pixel overlap between two fluorescence channels was calculated using Manders coefficients to determine colocalization.35, 36 Manders coefficients were calculated using the software ImageJ.37 Manders coefficients were calculated after removing background fluorescence; fluorescence above the thresholds was taken into account for Manders coefficients. In addition, Manders coefficients were calculated for a region of interest (ROI) surrounding the nuclei so that fluorescence outside of the nuclei did not influence Manders coefficients.
Isolation of soluble HeLa cytosolic proteins
Nuclei were isolated from HeLa cells as described in the previous section. After pelleting the nuclei, the supernatant was collected. Ice-cold acetone (4 × supernatant volume) was added to the supernatant, and the mixture stirred on ice for 1 hour. The mixture was distributed into 1.5 mL Eppendorf tubes, and the proteins were pelleted by centrifuging at 14,000 × g for 10 minutes. The supernatant was discarded and the pellets were resuspended in sterile import buffer (20 mM HEPES, 0.25 M sucrose, 100 mM potassium acetate, 2 mM magnesium acetate, and 1 mM EGTA), and protein concentrations were quantified using a Bio-Rad DC Protein Assay (Hercules, CA) according to manufacturer’s protocol.
Nuclear association in a cell-free system
In order to design a high-throughput method to determine the minimal requirements for pDNA-nuclear association, nuclei were isolated from HeLa cells via digitonin permeabilization and centrifugation as described above and deposited into 6 well plates at a concentration of 250,000 nuclei/well. The nuclei were incubated in a buffer solution consisting of 20 mM HEPES, 0.25 M sucrose, 100 mM potassium acetate, 2 mM magnesium acetate, and 1 mM EGTA as previously described by Munkonge et. al.34 The buffer solution was added with or without 50 µg HeLa cytosolic protein and with or without the ATP-generating system. Nuclei were also incubated in the presence or absence of HeLa cytosol extract and the presence or absence of an ATP-generating system (0.5 mM ATP, 0.2 mM GTP, 5 mM creatine phosphate, and 1 Unit creatine phosphokinase) as previously described by Hagstrom et. al.38 Polyplexes were formed as described above and added to the nuclei. After incubating for 4 hours at 37 °C and 5 % CO2, nuclei were pelleted, resuspended in 1 mL PBS to wash away unbound polyplexes/polymers, pelleted again, and resuspended in 500 µL of PBS before being analyzed by flow cytometry. In order to study mechanisms of pDNA nuclear import other than direct nuclear envelope permeabilization, nuclei with higher propidium iodide fluorescence intensity than the cells only and pDNA only controls were not included in these measurements. This was done to minimize the effect on Cy5 fluorescence for pDNA import through increased nuclear envelope permeability.
Confocal microscopy in a cell-free system
HeLa cells were seeded onto uncoated coverslips at a density of 50,000 cells/coverslip and allowed to incubate at 37 °C and 5 % CO2 for 24 hours. After incubation, cells were ~70 % confluent. The media was aspirated off the cells, and then the cells were washed with 1 mL of PBS/well, and 1 mL of ice-cold PBS containing 2 mM DTT, 1 µg/mL protease inhibitor cocktail and 40 µg/mL digitonin. After 5 minutes, this solution was aspirated off and 1 mL of ice-cold PBS containing 2 mM DTT and 1 µg/mL protease inhibitor cocktail was added to each well. The cells incubated in this solution for 5 minutes, after which this solution was aspirated off, the permeabilized cells were washed with PBS, and 1 mL of the aforementioned buffer solutions (20 mM HEPES, 0.25 M sucrose, 100 mM potassium acetate, 2 mM magnesium acetate, and 1 mM EGTA with/without 50 µg HeLa cytosolic protein and with/without an ATP-generating system) was added to each well. Polyplexes were formed by adding 50 µL of FITC-labeled polymer solution diluted to the appropriate N/P ratio to 50 µL of 0.02 mg/mL Cy5-labeled pDNA solution and allowed to incubate at room temperature for 1 hour. After one hour, the 100 µL of polyplex solution was added to the digitonin-permeabilized cells (1 µg pDNA/well). After 4 hours, the polyplex solution was aspirated off, the permeabilized cells were washed with 1 mL of PBS/well, and permeabilized cells were fixed in 4 % paraformaldehyde for 15 minutes at room temperature. Coverslips were mounted onto microscopy slides using ProLong Gold Antifade Reagent (Molecular Probes, Eugene, OR) according to manufacturer’s protocol. The slides were allowed to dry overnight in the dark before being sealed with clear nail polish and imaged on a Zeiss LSM 510 META confocal microscope (Carl Zeiss MicroImaging, LLC, Thornwood, NY). Images were taken using a slice thickness of 0.8 µm. Manders coefficients were used to calculate the amount of overlap between two channels to monitor colocalization.35, 36 Manders coefficients were calculated using the software ImageJ37 as described above.
Confocal microscopy in whole cells
HeLa cells were seeded onto poly-L-lysine (PLL)-coated coverslips at a density of 15,000 cells/coverslip and allowed to incubate under the aforementioned conditions for 48 hours to reach ~ 70 % confluency. Cells were then transfected using JetPEI-FluoF™ at an N/P ratio of 5 according to manufacturer’s protocol and FITC-labeled T4 and FITC-labeled G4 were used at an N/P of 20. A 0.02 mg/mL Cy5-labeled pDNA solution was used to make the polyplexes, and cells were transfected with a total of 1 µg of pDNA per coverslip. Transfection was carried out in serum-free media (OptiMEM), with 1 mL of OptiMEM/well. At the indicated time points, the cells were washed with PBS and fixed using 4 % paraformaldehyde. Cells were immunostained using a 1:200 dilution of anti-fibrillarin rabbit monoclonal antibody (Cell Signaling Technologies, Inc.) and Alexa Fluor 555 goat anti-rabbit secondary antibody (Molecular Probes, Eugene, OR) as previously described.17
Nuclear envelope permeability in a cell-free system
HeLa cells were grown to confluency and then trypsinized and pelleted at 200 × g, 4 °C for 10 minutes. Nuclei were isolated from HeLa cells as described above and added to a 12 well plate at a density of 50,000 nuclei/well. Polyplexes were formed as described above (50 µL of polymer solution was added to 50 µL of 0.02 mg/mL plasmid DNA solution) and added to the nuclei in 1 mL of OptiMEM. The nuclei were incubated at 37 °C and 5 % CO2 for 4 hours, after which 200 µL of a 0.4 % trypan blue solution (Gibco, Carlsbad, CA) was added to each well. The nuclei were then imaged using an EVOS fl digital inverted fluorescence microscope (Advanced Microscopy Group, Bothell, WA). The mean pixel intensity for individual nuclei was calculated using the software ImageJ.37 Only pixels inside of the nuclei were measured; the background was removed from the images using the “Threshold” tool to create a binary image with the regions of interest (ROIs) as white and background as black (in this case, ROIs are the nuclei). Next, the “set measurements” tool was used to redirect the binary image to the original for analysis. Then, the “Analyze Particles” tool was used to measure pixel intensity within each ROI (nucleus). Debris were excluded from these measurements by adjusting the circularity and size of the particles with the “Analyze Particles” tool. Measurements are reported as mean pixel intensity.
Statistical analysis
Dot plots are representative of a minimum of 50,000 events from flow cytometry experiments. Data in bar graphs are presented as means ± standard deviations. For statistical analysis of data, JMP software was used (SAS Institute Inc., Cary, NC) and means were compared using ANOVA followed by the Tukey-Kramer HSD method, with p < 0.05 being considered as statistically significant. Detailed statistical analyses can be found in the supporting information.
Results
Nuclear association
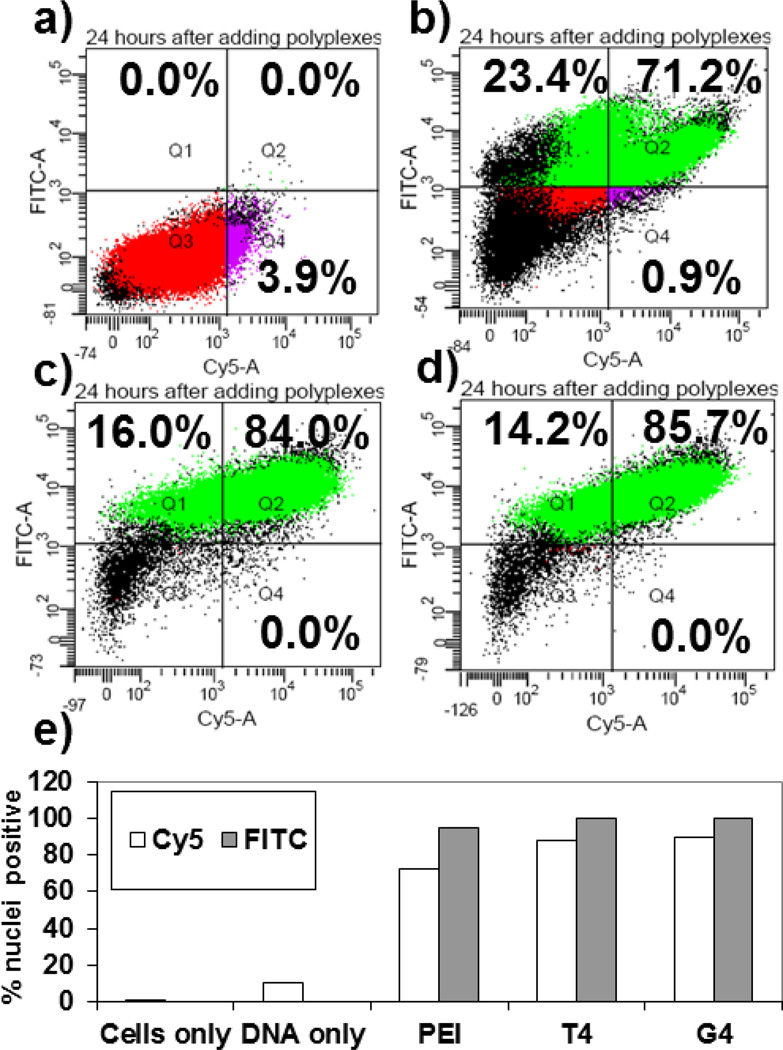
Polyplexes were prepared with a fluorescently- (FITC-) labeled preparation of each of the three polycations; the commercially-available JetPEI™ and Glycofect™ (G4), and T4 synthesized in our lab (polymer characterization results are shown in Table S1 in the supporting information). These labeled polymers were added to fluorescently-labeled (Cy5-) pDNA. To determine whether pDNA and/or polymers associated with nuclei, HeLa cells were transfected and their nuclei were isolated 24 hours after transfection. The nuclei were analyzed via flow cytometry, and dot plots of Cy5 and FITC fluorescence intensities are presented in Figure 1. A graphical representation of the dot plot data illustrating the percentage of nuclei positive for Cy5 and FITC is shown in Figure 1e. Histograms for the flow cytometry data are shown in Figure S1 in the supporting information.
Figure 1.
Flow cytometry analysis of isolated nuclei 24 hours after whole cell transfection. HeLa cell nuclei were analyzed for Cy5 (x-axis) and FITC (y-axis) fluorescence. Cells were transfected with polyplexes formed with (a) pDNA only; (b) JetPEI™ at an N/P ratio of 5; (c) T4; (d) Glycofect™ (G4). T4 and G4 were used at N/P ratios of 20. Colors represent the following: red = whole nuclei; purple = Cy5 positive nuclei; green = FITC positive nuclei; black = debris. Note: only whole nuclei were analyzed for FITC and Cy5 fluorescence, debris was gated out. All polyplex-treated nuclei exhibited increased pDNA association and polymer association as indicated by increased Cy5 and FITC fluorescence, respectively. (e) Bar graph representation of the flow cytometry data comparing the % nuclei positive for Cy5 and FITC fluorescence.
The majority of nuclei isolated from cells transfected with JetPEI™ (Figure 1b), T4 (Figure 1c), and Glycofect™ (G4) (Figure 1d) displayed high Cy5-pDNA-nucleus association compared to the pDNA only control (3.9 %) as indicated by the percent of nuclei positive for Cy5 fluorescence (> 70 % of polyplex-treated nuclei exhibited Cy5 fluorescence vs. 3.9 % for pDNA control, Figure 1a). In addition, > 90 % of nuclei isolated from polyplex-treated cells exhibited FITC fluorescence, indicating polymer-nucleus association. It should also be noted that a percentage of nuclei from polyplex-transfected cells displayed FITC fluorescence, but not Cy5 fluorescence, indicating that free polymers present in the polyplex solutions are able to traffic to the nucleus without their pDNA cargo. Based on these data, we are unable to observe whether the Cy5-pDNA/polymers were bound to the surface of the nuclei or whether the fluorescent signals were coming from inside of the nuclei, so we arbitrarily call these signals “nucleus association”, which has prompted further studies of nuclear internalization (vide infra). Increased Cy5 fluorescence indicates increased pDNA-nucleus association, while increased FITC fluorescence indicates increased polymer-nucleus association.
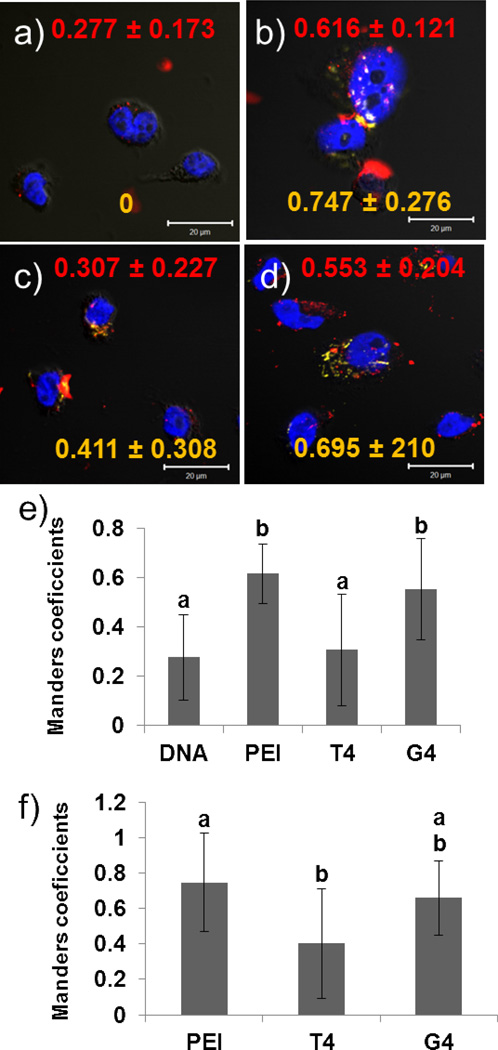
To determine direct nuclear envelope permeability as well as whether the Cy5 signal was coming from inside of the nuclei vs. the surface of the nuclei, cells were transfected with polyplexes formed with Cy5-labeled pDNA, and the nuclei were isolated 24 hours after transfection. Nuclei were incubated with 2 MDa rhodamine-labeled dextran and stained with DAPI before being mounted onto slides and imaged using confocal microscopy. Rhodamine (from the dextran) and DAPI (nuclear stain) colocalization were analyzed by calculating Manders coefficients35, 36 with the software ImageJ.37 Manders coefficients range from 0 to 1, with 0 being no overlap between the two channels (in this case, Cy5 or rhodamine and DAPI) and 1 corresponding 100 % overlap between Cy5/rhodamine and DAPI. In other words, for this study, if the Manders coefficient between Cy5 and DAPI or rhodamine and DAPI is 1, then all Cy5 or rhodamine pixels overlap with DAPI pixels. Cy5 (from the pDNA) and DAPI (from the nuclei) colocalization measurements were used to determine pDNA nuclear import. The confocal images and colocalization data are shown in Figure 2.
Figure 2.
Nuclei isolated from HeLa cells 24 hours after whole cell transfection. Cells were transfected with (a) pDNA only; (b) JetPEI™ (PEI) at an N/P ratio of 5; (c) T4 at an N/P ratio of 20, or (d) Glycofect™ (G4) at an N/P of 20. Colors represent the following: yellow = Cy5-labeled pDNA; blue = DAPI-labeled nucleus; red = rhodamine-labeled dextran. Scale bar = 20 µm. Red numbers in the upper portion of the images represent the colocalization data as determined by Manders coefficients between rhodamine-labeled dextran and DAPI-labeled nuclei ± standard deviation. Yellow numbers in the lower portion represent Manders coefficients between Cy5-labeled pDNA and DAPI-labeled nuclei ± standard deviation. (e) Bar graph representation of Manders coefficients between rhodamine-labeled dextran and DAPI-labeled nuclei. (f) Bar graph representation of Manders coefficients between Cy5-labeled pDNA and DAPI-labeled nuclei. Bars with different letters represent means that are statistically different from each other according to the Tukey-Kramer HSD method (p < 0.05, n = 10). Detailed statistical analysis can be found in the supporting information (Tables S2 and S3).
These results indicate that nuclei treated with JetPEI™-Cy5-pDNA polyplexes have the most Cy5-pDNA-nuclei internalization (Manders coefficient of Cy5-DAPI overlap = 0.747 ± 0.276). This was to be expected when taking into account the greater charge density of PEI, membrane permeation ability,26, 39 and generally high gene delivery and expression.15 Interestingly, similar nuclear envelope permeability was found for both JetPEI™ and G4 as indicated by dextran-nuclei association (Manders coefficients of 0.616 ± 0.121 and 0.553 ± 0.204, respectively); G4 also exhibited similar albeit slightly lower pDNA-nuclear association (Manders coefficient of Cy5 and DAPI overlap = 0.695 ± 0.210) when compared to JetPEI™ (for detailed statistical analyses, see Table S2 for rhodamine and DAPI Manders coefficients and Table S3 for Cy5 and DAPI Manders coefficients in the supporting information). However, T4, in addition to exhibiting the lowest gene expression efficiency (Figure 5), also induced the least nuclear envelope permeability; this finding is consistent with prior studies performed by our group linking high expression efficiency with the ability to permeabilize the nuclear envelope.30 Furthermore, even though the glycopolymer-based polyplexes formed with G4 and T4 were able to deliver an appreciable amount of Cy5-pDNA into the nucleus, the results are still slightly lower than JetPEI™, which could be related to the timing/kinetics of delivery and gene expression. After fixation, we were unable to see any FITC signal from commercially-labeled JetPEI-FluoF™ due to incompatibility with the dye and cell fixation, but were able to see FITC signal from T4 and G4 (Figure S2, Supporting Information). It is interesting to note that in these nuclei isolated from transfected cells, there are signs of nuclear envelope ruffling at sites of polyplex interaction, as denoted by white arrows in Figure S2. Furthermore, although all polymers were able to deliver the pDNA to the nucleus as indicated by the Cy5 signal in Figure 1, the confocal microscopy studies determined that JetPEI™ had the most pDNA nuclear uptake (Figure 2). There is also evidence of rhodamine and Cy5 fluorescence outside of the nuclei in Figure 2. Careful examination of the differential interference contrast (DIC) image reveal that there is still some cytoplasm/cellular components surrounding the nuclei even after isolation, and that the rhodamine and Cy5 particles appear to be “stuck” in this environment and could contribute to the fluorescence signal in the flow cytometry data, so Manders coefficients were used to determine the degree of fluorescence coming from inside of the nuclei. Rhodamine and Cy5 signals not associated with the nuclei or perinuclear region were not included in the Manders coefficient calculations.
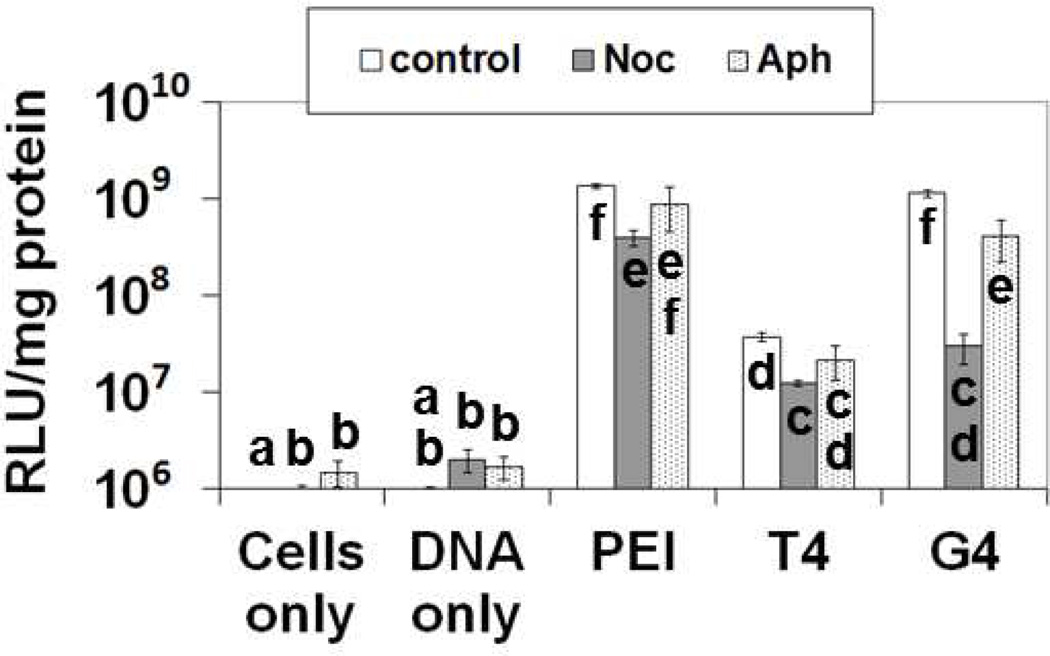
Figure 5.
Luciferase expression 24 hours after transfection as measured by relative light units (RLU) per milligram of protein in cell lysates. Cells were transfected with the indicated polymers in the presence or absence of nocodazole (Noc) or aphidicolin (Aph) to determine if microtubule disruption or cell cycle inhibition affected transfection efficiency. Control cells were treated with polyplexes in the absence of Noc and Aph. Bars with different letters represent means that are statistically different from each other according to the Tukey-Kramer HSD method (p < 0.05, n = 3).
Nuclear envelope permeability in a cell-free system
Based on the higher degree of dextran accumulation in nuclei isolated from JetPEI™ and G4-treated cells than from T4-treated cells, it was of interest to us to see whether the polyplexes themselves were responsible for the increased nuclear envelope permeability or if the increased nuclear envelope permeability was a result of apoptosis initiation. To test this, a cell-free system was used. Nuclei were first isolated from HeLa cells, treated with unlabeled polyplexes formed with the different polymers for 4 hours, and then a trypan blue assay was used to determine nuclear envelope permeability. Trypan blue is a small molecule able to diffuse through the nuclear pore complex. We hypothesized that if the nuclear envelope increased in permeability, more trypan blue would diffuse into the nucleus and, therefore, the nuclei with compromised nuclear envelopes would appear darker (Figure S3). We did in fact observe lower pixel intensities (indicated by darker color caused by increased trypan blue uptake) in nuclei treated with JetPEI™ polyplexes compared to nuclei treated with pDNA only. Mean pixel intensity was highest with T4 (indicating the least nuclear membrane permeability) and then followed by pDNA > G4 > JetPEI™ (Figure 3), which correlate well with the data in Figure 2. Detailed statistical analysis can be found in the supporting information (Table S4).
Figure 3.
Mean pixel intensities of isolated nuclei treated with the indicated polyplexes. Darker (lower) intensities indicate more trypan blue present in the nuclei, indicating increased nuclear envelope permeability. All means are statistically different from each other (p < 0.05, n ~ 300) according to the Tukey-Kramer HSD method. Pixel intensities are highest with nuclei treated with T4, and then pixel intensities decrease in the order pDNA (DNA) > Glycofect (G4) > JetPEI™ (PEI), indicating more trypan blue staining of the nuclei. JetPEI™ was used at an N/P of 5, and T4 and Glycofect™ (G4) were used at N/P ratios of 20.
Intracellular location of polyplexes
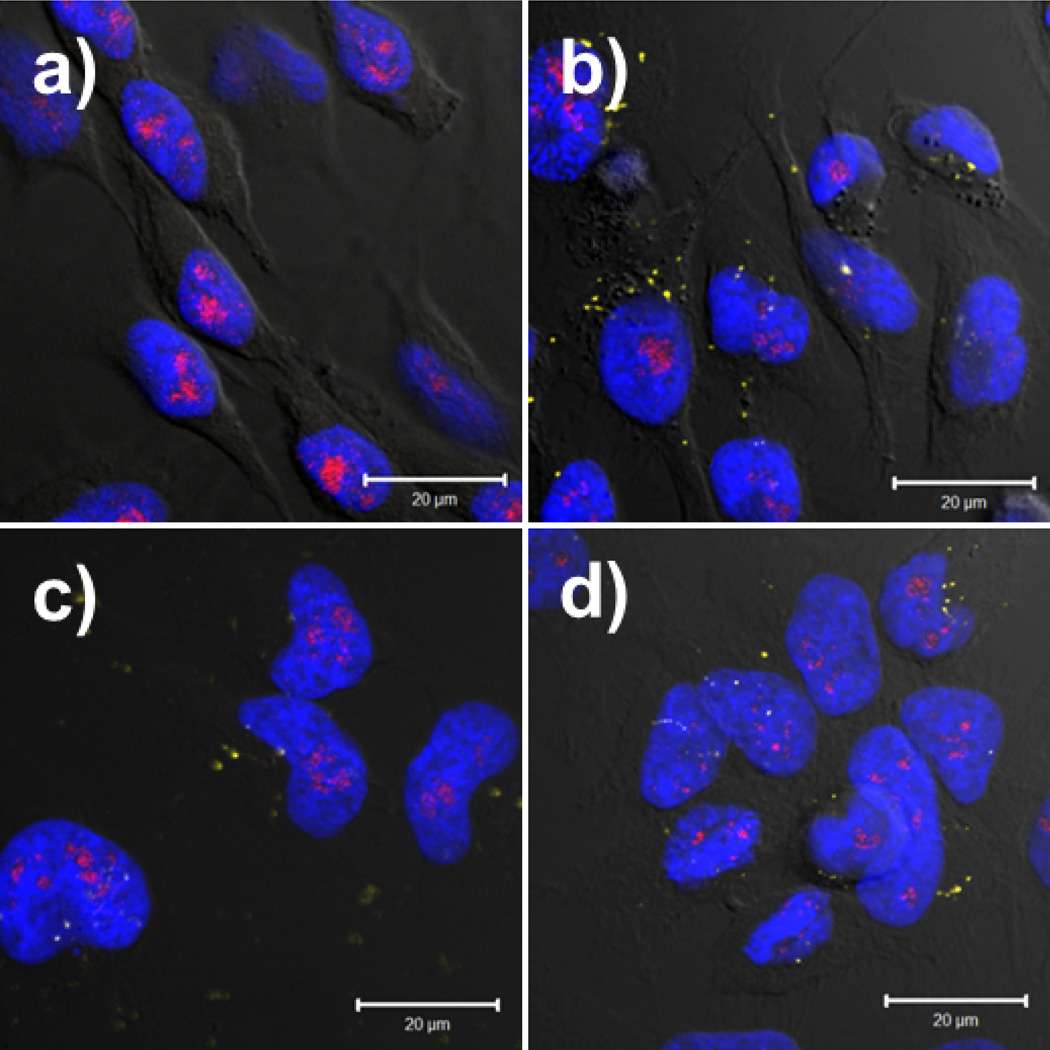
To further study polyplex location during transfection, HeLa cells were transfected with polyplexes formed with Cy5-labeled pDNA and unlabeled polymer and fixed 24 hours after transfection (Figure 4). Our results show that polyplexes formed with each of the polymers were able to deliver Cy5-pDNA to the nucleus by this time point. It is interesting to note that pDNA is able to reach the nucleus when complexed with all the polymers studied herein, as seen in Figure 4 and the flow cytometry results in Figure 1. However, we see marked differences in expression efficiency between these 3 polymers (Figure 5, JetPEI™ and G4 are similar but T4 is much lower). This may indicate that trafficking to the nucleus is not a limiting factor in efficient pDNA delivery; polyplexes also need an effective mechanism for the pDNA to physically enter the nuclei to be successful therapeutic agents. It is interesting to note that in digitonin-permeabilized cells, T4 and G4 seem to preferentially locate in the nucleoli (Figures S4 and S5 in the supporting information). In order to determine whether polyplexes in whole cells would colocalize with nucleoli, nucleoli were immunolabeled and imaged using confocal microscopy. Little colocalization was observed between the pDNA and nucleoli in whole cells at this time point (Figure 4).
Figure 4.
HeLa cells transfected for 24 hours with (a) pDNA only; and the following polymers: (b) JetPEI™ (PEI) at an N/P ratio of 5; (c) T4; or (d) Glycofect™ (G4). T4 and G4 were used at N/P ratios of 20. Polyplexes were formed with Cy5-labeled pDNA. Colors represent the following: yellow = pDNA; blue = nucleus; red = nucleoli. Scale bar = 20 µm. We observed little colocalization between the polyplexes (magenta) and the nucleoli (red) at this time point for all polymers tested.
Inhibition of polyplex intracellular trafficking
Prior studies indicate that microtubules play a role in polyplex trafficking to some extent, and that perhaps nuclear envelope breakdown during cell division enables polyplexes to enter the nuclei, thus also relating cell cycle to nuclear delivery.40 To test this, cells were treated for 4 hours with a microtubule-disrupting agent, nocodazole,41 or treated for 24 hours with a cell cycle inhibitor, aphidicolin.42 The results indicate that, for all polyplex types, disruption of the microtubules for the first 4 hours of transfection leads to statistically significant (p < 0.05) lower luciferase transgene expression (Figure 5). In particular, G4 polyplex trafficking appears to be the most affected by microtubule disruption. It is interesting to note that inhibition of the cell cycle slightly inhibits (p < 0.05) gene expression for G4, but not for T4 or JetPEI™ (Figure 5). The differences in gene expression for G4 and T4 in the presence of aphidicolin indicate how small changes in polymer structure impact intracellular mechanisms and that G4 appears to be more dependent on cell cycle than T4 and JetPEI™. These data support our hypothesis that both cell cycle and microtubules play a role in polyplex delivery to the nucleus, which are all appear to be impacted by polymer structure.
Nuclear association in isolated nuclei
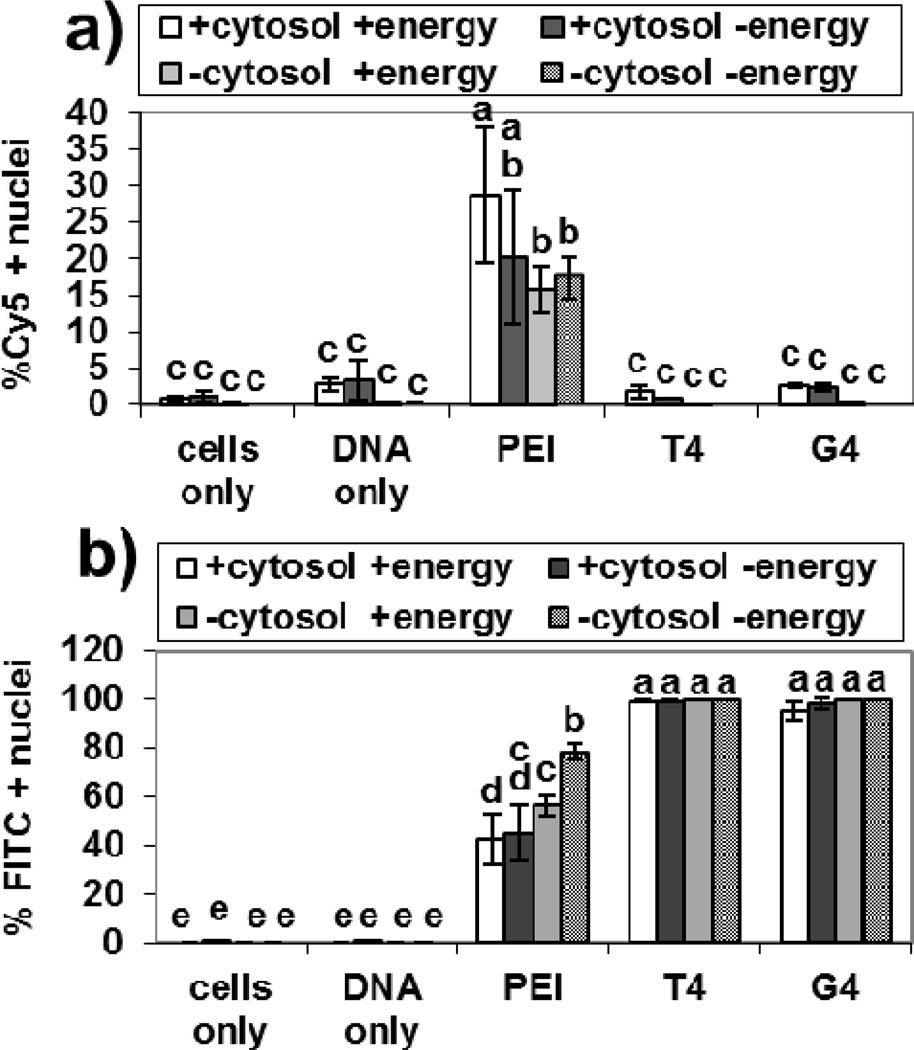
To determine whether nuclear association of the polyplexes could occur without cytosolic components, nuclei were first isolated from HeLa cells, and then treated with polyplexes formed with Cy5-labeled pDNA and JetPEI-FluoF™ (PEI) (N/P = 5), FITC-labeled-T4 (N/P = 20), or FITC-labeled-G4 (N/P = 20). The nuclei and polyplexes were incubated with or without cytosol extract and with or without ATP (“energy”). After 4 hours, the nuclei were analyzed for Cy5 and FITC fluorescence using flow cytometry. The results are illustrated in Figure 6. From these data, it seems that the PGAAs are more dependent on intracellular components than JetPEI™ for pDNA-nucleus association under all four conditions tested. Interestingly, under these conditions, nuclei incubated with T4 and G4 polyplexes exhibited Cy5-pDNA fluorescence (nuclear association of pDNA) that was not different from the cells only and pDNA only controls (Figure 6a). However, pDNA complexed with JetPEI™ was able to associate with the nuclei in this cell-free system under all conditions to a significantly higher (p < 0.05) extent than the other polyplexes (Figure 6a). For polyplexes formed with JetPEI™, the presence of ATP did not affect pDNA-nucleus association either in the presence or absence of cytosol. However, it is interesting to note that, for JetPEI™ polyplexes, the presence of both cytosol and ATP resulted in significantly higher (p < 0.05) pDNA-nucleus association when compared to JetPEI™ polyplexes incubated in the absence of cytosol (Figure 6a). Furthermore, we observed that nearly 100 % of the nuclei were positive for FITC-fluorescence when treated with polyplexes formed with FITC-G4 and FITC-T4 regardless of incubation conditions, indicating differences in pDNA nuclear association vs. polymer nuclear association (Figure 6b). We also see significantly higher (p < 0.05) polymer-nucleus association in the FITC-PGAAs compared to FITC-JetPEI™ (Figure 6b), so it would appear that the carbohydrate moieties in the PGAA repeat units could influence polymer-nucleus interactions. However, the increased polymer-nucleus association observed with FITC-T4 and FITC-G4 vs. FITC-JetPEI™ may also be due to the higher N/P ratios used for these two polymers; more free polymer in the polyplex solution may result in increased polymer-nucleus association. In addition, the different FITC signals observed could be due to different FITC-labeling between the polymers; T4 and G4 were labeled at a 3:1 FITC:polymer ratio, while FITC-labeled JetPEI™ (JetPEI-FluoF™) was purchased commercially and the FITC:polymer ratio is proprietary.
Figure 6.
Flow cytometry data from nuclei treated with polyplexes formed with different FITC-labeled polymers and Cy5-labeled plasmid DNA. Nuclei were isolated with polyplexes with or without HeLa cytosol extract (“cytosol”) and in the absence or presence of an ATP-generating system (“energy”). Nuclei were analyzed for (a) Cy5 fluorescence from the pDNA and (b) FITC fluorescence from the polymers 4 hours after polyplex treatment. Bars with different letters represent means that are statistically different from each other according to the Tukey-Kramer HSD method (p < 0.05, n = 3). Bars with matching letters represent means that are not statistically significant from each other. Polyplexes were formed at an N/P ratio of 5 for JetPEI™ (PEI) and N/P ratios of 20 for T4 and Glycofect™ (G4).
Nuclear import in digitonin-permeabilized HeLa cells
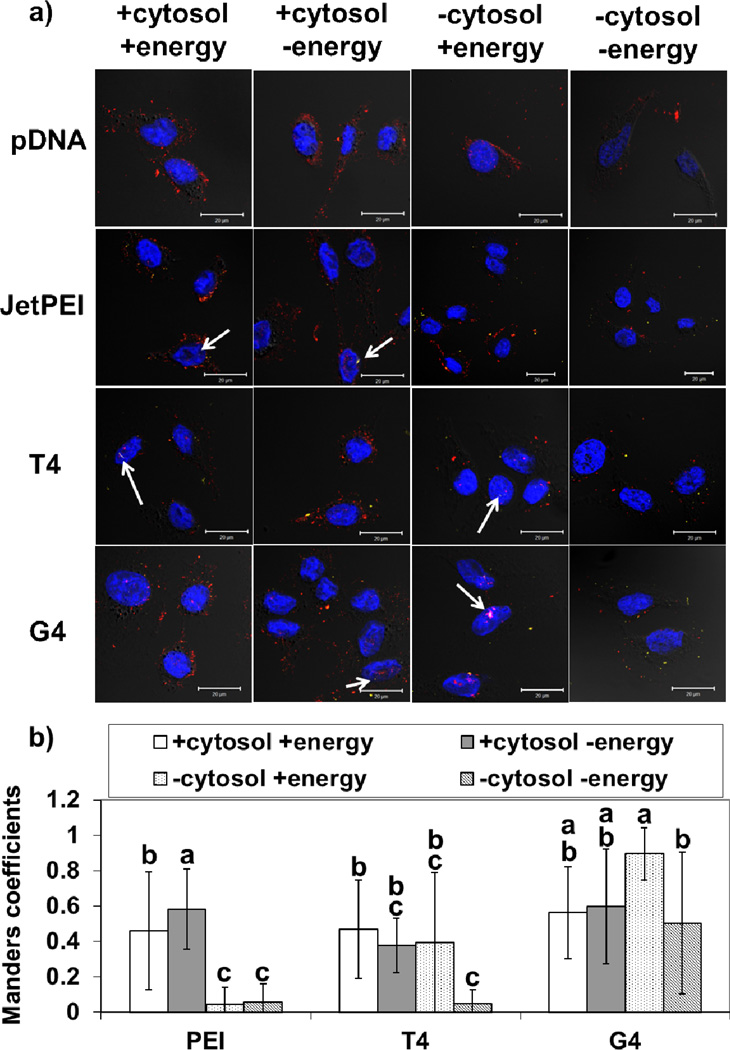
To further determine whether polymer-mediated pDNA import into nuclei required cytoplasmic factors, HeLa cells were grown on coverslips to confluency, then treated with DTT and digitonin to permeabilize the cytosol. The nuclei were isolated with or without ATP to determine whether plasmid DNA import in the absence of cytosol was energy-dependent. The nuclei were treated with polyplexes formed with Cy5-labeld pDNA and FITC-labeled polymers, incubated for 4 hours before being washed, fixed, and mounted onto microscopy slides. Confocal microscopy was used to determine colocalization between plasmid DNA and the nucleus. Colocalization (as determined using Manders coefficients) between 2 MDa rhodamine-labeled dextran and DAPI-labeled nuclei was used to assess nuclear membrane integrity. The images and colocalization data for digitonin-treated cells treated with the different polyplexes are shown in Figure 7.
Figure 7.
(a) Confocal images of digitonin-permeabilized HeLa cells. After permeabilization, the HeLa nuclei were incubated in buffer in the presence or absence of HeLa cytosol extract (“cytosol”) and in the presence or absence of an ATP-generating system (“energy”). The nuclei were treated with polyplexes formed with the indicated FITC-labeled polymers and Cy5-labeled pDNA for 4 hours before being fixed and imaged. JetPEI™ (PEI) was used at an N/P ratio of 5 and T4 and Glycofect™ (G4) were used at N/P ratios of 20. In these images, red represents rhodamine-labeled 2 MDa dextran, blue represents the nuclei, and yellow represents pDNA. Scale bar = 20 µM. Increases of red fluorescence in the nuclei are indicative of increased nuclear envelope permeability, and increases of yellow fluorescence in the nuclei are indicative of increased pDNA nuclear import. (b) Manders coefficients were used to measure colocalization between Cy5-labeled plasmid DNA and DAPI-labeled nuclei. Data is shown as the mean ± standard deviation for 10 samples. Bars with different letters represent means that are statistically significant from each other according to the Tukey-Kramer HSD method (p < 0.05, n = 10). No measureable Cy5 fluorescence was found in the nuclei treated with pDNA only or in the cells only controls. Bars with matching letters represent means that are not statistically different from each other. White arrows indicate some points of pDNA interaction.
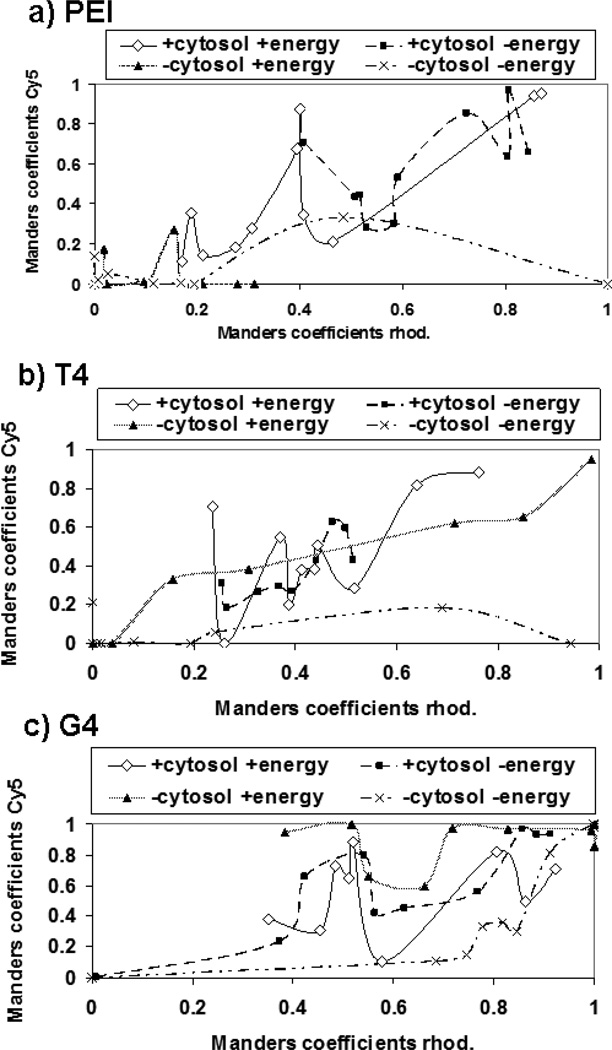
The data support the findings from the flow cytometry experiment in Figure 6a; the presence of cytosol increases pDNA-nucleus association when pDNA is complexed with JetPEI™. For T4 polyplexes, we observe statistically significant differences in pDNA-nucleus association in nuclei in the presence of both ATP and cytosol vs. in nuclei without ATP and cytosol. For G4 polyplexes, we see statistically significant differences between pDNA-nuclei colocalization in nuclei, in the absence of cytosol, with ATP versus without ATP. From these data, it appears that in the absence of cytosol, G4 polyplexes may be more dependent on the presence of ATP to get into nuclei. However, we noticed a high degree of variability in pDNA inside of the imaged nuclei. We hypothesized that the high variance could be due to multiple possible mechanisms of pDNA nuclear internalization occurring; one involving high nuclear envelope permeability and one not dependent on nuclear permeability. To further examine this, nuclei were also treated with rhodamine-labeled dextran to monitor nuclear permeabilization, and Manders coefficients of Cy5-labeled pDNA and DAPI-labeled nucleus were plotted against Manders coefficients of rhodamine-labeled dextran and DAPI-labeled nucleus. This analysis showed that for all polymers tested, there appeared to be a trend between nuclear permeability and pDNA nuclear internalization; nuclei with high dextran internalization also had high colocalization with pDNA (Figure 8). For T4 (Figure 8b) and G4 (Figure 8c), at low nuclear envelope permeability, only nuclei in the presence of ATP had high pDNA uptake, even in the absence of cytosol extract. For PEI, there did not appear to be a trend between pDNA nuclear uptake and the presence or absence of energy, but nuclei in the presence of cytosol extract exhibited higher pDNA internalization overall (Figure 8a).
Figure 8.
Colocalization data for nuclei treated with (a) JetPEI™ (PEI) polyplexes at an N/P ratio of 5; (b) T4 polyplexes at an N/P ratio of 20; and (c) Glycofect™ (G4) polyplexes at an N/P ratio of 20. Manders coefficients for Cy5 and DAPI are shown on the y-axis, and Manders coefficients for rhodamine (rhod.) and DAPI are shown on the x-axis. Data is plotted in an effort to discern trends between nuclear envelope permeability and pDNA nuclear import. In the absence of cytosol, the absence of energy results in overall lower pDNA import, unless the nuclear envelope has high permeability (as evidenced by high colocalization between rhodamine-labeled dextran and DAPI-labeled nuclei).
Discussion
Polymers are widely used to deliver various forms of nucleic acids for both research and therapeutic applications. However, the intracellular mechanisms in which they carry/traffic polynucleotides remain largely unknown. In an effort to study how polymer structure influences these mechanisms, studies were conducted on three polyamine-based polymers, JetPEI™ (PEI), Glycofect™ (G4), and T4. These studies emphasized how subtle differences in polymer structure influence pDNA import into the nucleus. Collectively, these data reveal interesting information about polymer-mediated pDNA delivery and delivery pathways to the nucleus. From nuclei isolated 24 hours after transfection, we observe that the polyplexes formed with all of the polymers displayed both higher pDNA-nucleus association and polymer-nucleus association (Figures 1b–d) when compared to pDNA only (Figure 1a). Further analysis using confocal microscopy revealed that, in the cases of T4 (Figure 2c) and G4 (Figure 2d), the pDNA was located around the periphery of the nucleus, whereas more pDNA within nuclei was observed from JetPEI™-transfected cells (Figure 2b). In addition, the nuclei isolated from PEI-treated and G4-treated cells had higher colocalization between dextran and the nucleus compared to T4, indicating increased nuclear envelope permeability. Interestingly, while T4 and G4 yielded similar Manders coefficients for Cy5-pDNA-nucleus colocalization, G4 appears to permeabilize the nucleus in a similar high manner to JetPEI™ and could indicate that the meso-galactose residues play a role in this effect. This is consistent with previous data in our group indicating that JetPEI™ and G4 are able to permeabilize the plasma membrane (JetPEI™ was higher).17 In addition, prior research in our group has shown that the meso-galactose residues influence pDNA binding compared to other carbohydrate-containing polymers, specifically in the ability to form hydrogen bonds between the polymer and the pDNA.43 The carbohydrate moieties in G4 and T4 have been shown to influence their mechanism of cellular uptake when compared to JetPEI™17, 44 and the meso-galactose residues in G4 also alter its degradation when compared to T4.45 Additionally, it would seem that the carbohydrate residues in G4 allow it to transfect primary and mesenchymal stem cells to a greater extent than JetPEI™.46 Taken together, the data from these prior experiments as well as the current data illustrate how slight changes in polymer structure (in this case, the number of hydroxyl groups per repeat unit) can impact their pDNA delivery mechanisms.
The data from Figures 1 and 2 indicate that all of the polymers are able to deliver pDNA to the nucleus within 24 hours after transfection, and that the polymers are able to enter the nucleus to a greater extent than pDNA, as about 90 % of the nuclei isolated from transfected cells were positive for FITC fluorescence (Figure 1b). These data suggest that free polymers may have different mechanisms of nuclear trafficking than polymers complexed with pDNA, but further study is needed to make any definite conclusions. In addition, it is possible that the addition of fluorescent tags to the polymers and pDNA may have affected their intracellular trafficking and nuclear uptake. Cells transfected with polyplexes formed with polymers T4 and G4 yielded a higher amount of nucleic positive for FITC-polymer than JetPEI™ (Figure 1), but this may be a result of the higher N/P ratio used for these polymers or differences in FITC labeling. In order to simulate typical transfection conditions, polymers were used at the N/P ratios that yielded the best expression/toxicity ratios. T4 and G4 were used at an N/P ratio of 20 to remain consistent with previously published data from our group, while JetPEI™ was used at the manufacturer’s recommended N/P ratio. Confocal microscopy images taken 24 hours after transfection (Figure 4) also confirmed that pDNA was indeed reaching the nucleus at this time point. After observing that the pDNA was capable of reaching the nucleus, we attempted to elucidate how the pDNA was trafficked to and subsequently entered the nucleus. To accomplish this, different trafficking inhibitors were used and their effects on luciferase expression were observed.
To examine the role that microtubules and mitosis play in the nuclear import of pDNA during polymer-mediated transfection, cells were transfected in the presence or absence of nocodazole (depolymerizes microtubules and inhibits microtubule-based polyplex trafficking) or aphidicolin (halts cell division). Our results indicate that cells treated with nocodazole during the first 4 hours of transfection exhibited lower luciferase expression compared to cells transfected in the absence of nocodazole with all polyplex types. This result indicates that microtubules may play a role in nuclear trafficking (Figure 5). Interestingly, the highest inhibition occurred during the transfections with G4, which indicates that this polyplex type may be more dependent on microtubules for nuclear trafficking. Also, during the nocodozole treatment, luciferase expression was not completely abolished with all of the polyplex types. This could be due, at least in part, to the microtubules reassembling after nocodazole was removed, and/or this could also be an indication that multiple mechanisms for polyplex-nuclear trafficking are occurring (i.e. via actin).17 G4 was also the only polymer that showed a decrease of luciferase expression in the presence of aphidicolin. This is an interesting result; our data indicate that G4 is able to permeabilize the nuclear envelope, so it would not be necessary for mitosis to allow pDNA entry into the nucleus when delivered by G4. Further studies are needed to make definite conclusions. However, because we still see relatively high luciferase expression in aphidicolin-treated cells (comparable to JetPEI™), we conclude that mitosis may not be necessary, but dividing cells do allow more luciferase expression than non-dividing cells when the pDNA is complexed with G4.
From the data obtained via measurement of colocalization between rhodamine-labeled dextran and DAPI-stained nuclei, we observed higher dextran accumulation in the nuclei isolated from cells treated with polyplexes formed with JetPEI™ and G4 compared to cells treated with pDNA only and polyplexes formed with T4 (Figure 2). It was unknown whether this increased nuclear envelope permeability was a result of direct polymer-induced permeabilization and/or apoptosis initiation. Thus, studies were performed in a cell-free system to investigate this further. The trends in nuclear envelope permeability (as measured by pixel intensity of trypan blue-treated nuclei examined by light microscopy) illustrated in Figure 3 followed the trends observed in Figure 2. In Figure 2, nuclear envelope permeability as indicated by dextran fluorescence was the highest with polyplexes formed with JetPEI™ and G4. Polyplexes formed with T4 yielded similarly low colocalization as the pDNA only control (to the dextran and nuclei). In the case of the trypan blue assay, mean pixel intensity was highest with T4, indicating the least trypan blue uptake and lowest nuclear permeability, and then followed by pDNA > G4 > JetPEI™ (highest nuclear permeability). It is interesting to note that the polymers able to induce the most nuclear envelope permeability (JetPEI™ and G4, Figs. 2 and 3) are also the polymers with the highest transfection efficiency (Figure 5). These data illustrate that the polyplexes are able to induce nuclear envelope permeability in a cell-free system that has no apoptotic signaling. In addition, prior work in our group illustrate that expression efficiency is not enhanced in the presence of an apoptosis-inducing agent.30
Next, we wanted to determine the minimal requirements necessary for pDNA import into the nucleus when complexed with the different polymer vehicles. For this, we used a cell-free system and isolated nuclei in buffers with or without an ATP-generating system and/or HeLa cytosol extract. First, we isolated nuclei, incubated them in the aforementioned buffers, and then treated them with the different polyplex solutions for 4 hours before flow cytometric analysis. The 4 hour time point was chosen based on previous studies in our group30 as well as other research on polyplexes47, 48 and pDNA-nuclear import.18, 34 Our results indicate that JetPEI™ polyplexes were able to reach the nucleus in all conditions, but yielded more pDNA association with the nucleus in the presence of cytosol (Figure 6a). Interestingly, pDNA-nucleus association was not observed in the cell-free system when the pDNA was delivered by T4 or G4 polyplexes (Figure 6a). Moreover, despite having low pDNA-nucleus association, the polyplexes formed with T4 and G4 exhibited high polymer only-nucleus association in all conditions tested (Figure 6b). This indicates that free polymer is able to enter the nucleus without pDNA. The difference between the results obtained for JetPEI™ and the PGAA polymer-nucleus association could be due to the presence of a higher fraction of free polymer in the transfection solution; T4 and G4 polyplexes were formed at higher N/P ratios (N/P=20) than JetPEI™ (N/P=5) in order to remain consistent with previously published transfection conditions, thus, more free polymer is present in nuclei treated with these polyplexes. Furthermore, JetPEI™ exhibited lower nucleus association in the presence of cytosol and energy (Figure 6b), contradictory to the Cy5 data for JetPEI™ (Figure 6a). Taken together, these results suggest that different mechanisms of nuclear trafficking could be occurring for free polymers vs. polyplexes. This is evident in the Cy5-pDNA data, where we observe less Cy5-pDNA fluorescence in nuclei with the cell-free system (Figure 6a) when compared to nuclei isolated from transfected whole cells (Figure 1), indicating that intracellular components (cytoplasm-based trafficking mechanisms) play a large role in polyplex trafficking. However, cytoplasmic factors may not necessarily play a role in trafficking of polymer only for T4 and G4, as there was little difference in the amount of FITC-positive nuclei isolated from transfected cells (Figure 1b) vs. nuclei in a cell-free system (Figure 6b). In the case of JetPEI™, we do notice less polymer-nucleus association in the presence of cytosol; this is perhaps indicative of the free polymer interacting with anionic cytosolic components but also is likely related to the lower N/P ratio used for this polymer. The reason(s) why JetPEI™ is more affected by the presence of cytosol than the PGAAs remain to be determined and is an interesting topic for future study. Interestingly, we do see evidence of polymer-cytosol interaction in confocal microscopy images of FITC-T4 and FITC-G4 polyplexes. It should be noted here that we were unable to observe any FITC signal from JetPEI™ in the fixed cells; the commercially-available JetPEI-FluoF™ is not compatible with cell fixation per the manufacturers specifications.
When nuclei were isolated after transfection of whole cells with G4 and T4 polyplexes, punctuate FITC fluorescence was observed as was colocalization between the polymer and pDNA (indicated by white pixels) (Supporting Information Figure S2). However, in the cell-free system (nuclei only), we see less colocalization between the polymer and pDNA, and significant free polymer fluorescence in the nucleus, in particular, free polymer colocalized with the nucleoli for both T4 (Figure S3) and G4 (Figure S4). In the presence of HeLa cytosol extract and ATP, we see some aggregates of polymer fluorescence outside of the nuclei (Figures S3a and S4a) that is not present in the images without cytosol extract (Figures S3 and S4 c and d). These results are intriguing and may indicate that the PGAAs are able to interact with cytosolic proteins, similar to the flow cytometry data observed for PEI (Figure 6b). In addition, in the absence of cytosol but in the presence of ATP, we also notice more Cy5-pDNA present within G4-treated nuclei (Figure S4c) vs. the T4-treated nuclei (Figure S3c). This corresponds with the higher luciferase expression efficiency observed with G4 polyplexes (Figure 5) as well as the possibility of an ATP-dependent mechanism of nuclear entry in the absence of cytosol, as observed in Figure 8.
In these studies, we saw nearly 100 % of the nuclei were positive for FITC-fluorescence when treated with polyplexes formed with G4 or T4, regardless of incubation conditions, possibly indicating differences in the degree of nuclear association of pDNA and polymer (Figure 6b). We also see significantly higher (p < 0.05) polymer-nucleus association in the PGAAs compared to JetPEI™ (Figure 6b), which may be due to the presence of more free polymer in these polyplex solutions compared to JetPEI™ but may also be influenced by the presence of carbohydrate moieties in the polymer backbone. Previous research has shown that the addition of carbohydrate groups to DNA delivery polymers enhances nuclear trafficking and pDNA nuclear import.32, 49 It is possible that, within free polymer molecules, the carbohydrate groups are more accessible to carbohydrate receptors found on the nuclear envelope32 compared to polymers interacting with pDNA within a polyplex; this could explain why free PGAA molecules appear to interact with the nucleus even in a cell-free system. It is also possible that nuclear association is dependent on charge. Since the PGAAs were formulated with pDNA at a higher N/P ratio (20) than JetPEI™ (5), there may be more free polymer/ positively-charged moieties able to interact with negatively-charged moieties found on the nuclear envelope, such as sialic acid, and in the nucleus.50, 51 In addition, many nuclear shuttle proteins that aid in nuclear envelope translocation through the nuclear pore complexes (NPCs) are negatively-charged.52 Interestingly, for the JetPEI™ polyplex-treated nuclei, FITC-positive nuclei were highest in the absence of both cytosol and energy (Figure 6b); this data exhibits nearly an opposite trend from that of the Cy5-positive nuclei in Figure 6a. The difference in cytosolic interactions of JetPEI™ vs. the PGAAs may not only be influenced by the presence of the sugar moiety but also by polymer length; JetPEI™ is much longer than the PGAAs (PEI ~ 22,000 Da vs. PGAA ~ 4,000 Da).
Using these flow cytometry data, we were unable to identify whether the pDNA is inside of the nucleus or on the nuclear surface. In order to determine if pDNA was being imported into the nuclei and not just bound to the nuclear envelope, confocal microscopy was used to image the transfected nuclei (Figure 7). When studying digitonin-permeabilized cells in this manner, we observed significant variations in Cy5-pDNA fluorescence among the polymer vehicles tested (Figure 7b). We hypothesized that there are likely multiple mechanisms for pDNA import occurring, with at least one mechanism dependent on nuclear envelope permeability. If this is the case, then we would expect to see cells with high rhodamine-dextran fluorescence, indicating high nuclear envelope permeability, also exhibiting high Cy5-pDNA fluorescence. To examine this, we plotted the Manders coefficients for pixel overlap of rhodamine-dextran and DAPI fluorescence vs. the Manders coefficients for the pixel overlap of Cy5-pDNA and DAPI fluorescence. For the PGAA vehicles, in the absence of cytosol, there is evidence of an energy-dependent mechanism for pDNA import occurring (Figs. 8b,c) due to the observation that there is more pDNA-nucleus association in the presence of ATP vs. the absence of ATP, while in the presence of cytosol there is less dependence on ATP for internalization. The nuclear import of pDNA delivered by JetPEI™ (Figure 8a) seems to be more cytosol-dependent than energy-dependent. Interestingly, in all cases, high nuclear permeability appeared to correlate to an increase in pDNA nuclear import (Figure 8), except for JetPEI™ and T4 in the absence of both cytosol and an ATP-generating system. Interestingly, the polymers with the highest expression efficiency (JetPEI™ and G4, Figure 5) are able to induce the most nuclear envelope permeability as assessed by both rhodamin-dextran (Figure 2) and trypan blue (Figure 3) permeability.
Overall, we demonstrate that polymer structure can influence the mechanisms of pDNA nuclear import. The hydroxyl-containing polymers exhibited more dependence on cytosolic presence for nuclear trafficking than JetPEI™, as evidenced by the dramatic decrease in pDNA-nucleus association in a cell-free system vs. nuclei isolated from whole cells. We have also found that nuclear envelope permeability by the polymeric delivery vehicles impacts pDNA nuclear import, and that nuclei isolated from JetPEI™-transfected cells display greater nuclear envelope permeability than nuclei isolated from T4 or G4-treated cells. Direct nuclear envelope permeabilization presents a possible link between high expression efficiency and high toxicity observed in many polymer-based DNA delivery systems. These data provide a parameter of investigation that could direct the development of safer and more potent polymers for nucleic acid delivery.
Supplementary Material
Acknowledgements
The authors acknowledge Anton Sizovs and Dustin Sprouse for useful scientific discussion, Anton Sizovs for assistance in labeling the polymers, and Marion Ehrich and Stephen Werre for assistance with statistical analyses. Giovanna Grandinetti would like to thank the NSF MILES-IGERT program for financial support. This work was supported by the NIH New Innovator Award Program (1 DP2 OD006669).
Footnotes
Supporting Information Available: Polymer characterization results for T4, histograms of flow cytometry data for nuclei isolated from transfected cells, detailed statistical analyses for average Manders coefficients and pixel intensity, confocal microscopy images of nuclei isolated from cells transfected with T4 and G4, confocal microscopy images of permeabilized cells treated with T4 and G4 polyplexes, and microscopy images of trypan-blue stained nuclei can be found in the supporting information. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Hunter AC. Molecular hurdles in polyfectin design and mechanistic background to polycation induced cytotoxicity. Advanced Drug Delivery Reviews. 2006;58:1523–1531. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. Journal of Controlled Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 3.Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24:1121–1131. doi: 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- 4.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godbey WT, Wu KK, Mikos AG. Poly(ethylenimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials. 2001;22:471–480. doi: 10.1016/s0142-9612(00)00203-9. [DOI] [PubMed] [Google Scholar]
- 6.Ira Y, Mely Y, Krishnamoorthy G. DNA vector polyethyleneimine affects cell pH and membrane potential: A time-resolved fluorescence microscopy study. Journal of Fluorescence. 2003;13:339–347. [Google Scholar]
- 7.Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethyleneimine)-mediated cytotoxicity: implications for gene transfer/therapy. Molecular Therapy. 2005;11:990–995. doi: 10.1016/j.ymthe.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 8.Brownlie A, Uchegbu IF, Shatzlein AG. PEI-based vesicle-polymer hybrid gene delivery system with improved biocompatibility. International Journal of Pharmaceutics. 2004;274:41–52. doi: 10.1016/j.ijpharm.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 9.Wong K, Sun G, Zhang, Dai H, Liu Y, He, Leong KW. PEI-g-chitosan, a novel gene delivery system with transfection efficiency comparable to polyethylenimine in vitro and after liver administration in vivo. Bioconjugate Chemistry. 2005;17:152–158. doi: 10.1021/bc0501597. [DOI] [PubMed] [Google Scholar]
- 10.Twaites BR, de las Heras Alarcón C, Lavigne M, Saulnier A, Pennadam SS, Cunliffe D, Górecki DC, Alexander C. Thermoresponsive polymers as gene delivery vectors: Cell viability, DNA transport and transfection studies. Journal of Controlled Release. 2005;108:472–483. doi: 10.1016/j.jconrel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Forrest ML, Gabrielson N, Pack DW. Cyclodextrin-polyethylenimine conjugates for targeted in vitro gene delivery. Biotechnology and Bioengineering. 2005;89:416–423. doi: 10.1002/bit.20356. [DOI] [PubMed] [Google Scholar]
- 12.Pun SH, Bellocq NC, Liu A, Jensen G, Machemer T, Quijano E, Schluep T, Wen S, Engler H, Heidel J, Davis ME. Cyclodextrin-modified polyethylenimine polymers for gene delivery. Bioconjugate Chem. 2004;15:831–840. doi: 10.1021/bc049891g. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Wenning L, Lynch M, Reineke TM. Gene delivery with novel poly(L-tartaraminoamine)s. In: Svenson S, editor. Polymeric Drug Delivery Volume I - Particulate Drug Carriers. Vol. 923. Washington, D.C: American Chemical Society; 2006. pp. 217–227. [Google Scholar]
- 14.Liu Y, Reineke TM. Poly(glycoamidoamine)s for gene delivery: Stability of polyplexes and efficacy with cardiomyoblast cells. Bioconjugate Chem. 2006;17:101–108. doi: 10.1021/bc050275+. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Reineke TM. Hydroxyl Stereochemistry and amine number within poly(glycoamidoamine)s affect intracellular DNA delivery. J. Am. Chem. Soc. 2005;127:3004–3015. doi: 10.1021/ja0436446. [DOI] [PubMed] [Google Scholar]
- 16.Lee CC, Liu Y, Reineke TM. General structure-activity relationship for poly(glycoamidoamine)s: The effect of amine density on cytotoxicity and DNA delivery efficiency. Bioconjugate Chem. 2008;19:428–440. doi: 10.1021/bc7001659. [DOI] [PubMed] [Google Scholar]
- 17.McLendon PM, Fichter KM, Reineke TM. Poly(glycoamidoamine) vehicles promote pDNA uptake through multiple routes and efficient gene expression via caveolae-mediated endocytosis. Mol. Pharmaceutics. 2010;7:738–750. doi: 10.1021/mp900282e. [DOI] [PubMed] [Google Scholar]
- 18.Dean DA. Import of plasmid DNA into the nucleus is sequence specific. Exp. Cell. Res. 1997;230:293–302. doi: 10.1006/excr.1996.3427. [DOI] [PubMed] [Google Scholar]
- 19.Dean DA, Strong DD, Zimmer WE. Nuclear entry of nonviral vectors. Gene Therapy. 2005;12:881–890. doi: 10.1038/sj.gt.3302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dean DA, Dean BS, Muller S, Smith LC. Sequence requirements for plasmid nuclear transport. Exp. Cell. Res. 1999;253:713–722. doi: 10.1006/excr.1999.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li S, MacLaughlin FC, Fewell JG, Gondo M, Wang J, Nicol F, Dean DA, Smith LC. Muscle-specific enhancement of gene expression by incorporation of SV40 enhancer in the expression of plasmid. Gene Therapy. 2001;8:494–497. doi: 10.1038/sj.gt.3301419. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Sahay G, Sriadibhatla S, Kabanov AV. Amphiphilic block copolymers enhance cellular uptake and nuclear entry of polyplex-delivered DNA. Bioconjugate Chem. 2008;19:1987–1994. doi: 10.1021/bc800144a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeong JH, Kim SH, Christensen LV, Feijen J, Kim SW. Reducible poly(amido ethylenimine)-based gene delivery system for improved nucleus trafficking of plasmid DNA. Bioconjugate Chem. 2010;21:296–301. doi: 10.1021/bc9003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duverger E, Pellerin-Mendes C, Mayer R, Roche A-C, Monsigny M. Nuclear import of glycoconjugates is distinct from the classical NLS pathway. J. Cell Sci. 1995;108:1325–1332. doi: 10.1242/jcs.108.4.1325. [DOI] [PubMed] [Google Scholar]
- 25.Pollard H, Remy J-S, Loussouarn G, Demolombe S, Behr J-P, Escande D. Polyethylenimine but not cationic lipids promote delivery to the nucleus in mammalian cells. J. Biol. Chem. 1998;273:7507–7511. doi: 10.1074/jbc.273.13.7507. [DOI] [PubMed] [Google Scholar]
- 26.Hong S, Leroueil PR, Janus EK, Peters JL, Kober M-M, Islam MT, Orr BG, Baker JR, Holl MMB. Interaction of polycationic polymers with supported lipid bilayers and cells: Nanoscale hole formation and enhanced membrane permeability. Bioconjugate Chem. 2006;17:728–734. doi: 10.1021/bc060077y. [DOI] [PubMed] [Google Scholar]
- 27.Prevette LE, Mullen DG, Banaszak Holl MM. Polycation-induced cell membrane permeability does not enhance cellular uptake or expression efficiency of delivered DNA. Mol. Pharmaceutics. 2010;7:870–883. doi: 10.1021/mp100027g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumhover NJ, Anderson K, Fernandez CA, Rice KG. Synthesis and in vitro testing of new potent polyacridine-melittin gene delivery peptides. Bioconjugate Chem. 2010;21:74–83. doi: 10.1021/bc9003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C-p, Kim J-s, Liu D, Rettig GR, McAnuff MA, Martin ME, Rice KG. Synthetic PEGylated glycoproteins and their utility in gene delivery. Bioconjugate Chem. 2007;18:371–378. doi: 10.1021/bc060229p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grandinetti G, Smith AE, Reineke TM. Direct nuclear permeabilization by polymeric pDNA vehicles: efficient method for delivery or mechanism of cytotoxicity? Mol. Pharmaceutics. 2012;9:523–538. doi: 10.1021/mp200368p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boutin V, Legrand A, Mayer R, Nachtigal M, Monsigny M, Midoux P. Glycofection: The ubiquitous roles of sugar bound on glycoplexes. Drug Delivery. 1999;6:45–50. [Google Scholar]
- 32.Monsigny M, Rondanino C, Duverger E, Fajac I, Roche A-C. Glyco-dependent nuclear import of glycoproteins, glycoplexes and glycosylated plasmids. Biochimica et Biophysica Acta. 2004;1673:94–103. doi: 10.1016/j.bbagen.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Duverger E, Carpentier V, Roche A-C, Monsigny M. Sugar-dependent nuclear import of glycoconjugates from the cytosol. Exp. Cell. Res. 1993;207:197–201. doi: 10.1006/excr.1993.1181. [DOI] [PubMed] [Google Scholar]
- 34.Munkonge FM, Amin V, Hyde SC, Green A-M, Pringle IA, Gill DR, Smith JWS, Hooley RP, Xenariou S, Ward MA, Leeds N, Leung K-Y, Chan M, Hillery E, Geddes DM, Griesenbach U, Postel EH, Dean DA, Dunn MJ, Alton EWFW. Identification and functional characterization of cytoplasmic determinants of plasmid DNA nuclear import. J. Biol. Chem. 2009;284:26978–26987. doi: 10.1074/jbc.M109.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manders EMM, Stap J, Brakenhoff GJ, Driel RV, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 1992;103:857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- 36.Manders EMM, Verbeek FJ, Aten JA. Measurement of co-localizatin of objects in dual-colour confocal images. J. Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 37.Rasband WS. ImageJ. Bethseda, MD: U.S. National Institutes of Health; pp. 1997–2009. [Google Scholar]
- 38.Hagstrom JE, Ludtke JJ, Bassik MC, Sebestyen MG, Adam SA, Wolff JA. Nuclear import of DNA in digitonin-permeabilized cells. J. Cell Sci. 1997;110:2323–2331. doi: 10.1242/jcs.110.18.2323. [DOI] [PubMed] [Google Scholar]
- 39.Chen J, Hessler JA, Putchakayala K, Panama BK, Khan DP, Hong S, Mullen DG, DiMaggio SC, Som A, Tew GN, Lopatin AN, Baker JR, Holl MMB, Orr BG. Cationic nanoparticles induce nanoscale disruption in living cell plasma membranes. J. Phys. Chem. B. 2009;113:11179–11185. doi: 10.1021/jp9033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grosse S, Thevenot G, Monsigny M, Fajac I. Which mechanism for nuclear import of plasmid DNA complexed with polyethylenimine derivatives? J. Gene Med. 2006;8:845–851. doi: 10.1002/jgm.915. [DOI] [PubMed] [Google Scholar]
- 41.Barua S, Rege K. The influence of mediators of intracellular trafficking on transgene expression efficacy of polymer-plasmid DNA complexes. Biomaterials. 2010;31:5894–5902. doi: 10.1016/j.biomaterials.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 42.Chida K, Sueyoshi R, Kuroki T. Efficient and stable gene transfer following microinjection into nuclei of synchronized animal cells progressing from G1/S boundary to early S phase. Biochemical and Biophysical Research Communications. 1998;249:849–852. doi: 10.1006/bbrc.1998.9245. [DOI] [PubMed] [Google Scholar]
- 43.Prevette LE, Kodger TE, Reineke TM, Lynch ML. Deciphering the role of hydrogen bonding in enhancing pDNA-polycation interactions. Langmuir. 2007;23:9773–9784. doi: 10.1021/la7009995. [DOI] [PubMed] [Google Scholar]
- 44.McLendon PM, Buckwalter DJ, Davis EM, Reineke TM. Interaction of poly(glycoamidoamine) DNA delivery vehicles with cell-surface glycosaminoglycans leads to polyplex internalization in a manner not solely dependent on charge. Mol. Pharmaceutics. 2010;7:1757–1768. doi: 10.1021/mp100135n. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Reineke TM. Degradation of poly(glycoamidoamine) DNA delivery vehicles: Polyamide hydrolysis at physiological conditions promotes DNA release. Biomacromolecules. 2010;11:316–325. doi: 10.1021/bm9008233. [DOI] [PubMed] [Google Scholar]
- 46.Kizjakina K, Bryson JM, Grandinetti G, Reineke TM. Cationic glycopolymers for the delivery of pDNA to human dermal fibroblasts and rat mesenchymal stem cells. Biomaterials. 2012;33:1851–1862. doi: 10.1016/j.biomaterials.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc. Natl. Acad. Sci. USA. 1999;96:5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc. Natl. Acad. Sci. USA. 2002;100:3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roche AC, Fajac I, Grosse S, Frison N, Rondanino C, Mayer R, Monsigny M. Glycofection: facilitated gene transfer by cationic glycopolymers. Cellular and Molecular Life Sciences. 2003;60:288–297. doi: 10.1007/s000180300024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gagliardi LJ. Electrostatic considerations in nuclear envelope breakdown and reassembly. Journal of Electrostatics. 2006;64:843–849. [Google Scholar]
- 51.Mazzanti M, Bustamente JO, Oberleithner H. Electrical dimension of the nuclear envelope. Physiological Reviews. 2001;81:1–19. doi: 10.1152/physrev.2001.81.1.1. [DOI] [PubMed] [Google Scholar]
- 52.Colwell LJ, Brenner MP, Ribbeck K. Charge as a selection criterion for translocation through the nuclear pore complex. PLOS Computational Biology. 2004;6:1–8. doi: 10.1371/journal.pcbi.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.