Abstract
Exposure to ionizing radiation (IR) elicits a set of complex biological responses involving gene expression and protein turnover that ultimately manifest as dysregulation of metabolic processes representing the cellular phenotype. Although radiation biomarkers have been reported in urine and serum, they are not informative about IR mediated tissue or organ specific injury. In the present study we report IR induced metabolic changes in gastrointestinal (GI) tissue of CD2F1 mice using ultra-performance liquid chromatography (UPLC) coupled with electrospray time-of-flight mass spectrometry. Post-radiation GI injury is a critical determinant of survival after exposure to IR. Our results show a distinct dose and time dependent response to GI tissue injury.
Introduction
Increased probability of radiological or nuclear incidents due to detonation of nuclear weapons by terrorists, accidents/sabotage of nuclear facilities, exposure/dispersal of radioactive materials and accidents during handling of radioactive materials enhances overall radiation risks for the civilian population. With the increasing likelihood of radiation exposure, the first step in medical management including triage is high-throughput assessment of the radiation dose received. Cytogenetic analysis, particularly dicentric chromosome aberration assay of peripheral blood lymphocytes is a gold standard technique for estimating the extent of ionizing radiation (IR) exposure; however, it is time-consuming and labor intensive. Alternate methods including assessment of DNA damage and repair 1, DNA-protein cross links 2, polyamine levels in red blood cells 3, serum proteomic profiles and gene expression profiles 4, 5 have been developed to estimate the absorbed radiation dose.
Biomarkers that can help identify exposed individuals are critically important in the event of mass casualty incidents 6. Although early physical symptoms of acute radiation syndrome in humans include nausea (1.4 Gy), vomiting (1.8 Gy) and erythema (3–4 Gy) 7, there is a latent period between the end of prodromal symptoms and the onset of physiological complications. A metabolomics approach offers to identify and quantitate global changes in the relative levels of small molecule metabolites as a readout of the physiological status of an individual 8. Recent technological improvements in liquid chromatography and time of flight mass spectrometry (TOF-MS) have enabled researchers to use a metabolomics approach to discover and characterize predictive biomarkers for radiation dosimetry, which has been comprehensively reviewed by us and others 9, 10. In the present study we report IR induced metabolite changes in gastrointestinal (GI) tissue o f mice, using ultra-performance liquid chromatography (UPLC) coupled with electrospray TOF-MS. The radiation-induced GI syndrome results due to whole body radiation exposure to IR doses in excess of 6 Gy, causing the death of epithelial stem and progenitor cells of the crypts. This is followed by the shrinkage of the villus and involution of the mucosa due to metabolite and electrolyte imbalance, resulting in severe diarrhoea, dehydration, and ultimate lethality 11. Additionally, sepsis caused byenteric bacteria (bacterial translocation from the intestinal lumen through the damaged mucosal barrier and into the blood stream) is an important cause of lethality after radiation exposure 11. Hence, it is important to identify biomarkers of radiation-induced GI injury not only to facilitate triage but also to understand organ specific response to IR exposure.
The primary goal of the study therefore, was to identify metabolite markers of GI tissue injury in response to two sub-lethal IR doses of 4 and 8 Gy at 1 and 4 days post-IR exposure using a murine model. Mass spectrometry based metabolomic profiling and subsequent multivariate analysis facilitated the identification of novel metabolites of GI injury. These include lipids, glutamate, tryptophan, taurocholate and the dipeptide Cys-Gly. To our knowledge; this is a first report on identification of small molecule markers of GI tissue injury. Future follow up studies with bio-fluids will augment the development of minimally invasive assays for evaluation of IR mediated tissue specific injury.
Materials and methods
Chemicals and reagents
LC/MS-grade acetonitrile (ACN), water and methanol were purchased from Fisher Scientific (NJ, USA). High purity formic acid (99%) was purchased from Thermo Scientific (Rockford, IL). Cys-Gly, hypoxanthine, methionine, inosine, glutamate, prostaglandin E2, tryptophan, 5-AMP, spermidine, citrulline, tyrosine, alpha-methyl-tyrosine, taurocholic acid, UDP-N-Acetyl-glucosamine, debrisoquine, 4-nitrobenzoic acid (4-NBA) were purchased from Sigma Aldrich St. Louis MO (USA). 2-deoxyionsine was purchased from MP Biomedicals (USA). Lipid standards were purchased from Avanti Polar Inc. All the reagents and chemicals used were LC/MS grade.
Animals
Six to eight week old specific pathogen free (SPF) male CD2F1 mice were purchased from Harlan Laboratories (Indiana, USA) and were housed (8 per cage) in an air-conditioned facility at the Armed Forces Radiobiology Research Institute (AFRRI), which is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) as described previously 12. Mice were held in quarantine for 1 week and were used after microbiology, serology, and histopathology examination to ensure the absence of Pseudomonas aeruginosa and common murine diseases. Mice were provided with certified rodent rations (Harlan Teklad Rodent Diet #8604, Harlan Teklad, WI, USA) and acidified water (with HCl, pH 2.5–3.0) ad libitum13–16. All mice were kept in rooms with a 12 hour light/dark cycle with lights on from 0600 to 1800 h. All animal procedures were performed in accordance with a protocol approved by the AFRRI’s Institutional Animal Care and Use Committee. Research was conducted according to the Guide for the Care and Use of Laboratory Animals, prepared by the Institute of Laboratory Animal Resources, the National Research Council, and U.S. National Academy of Sciences. All mice were 10–11 weeks old at the time of experiment.
Radiation Exposure
Mice were placed in Plexiglas boxes with 8 compartments to accommodate 8 mice per box and irradiated bilaterally at the AFRRI’s cobalt-60 gamma radiation facility to two sub-lethal doses, 4 and 8 Gy. The mid-line tissue dose to the animals was 3–12 Gy at a dose rate of 0.6 Gy/min. LD50/30 (lethal dose of 50% animals in 30 days) of CD2F1 mice was found to be 8.6 Gy (0.6 Gy/min) 17. An alanine ESR (electron spin resonance) dosimetry system (American Society for Testing and Material Standard E 1607) was used to measure dose rates (to water) in the cores of acrylic mouse phantoms 18, 19. Phantoms were 3 inches long and 1 inch in diameter and were located in all other compartments of the exposure rack. The ESR signals were measured with a calibration curve based on standard calibration dosimeters provided by the National Institute of Standard and Technology (NIST). The accuracy of the calibration curve was verified by inter-comparison with the National Physical Laboratory (NPL), United Kingdom. The only corrections applied to the dose rates in phantoms were for the decay of cobalt-60 source and for a small difference in mass energy-absorption coefficients for water and soft tissue. The radiation field was uniform within ±2%. After radiation, mice were returned to the cages for free access of food and water and monitored for any sign of radiation sickness (ruffled fur, weight loss etc).
Blood and Organ Collection
Blood (0.6–1.0 ml) was drawn under anesthesia (as a terminal procedure) using isoflurane (Abbott Laboratories, Chicago, IL) from the inferior vena cava (IVC) with a 23-G needle and collected in serum separator tubes. It was allowed to clot for 30 min at room temperature. After centrifugation at 1500 g for 10 min, serum was transferred to a micro-centrifuge tube and stored at 80 °C until used20.
For organ collection, mice were humanely euthanized after blood collection. Jejunum (two 5 cm sections) were harvested on day 1 and day 4 from animals exposed to 4 and 8 Gy of IR and from the sham treated groups (0 Gy) and cleaned by flushing with PBS. Samples were snap frozen in liquid nitrogen and stored at −80 °C until use.
Sample preparation
Metabolomics
GI tissue samples were sectioned uniformly and 10 mg of frozen tissue samples were placed in sterile glass vials and homogenized in 250 μl of 50% chilled methanol in water containing internal standards. Subsequently, 600 μl of acetonitrile was added and the samples were centrifuged after incubation on ice for 15 min, to facilitate protein precipitation. The supernatant was transferred to a fresh tube and dried under vacuum and re-suspended in solvent containing 98% water for MS analysis. The pellets were re-suspended in Ripa buffer, incubated on ice for 15 min and centrifuged. Protein estimation was performed using Bradford method 21. Total protein concentration was used to normalize the raw data.
Metabolite extraction from serum samples was performed by mixing 5μl of sample in 195 μl of 40% ACN, 25% CH3OH, 35% H2O, and containing internal standards. The samples were centrifuged at 13,000 rpm for 20 minutes after incubation on ice for 10 minutes. The supernatant was transferred to a fresh tube and dried under vacuum and re-suspended in 200 μl solvent containing 5% CH3OH, 1% ACN, 94% H2O for MS analysis.
UPLC-TOFMS based Metabolomics Analysis
Metabolomic profiling was performed using UPLC-QTOF-MS as described previously 22. Briefly, 5 μl of each sample was injected onto a reverse-phase 2.1 × 50 mm Acquity 1.7 μm C18 column (Waters Corporation, Milford, MA) using an Acquity UPLC system with a gradient mobile phase consisting of 2% acetonitrile in water containing 0.1% formic acid (A) and 2% water in acetonitrile containing 0.1% formic acid (B). Each sample was resolved for 10 min at a flow rate of 0.5 ml/min. The gradient consisted of 100% A for 0.5 min then a ramp of curve 6 to 60% B from 0.5 min to 4.0 min, then a ramp of curve 6 to 100% B from 4.0–8.0 min, hold at 100% B until 9.0 min, then a ramp of curve 6 to 100% A from 9.0 min to 9.2 min, followed by a hold at 100% A until 10 min. The column eluent was introduced directly into the mass spectrometer by electrospray. Mass spectrometry was performed on a Q-TOF instrument (QTOF Premiere, Waters), operating in either negative (ESI−) or positive (ESI+) electrospray ionization mode with a capillary voltage of 3200 V and a sampling cone voltage of 20 V in negative mode and 35 V in positive mode. The desolvation gas flow was set to 800 liters/h and the temperature was set to 350 °C. The cone gas flow was 25 liters/h, and the source temperature was 120 °C. Accurate mass was maintained by introduction of lock spray interface of sulfa-dimethoxine (m/z = 311.0814 [M+H]+ or 309.0658 [M-H]−) at a concentration of 250 pg/μl in 50% aqueous acetonitrile and a rate of 150 μl/min. Data were acquired in centroid mode from 50 to 850 m/z in MS scanning.
UPLC-QTOF-MS based metabolomic profiling of serum samples was performed by injecting 5 μl of each sample onto a reverse-phase 2.1 × 50 mm Acquity 1.7 μm C18 column (Waters Corporation, Milford, MA) using an Acquity UPLC system with a gradient mobile phase consisting of 100% water containing 0.1% formic acid (A) and 100% ACN (B). Each sample was resolved for 13 min at a flow rate of 0.5 ml/min. The gradient consisted of 99% A for 0.5 min then a ramp of curve 6 to 60% B from 0.5 min to 4.0 min, then a ramp of curve 6 to 99% B from 4.0–9.0 min, hold at 99% B until 10.0 min, then a ramp of curve 6 to 99% A from 11.0 min to 12.0 min, followed by a hold at 99% A until 13 min. The column eluent was introduced directly into the mass spectrometer by electrospray. The other Q-TOF instrument operating were as described previously.
Lipidomics
GI tissue samples were homogenized with extraction buffer containing 50% methanol and 20% IPA in water with 10 pmoles of 14:0 LPA and 5 pmoles of 17:0 Ceramide as internal standards. To facilitate protein precipitation, 600 μl of chilled acetonitrile was added and the samples were incubated on ice for 15 min. The samples were centrifuged at 13,000 rpm at 4 °C for ten minutes. The supernatant was transferred to a fresh tube and dried under vacuum and re-suspended in solvent containing 25:30:45 (water: acetonitrile: IPA v/v) for MS analysis.
G2-TOFMS based Lipidomics Analysis
Samples were injected onto a reverse-phase 100 × 2.1 mm Acquity 1.7 μm C18 CSH column (Waters Corp, Milford, MA) using an Acquity H Class UPLC system (Waters) with a gradient mobile phase consisting of 50% acetonitrile in water containing 0.1% formic acid 10 mM ammonium formate (A) and 10% acetonitrile in isopropanol containing 0.1% formic acid and 10 mM of ammonium formate (B). Each sample was resolved for 10 min at a flow rate of 0.5 ml/min. The gradient consisted of 60% A for 0.5 min then a ramp of curve 6 to 100% B from 0.5 min to 8 min, then the gradient held at 100% B for 0.5 min and back to 60% A at 10 min. The column eluent was introduced directly into the mass spectrometer by electrospray. Mass spectrometry was performed on a Q-TOF instrument (Xevo G2 QTOF, Waters Corp, USA) operating in either negative (ESI−) or positive (ESI+) electrospray ionization mode with a capillary voltage of 3200 V in positive and 2600 V in negative, and a sampling cone voltage of 40 V in negative mode and 30 V in positive mode. The desolvation gas flow was set to 700 liters/h and the temperature was set to 300 °C and the source temperature was 120 °C. Accurate mass was maintained by introduction of lock spray interface of leucine enkephalin (556.2771 [M+H] + or 554.2615 [M-H]−) at a concentration of 2 pg/μl in 50% aqueous acetonitrile and a rate of 2 μl/min. Data were acquired in continuum MSe mode from 50 to 1200 m/z. The high energy employed a 10–40 volts collision energy ramp and the low energy without collision energy as a MS scan.
Data pre-processing and Data Analysis
LC-MS data were pre-processed using XCMS software 23. The data were normalized to the ion intensity of the internal standards and protein concentration. The normalized data sets were analyzed by Principal Component Analysis (PCA) and Orthogonal Projections to Latent Structures (OPLS). OPLS analysis was considered best fit to determine the separation in the sham and irradiated mice. Candidate features with high correlation values and positioned furthest from the point of origin in the upper right and lower left quadrants of the S-plot were deemed significantly altered in the two groups and were chosen for further characterization. Clustering analysis using Radom Forests (RF) was also performed to interrogate the top 50 discriminant features in the two study groups 24. The OPLS-DA models were run using SIMCA-P version 12 (Umetrics, Inc.) and the Random Forests methods were run using several R packages with specific use of the G plot library for heat map graphics within R. 2.11.0 25. Quantitative descriptors of model quality for the OPLS-DA models included R2 (explained variation of the binary outcome: sham vs irradiated) and Q2 (cross-validation based predicted variation of the binary outcome). We used score plots to visualize the discriminating properties of the OPLS-DA models, and also S-plots for putative biomarker identification by visualization of the OPLS-DA loadings on the predictive score. Selection of features based on the OPLS-DA model used a p (corr) cut-off of 0.8, as reported previously26, 27. The features selected via OPLS-DA together with those selected by the RF method were used for accurate mass based data base search.
Verification and validation of metabolite identification
For metabolite identification, initially accurate mass based search was performed using online database Madison Metabolomics Consortium Database (MMCD) while the MSe data from the lipidomics experiment was used to perform s spectral library using SimLipid v3.0, (Premier Biosoft Intl, Palo Alto CA). This enabled putative identification of the compounds that corresponded to the accurate monoisotopic mass measurements detected using UPLC-TOFMS analysis. The mass tolerance was kept at 5 ppm to minimize false positive identifications.
Subsequently, the mass based putative identity of the metabolites was validated by matching the fragmentation pattern of the parent ion to that of the standard metabolite using tandem mass spectrometry (UPLC-TOFMS/MS) and also by comparing the retention time under the same chromatographic conditions.
Metabolomic analysis using GEDI
The Gene Expression Dynamic Inspector (GEDI) uses Self-organizing Maps (SOMs) to reduce data dimensionality with respect to gene number and to create characteristic visual representations for each sample 28. We used features exhibiting the highest intensity in all of the binary-comparison sub-sets for GEDI analysis. Each tile in the SOM corresponds to a cluster of biomarkers that share a similar intensity pattern across conditions. Different colors reflect the biomarker intensity of a centroid in each group29, 30.
Results
Metabolomic and lipidomic profiling for identification of markers of acute response (day 1) to IR
We performed metabolomic profiling of GI tissue obtained from sham treated mice or those exposed to 4 or 8 Gy of IR, with six mice per treatment group. Pre-processing using XCMS resulted in a three dimensional data matrix consisting of 3851 & 3333 features in the respective modes. Lipidomic profiling yielded 1697 & 815 features respectively. A total of 6421 (4 Gy) and 6478 (8 Gy) features were used for multivariate analyses (Table 1). Initially, we performed principal component analysis which did not result in a clear group separation. Hence, Orthogonal Partial Least Square Discriminant Analysis (OPLS-DA) was performed to identify discriminating features between sham treated mice and those exposed to 4 Gy after 1 day of IR exposure. The R2 and Q2 for the OPLS model were 0.98 and 0.70 respectively, supporting the quality of the model.
Table 1.
Statistically significant features detected for different IR doses and time points.
| Radiation dose | Time | No of peaks |
|---|---|---|
| Total number of features post XCMS processing | ||
|
| ||
| 4Gy | Day 1 | 7184 |
| Day 4 | 7184 | |
| 8Gy | Day 1 | 7184 |
| Day 4 | 7184 | |
|
| ||
| Total number of features selected by PLS-DA based on Standard Deviation* | ||
|
| ||
| 4Gy | Day 1 | 6421* |
| Day 4 | 6588* | |
| 8Gy | Day 1 | 6478* |
| Day 4 | 6378* | |
|
| ||
| Total number of significantly dysregulated features for different IR doses | ||
|
| ||
| 4Gy | Day 1 | 133 |
| Day 4 | 147 | |
| 8Gy | Day 1 | 141 |
| Day 4 | 111 | |
|
| ||
| Total number of features used for pathway analysis by IPA** | ||
|
| ||
| 4Gy | Day 1 | 56 |
| Day 4 | 37 | |
| 8Gy | Day 1 | 28 |
| Day 4 | 41 | |
Feature selection based on standard deviation cut off of 10%.
Top 50 metabolites from each set were putatively identified via accurate mass based search with MMCD.
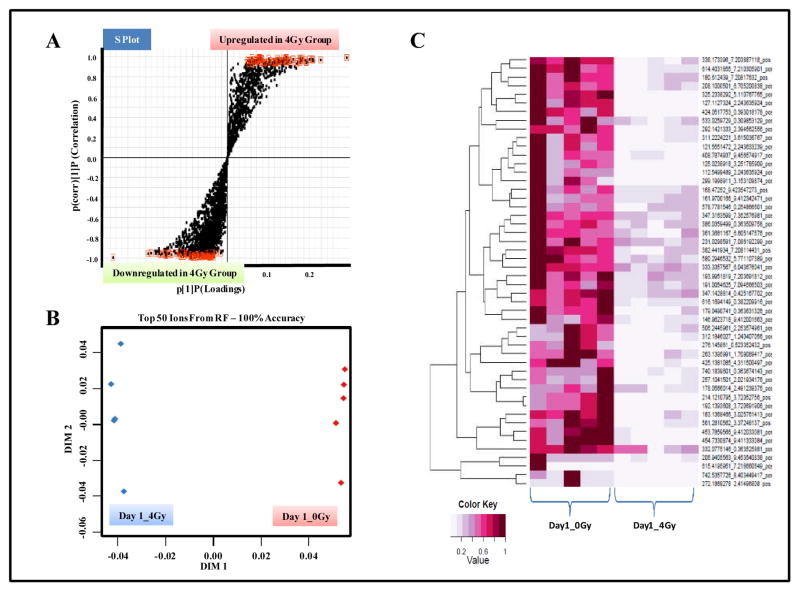
The highlighted features exhibited high correlation values and were selected for further characterization (Figure 2, Panel A). Additionally, RF analysis resulted in an unambiguous separation of the two groups with 100% accuracy (Figure 2, Panel B). The heat map depicts the top 50 discriminating features between the two study groups (Figure 2, Panel C).
Figure 2. UPLC-ESI-TOFMS profiling of GI tissue in CD2F1 mice.
Animals were either sham irradiated or exposed to 4Gy of γ radiation. The mice were euthanized 1 day, post-IR exposure for blood and organ collection. Comparative metabolomics analyses were performed as described in methods. Panel A. OPLS loadings S-plot displaying dysregulated features in irradiated tissue samples as compared to those from sham exposed group. Panel B. Two dimensional accuracy plot for top 50 features interrogated using Random Forests. The X-axis (dimension 1) denotes the interclass separation while Y-axis (dimension 2) displays the intra-class variability. Panel C. Heat map visualization of the feature rankings comparing relative levels of metabolites in control and irradiated GI tissue samples. Each row represents a unique feature with a characteristic mass to charge and retention time value.
A similar analysis was performed to compare the metabolomic profiles of sham treated mice with those that were exposed to 8 Gy of IR and Figure S1). A total of 174 features were selected for accurate mass based putative identifications using the MMCD data base. Taken together, the compound identification of nine metabolites was confirmed by matching the MS/MS fragmentation patterns with those of standard compounds (Table 2 and Supplementary Figures S2–S8), while those without any identification are listed in Supplementary Table ST1. The trend plot showed a time and dose dependent tissue injury response for these markers (Figure 3 and Supplementary Figure S9).
Table 2. Putative biomarkers of radiation injury of GI tissue of CD2F1 mice at day 1.
The features that were significantly altered in the irradiated GI tissue (4 & 8 Gy) at day 1 are listed. These metabolites were unambiguously identified using tandem mass spectrometry.
| S. No | Metabolites | Kegg/PubChem/Lipid ID | ESI Mode | m/z* | RT* | *Fold Change
|
Major CID fragments | |
|---|---|---|---|---|---|---|---|---|
| 4 Gy | 8 Gy | |||||||
| P-value | P-value | |||||||
| 1 | Cys-Gly | C01419 | POS | 179.049 | 0.3636 | ↓(0.01) | ↓(0.03) | 76.0229, 116.0172, 119.0599 |
| 2 | Glutamic acid | C00025 | POS | 148.061 | 0.3308 | ↓(0.03) | ↓(0.02) | 84.0453, 102.0559 |
| 3 | D-Tryptophan | C00525 | NEG | 203.082 | 1.4182 | ↑(0.02) | ↑(0.01) | 116.0503, 74.0260, 142.0665 |
| 4 | Methionine | C01733 | POS | 150.059 | 0.3822 | ↓(0.04) | ↑(0.02) | 133.0332, 104.0550 |
| 5 | PE(16:0/0:0) | LMGP02050002 | NEG | 452.285 | 1.2883 | ↑(0.05) | ↓(0.04) | 140.0131, 196.0404 |
| 6 | PS(16:0/0:0) | LMGP03050002 | NEG | 496.275 | 1.0176 | ↓(0.05) | ↓(0.03) | 255.2361, 409.2421, 496.2751 |
| 7 | PE(20:1/0:0) | LMGP02050020 | NEG | 506.334 | 1.3263 | ↑(0.05) | ↓(0.06) | 140.0131, 196.0404 |
| 8 | PC(16:0/2:0) | LMGP01010612 | POS | 538.387 | 2.2977 | ↓(0.04) | ↓(0.05) | 104.1063, 184.0734 |
| 9 | PC(8:2/8:2) | LMGP01011254 | POS | 502.315 | 0.6916 | ↓(0.03) | ↓(0.02) | 184.0736 |
Figure 3. Gamma radiation exposure in CD2F1 mice induces dose dependent alterations in metabolite levels in the GI tissue.
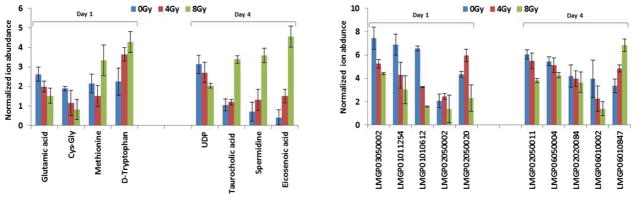
Mean normalized ion abundance for each metabolite is plotted for groups of CD2F1 mice that were exposed to sham, 4 Gy or 8 Gy of IR at days 1 and 4.
Tryptophan was found to be up regulated in GI tissue, 1 day after IR exposure in a dose dependent manner. Dysregulation of tryptophan metabolism has been reported to result in gastrointestinal malfunction as well as the disruption of serotonin and kynurenine pathways 31. Glutamic acid, Cys-Gly & methionine were also found to be down regulated 1 day post-IR exposure. The dipeptide Cys-Gly is a precursor of the anti-oxidant glutathione which plays an important role in the detoxification of the reactive oxygen species in cells and tissues 32. Depletion of Cys-Gly post-IR may thus result from or contribute to increased oxidative stress that is a hallmark of radiation exposure 33. We also found a concomitant decrease in the levels of glutamic acid in response to increasing doses of IR. Glutamate (Glu) is formed by the hydrolysis of the amino group of glutamine (Gln) via the enzyme glutaminase. It has been reported that the GI tract accounts for almost 80% of the total glutaminase activity 34. Hence, decrease in the levels of glutamate in the GI tissues, post-IR may be due to a decrease in the total enzyme activity of this enzyme. Thus GI injury is likely to have serious implications on the overall energy metabolism and hence, availability of markers directly indicative of tissue injury could not only aid population screening but also aid the development of therapeutics to combat tissue specific radiation injury 35.
Additionally, we found a significant dysregulation of lipid panel after IR exposure which is consistent with previous reports 36–39. PS (16:0/0:0), PC (8:2/8:2), PC (20:1/0:0) showed a down regulation in response to IR exposure in a dose dependent manner. Our results are consistent with studies that have reported a depletion of phospholipids in rat thymus after fractionated gamma irradiation 40.
Metabolomic and Lipidomic analysis for the identification of markers of GI injury at 4 days post-IR
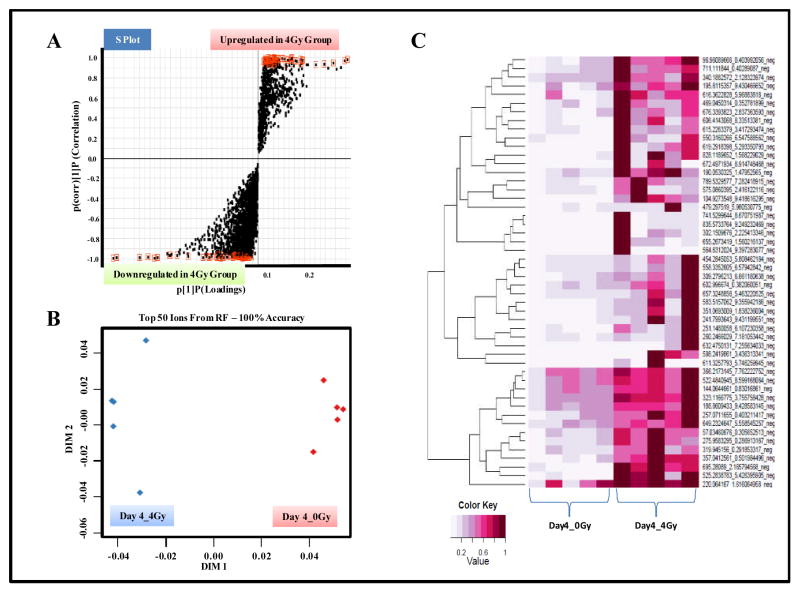
In order to characterize delayed GI tissue injury response, we selected 4 days as a post-IR exposure time point. GI tissues were profiled using UPLC-QTOF-MS in electrospray positive and negative modes. Data pre-processing by XCMS resulted in the detection of 3851 & 3333 features for metabolomics profiling and 1697 & 815 for lipidomic profiling respectively in the positive mode (Table 1). Feature rankings obtained by comparing profiles of sham irradiated samples showed a clear separation at both IR doses (Figure 4 and Supplementary Figure S10). Of these the metabolites that were unambiguously identified by matching fragmentation patterns and retention times with standard compounds are detailed in Table 3 and Supplementary Figure S11–S18. The unidentified markers with significant fold change and p-value are listed in Supplementary Table T2. For many of the identified metabolites a distinct dose dependent response was observed (Figure 3 and Supplementary Figure S19).
Figure 4. GI metabolomics shows a distinct metabolic response, 4 days post-IR exposure.
Panel A. OPLS loadings S-plot displaying significantly altered features in GI tissue samples obtained from irradiated mice as compared to sham exposed mice. Panel B. Two dimensional accuracy plot for top 50 features interrogated using Random Forests. Panel C. Heat map visualization of the feature rankings comparing relative levels in GI tissue samples obtained from sham and irradiated mice.
Table 3. Putative biomarkers of radiation injury of GI tissue of CD2F1 mice (Day 4).
The features that were significantly altered in the irradiated GI tissue after 4 days of IR exposure are listed. These metabolites were validated using tandem mass spectrometry.
| S. No | Metabolites | Kegg/PubChem/Lipid ID | ESI Mode | m/z* | RT* | *Fold Change
|
Major CID fragments | |
|---|---|---|---|---|---|---|---|---|
| 4 Gy | 8 Gy | |||||||
| P-value | P-value | |||||||
| 1 | Spermidine | C00315 | POS | 146.166 | 0.2437 | ↑(0.02) | ↑(0.03) | 72.0819, 112.1130, 84.0822 |
| 2 | Eicosenoic acid | 6438157 | POS | 311.169 | 0.1542 | ↑(0.04) | ↑(0.02) | 183.0119, 119.0515, 79.9594 |
| 3 | Taurocholic acid | C05122 | NEG | 514.285 | 3.1882 | ↑(0.03) | ↑(0.01) | 124.0081, 79.9595, 496.2741 |
| 4 | UDP-N-Acetyl-glucosamine | C00043 | NEG | 606.077 | 0.3992 | ↓(0.02) | ↓(0.03) | 384.9803, 158.9243,78.9694 |
| 5 | PE(O-20:0/20:5) | LMGP02020084 | NEG | 778.574 | 5.0338 | ↓(0.04) | ↓(0.02) | 742.5486 |
| 6 | PI(18:2/16:0) | LMGP06010847 | NEG | 833.534 | 4.2999 | ↑(0.03) | ↑(0.01) | 241.0149, 255.2363, 279.237 |
| 7 | PI(18:1/18:1) | LMGP06010002 | NEG | 861.565 | 4.7912 | ↓(0.02) | ↓(0.05) | 241.0153, 281.2524 |
| 8 | PE(18:2/0:0) | LMGP02050011 | NEG | 476.287 | 1.0965 | ↓(0.01) | ↓(0.05) | 279.2378 |
| 9 | PI(18:0/0:0) | LMGP06050004 | NEG | 599.331 | 1.4042 | ↓(0.02) | ↓(0.04) | 283.2679, 437.2751, 599.3305 |
The endogenous levels of metabolites spermidine, eicosenoic acid, PI (18:2/16:0) and taurocholic acid were found to be elevated in response to 4 Gy and 8 Gy IR exposures. The levels of UDP-N-acetyl-D-glucosamine, PE (18:2/0:0), PE (0–20:0/20:5) on the other hand were depleted post-IR exposure.
An increase in serum levels of spermidine has been reported in mice irradiated with 6 Gy of gamma radiation 41. Elevated bile acid levels, specifically taurocholate, have been implicated in inducing gastric mucosal ulcers 42. The up-regulation of lipid molecules in the present study are consistent with previous reports, wherein GC-MS analysis was performed on rat serum collected at 1 and 24 h post-irradiation at 3 Gy radiation dose 43.
In order to obtain a global metabolome visualization of dose and time dependent response in GI tissue, we used the gene expression dynamics inspector to plot self-organizing maps. The results depict clusters of metabolites that exhibit differential response in a dose and time dependent manner (Supplementary Figure S20).
Functional Pathway analysis
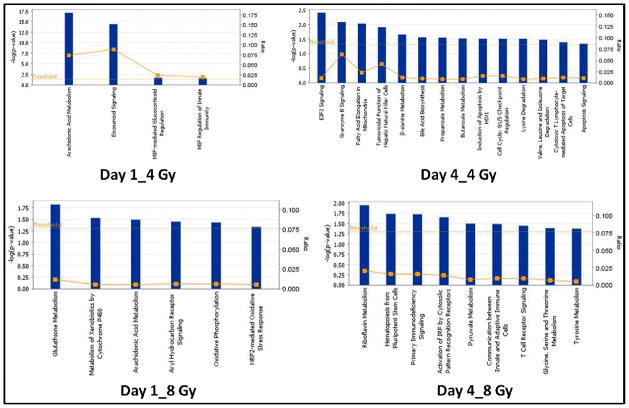
The Comparative canonical pathway analysis associated with GI tissue injury using murine model at different times and doses of IR exposure included arachidonic acid metabolism, eicosanoid signaling, and oxidative phosphorylation after 1 day of IR exposure. The pre-dominant pathways that were dysregulated at the four day time point included glycine, serine and threonine metabolism. Malfunction in arachidonic acid (AA) metabolism has been reported in rat platelets after 24 h whole body radiation 44. Hahn et al 1983 45 reported AA dysfunction in endothelial cells after gamma radiation exposure which resulted in elevated levels of COX-2. The elevation of COX-2 is well studied in promoting inflammation 46. Pyruvate metabolism was found to be perturbed four days post radiation exposure, which is known to be a major indicator of disrupted central carbon metabolism 47, 48. The participating molecules are listed in Supplementary Table ST3.
Discussion
Exposure to IR is associated with induction of acute radiation syndrome (ARS) which can include depletion of bone marrow, internal bleeding, massive fluid and electrolyte loss from the GI tract and death. Depending on the dose, the symptoms of ARS can appear within hours to weeks. It is a challenge to triage individuals who are minimally exposed compared to those who received high radiation dose and need immediate treatment 49. The current strategy to assess individual radiation exposure is based on symptoms developed over time 1. These methods are time-consuming and are not practical in a real time scenario. In addition, the latent period (from radiation exposure to symptom onset) is a critical time for therapeutic intervention which can help save lives.
Therefore, there is an urgent need to develop biochemical or molecular biomarkers that can help implement effective countermeasures of radiation injury to affected individuals and provide basis for treatment decisions. Previous bio-dosimetry studies with urine or serum samples have reported purines and pyrimidines as major markers of radiation exposure 50–52.
Although urine and serum biomarkers of metabolomic response to gamma radiation have been reported, these are not informative of organ specific changes 50, 53. In this study, we report the identification of acute and delayed metabolomic profiles of GI tissue specific radiation injury responses in CD2F1 mice exposed to different doses of gamma radiation. To our knowledge, this is the first report on bio-dosimetry of GI tissue metabolomics. Intestinal mucosal surface area is a well-validated, sensitive parameter of intestinal radiation injury. Authors demonstrated nadir of mucosal surface area at 3.5 days post radiation at 8 Gy 54. Even at lower doses of radiation, the GI tract plays a major role in the pathophysiology of toxicity and clinical outcome. We demonstrated that intestinal epithelial cells undergo massive apoptosis within 24 h post-radiation (6 Gy) due to severe damage to the functioning villi14. Whole-body irradiation of mice at 4 Gy exhibited altered structures of mucosa and sub-mucosa layers 55. GI damage responses in 4 Gy irradiated mice was evident from epithelial crypt cell necrosis, blunting of the villi and diffused lymphatic and plasma cellular infiltration 55. The underlying hypothesis was that the IR exposure would elicit an organ specific injury response. Hence, we compared the metabolomics profiles of kidney and GI tissue after 1 day of 4 Gy IR exposures. As expected the profiles were markedly different for each tissue type (Supplementary Figure S21). The IR induced metabolomics response in GI tissue was studied as a function of time and dose. GI tissue metabolomics profiles provided an information rich matrix that enabled the identification of metabolites like lipids, organic acids as well as amino acids which can impact the overall GI metabolism and in part explain IR induced GI toxicity. In addition, our study has delineated IR mediated markers of GI injury for the first time. Although we used 4 and 8 Gy for this initial study in order to ensure a robust tissue response, future studies will use a larger range of radiation doses as well as bone marrow shielding irradiation models to investigate qualitative and quantitative differences in the metabolomic profiles and threshold doses. In order to evaluate these markers in a minimally invasive matrix we examined the relative levels of the putative GI markers in serum obtained from the same group of mice. We detected seven markers displaying similar trend as that seen in GI metabolomics while those with an opposite pattern are listed in Supplementary Table ST5. These results emphasize the applicability of this approach to validate markers in minimally invasive matrices that were originally discovered in a specific tissue matrix.
Conclusion
Metabolomics is a powerful tool to study post-radiation changes in metabolism even before the onset of clinical symptoms and can augment population screening in real life scenarios. Improvements in metabolite identification databases are likely to facilitate the development of robust, multi-metabolite panels for potential clinical use.
Supplementary Material
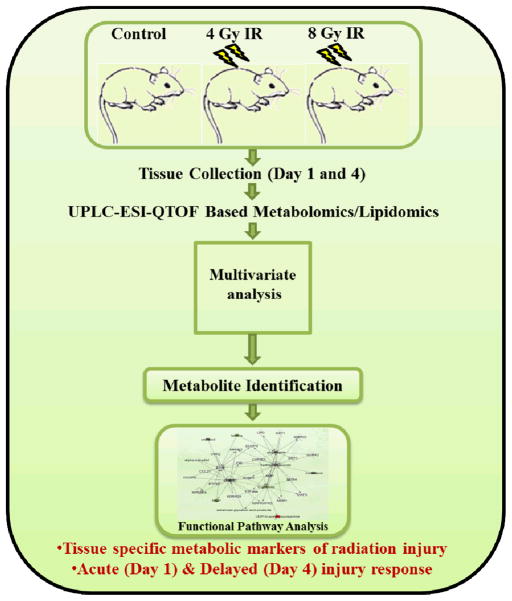
Figure 1.
Schema for GI metabolomics
Figure 5. Canonical pathways associated with response of gamma radiation in GI tissue of CD2F1 mice.
The four groups were compared viz. Day1_4 Gy, Day1_8 Gy, Day4_4 Gy & Day4_8 Gy. Each bar represents type of metabolism in particular pathway dys-regulated after radiation exposure.
Table 4.
Putative serum biomarkers of radiation injury after 1 and 4 days of irradiation with 4 & 8 Gy in CD2F1 mice.
| S. No | Metabolite | m/z | RT | ESI Mode | Fold Change
|
p-value
|
||
|---|---|---|---|---|---|---|---|---|
| 4 Gy | 8 Gy | 4 Gy | 8 Gy | |||||
| Day 1 | ||||||||
|
| ||||||||
| 1 | Cys-Gly | 179.065 | 1.2146 | POS | ↓ | ↓ | 0.05 | 0.04 |
| 2 | Methionine | 150.036 | 0.3198 | POS | ↓ | ↑ | 0.01 | 0.02 |
| 3 | PC(16:0/2:0) | 538.389 | 3.8174 | POS | ↓ | ↓ | 0.05 | 0.03 |
|
| ||||||||
| Day 4 | ||||||||
|
| ||||||||
| 4 | Taurocholic acid | 514.283 | 3.1747 | NEG | ↑ | ↑ | 0.02 | 0.05 |
| 5 | UDP-N-Acetyl-glucosamine | 606.071 | 0.3313 | NEG | ↓ | ↓ | 0.04 | 0.04 |
| 6 | PE(18:2/0:0) | 476.277 | 4.5347 | NEG | ↓ | ↓ | 0.03 | 0.03 |
| 7 | PE(O-20:0/20:5) | 778.563 | 8.4751 | NEG | ↓ | ↓ | 0.03 | 0.02 |
Acknowledgments
This study was supported by US Department of Defense Threat Reduction Agency grant H. 10016_09_AR_R, administered by The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc.) and by the National Institutes of Health (U19 AI67798). The authors would like to acknowledge the Proteomics and Metabolomics shared resource partially supported by NIH/NCI grant P30-CA051008.
Footnotes
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
References
- 1.Waselenko JK, MacVittie TJ, Blakely WF, Pesik N, Wiley AL, Dickerson WE, Tsu H, Confer DL, Coleman CN, Seed T, Lowry P, Armitage JO, Dainiak N. Ann Intern Med. 2004;140:1037–1051. doi: 10.7326/0003-4819-140-12-200406150-00015. [DOI] [PubMed] [Google Scholar]
- 2.Budhwar R, Bihari V, Mathur N, Srivastava A, Kumar S. Biomarkers : biochemical indicators of exposure, response, and susceptibility to chemicals. 2003;8:162–166. doi: 10.1080/1354750031000067495. [DOI] [PubMed] [Google Scholar]
- 3.Porciani S, Lanini A, Balzi M, Faraoni P, Becciolini A. Phys Med. 2001;17(Suppl 1):187–188. [PubMed] [Google Scholar]
- 4.Amundson SA, Grace MB, McLeland CB, Epperly MW, Yeager A, Zhan Q, Greenberger JS, Fornace AJ., Jr Cancer Res. 2004;64:6368–6371. doi: 10.1158/0008-5472.CAN-04-1883. [DOI] [PubMed] [Google Scholar]
- 5.Kang CM, Park KP, Song JE, Jeoung DI, Cho CK, Kim TH, Bae S, Lee SJ, Lee YS. Radiat Res. 2003;159:312–319. doi: 10.1667/0033-7587(2003)159[0312:pbfire]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 6.Rothman N, Stewart WF, Schulte PA. Cancer Epidemiol Biomarkers Prev. 1995;4:301–311. [PubMed] [Google Scholar]
- 7.Horan JR, Gammill WP. Health Phys. 1963;9:177–186. doi: 10.1097/00004032-196302000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Johnson CH, Gonzalez FJ. J Cell Physiol. doi: 10.1002/jcp.24002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coy SL, Cheema AK, Tyburski JB, Laiakis EC, Collins SP, Fornace A., Jr Int J Radiat Biol. 87:802–823. doi: 10.3109/09553002.2011.556177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patterson AD, Lanz C, Gonzalez FJ, Idle JR. Mass Spectrom Rev. 29:503–521. doi: 10.1002/mas.20272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauer-Jensen M, Kumar KS, Wang J, Berbee M, Fu Q, Boerma M. In: Global Terrorism Issues and Developments. Larche RA, editor. Nova Science Publishers. Inc; New York: 2008. pp. 61–100. [Google Scholar]
- 12.Ghosh SP, Perkins MW, Hieber K, Kulkarni S, Kao TC, Reddy EP, Reddy MV, Maniar M, Seed T, Kumar KS. Radiat Res. 2009;171:173–179. doi: 10.1667/RR1367.1. [DOI] [PubMed] [Google Scholar]
- 13.Flynn RJ. Laboratory animal care. 1963;13(SUPPL):126–129. [PubMed] [Google Scholar]
- 14.Ghosh SP, Kulkarni S, Perkins MW, Hieber K, Pessu RL, Gambles K, Maniar M, Kao TC, Seed TM, Kumar KS. Journal of radiation research. 2012;53:526–536. doi: 10.1093/jrr/rrs001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh PK, Wise SY, Ducey EJ, Fatanmi OO, Elliott TB, Singh VK. Radiat Res. 2012;177:133–145. doi: 10.1667/rr2627.1. [DOI] [PubMed] [Google Scholar]
- 16.Wensinck F, Van Bekkum DW, Renaud H. Radiat Res. 1957;7:491–499. [PubMed] [Google Scholar]
- 17.Landauer MR, Castro CA, Benson KA, Hogan JB, Weiss JF. J Appl Toxicol. 2001;21:25–31. doi: 10.1002/jat.728. [DOI] [PubMed] [Google Scholar]
- 18.Nagy VV. Applied radiation and isotopes : including data, instrumentation and methods for use in agriculture, industry and medicine. 2000;52:1039–1050. doi: 10.1016/s0969-8043(00)00052-x. [DOI] [PubMed] [Google Scholar]
- 19.ISO-ASTM. Standard practice for use of an alanine dosimetry. ISO/ASTM International Standard; 2004. [Google Scholar]
- 20.Kulkarni SS, Cary LH, Gambles K, Hauer-Jensen M, Kumar KS, Ghosh SP. Int Immunopharmacol. 2012;14:495–503. doi: 10.1016/j.intimp.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 22.Kaur P, Sheikh K, Kirilyuk A, Kirilyuk K, Singh R, Ressom H, Cheema A. International Journal of Mass Spectrometry. 2012;310:44–51. [Google Scholar]
- 23.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. Anal Chem. 84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan J, Nunn ME, Su X. Comput Stat Data Anal. 2009;53:1110–1121. doi: 10.1016/j.csda.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R. D. C. Team. R; A Language and Envirinment for Statistical Computing. http://www.r-project.org.
- 26.Johnson CH, Patterson AD, Krausz KW, Lanz C, Kang DW, Luecke H, Gonzalez FJ, Idle JR. Radiat Res. 2011;175:473–484. doi: 10.1667/RR2437.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sieber M, Hoffmann D, Adler M, Vaidya VS, Clement M, Bonventre JV, Zidek N, Rached E, Amberg A, Callanan JJ, Dekant W, Mally A. Toxicol Sci. 2009;109:336–349. doi: 10.1093/toxsci/kfp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patterson AD, Li H, Eichler GS, Krausz KW, Weinstein JN, Fornace AJ, Jr, Gonzalez FJ, Idle JR. Anal Chem. 2008;80:665–674. doi: 10.1021/ac701807v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eichler GS, Huang S, Ingber DE. Bioinformatics. 2003;19:2321–2322. doi: 10.1093/bioinformatics/btg307. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Zheng Y, Li D, Zhong Y. Comput Biol Chem. 2011;35:211–217. doi: 10.1016/j.compbiolchem.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 31.Keszthelyi D, Troost FJ, Masclee AA. Neurogastroenterol Motil. 2009;21:1239–1249. doi: 10.1111/j.1365-2982.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 32.Ralf Dringen BP, Hamprecht Bernd. The Journal of Neuroscience. 1999;19:562–569. doi: 10.1523/JNEUROSCI.19-02-00562.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datta K, Suman S, Kallakury BV, Fornace AJ., Jr PLoS ONE. 7:e42224. doi: 10.1371/journal.pone.0042224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James LA, Lunn PG, Elia M. Br J Nutr. 1998;79:365–372. doi: 10.1079/bjn19980061. [DOI] [PubMed] [Google Scholar]
- 35.Klimberg VS, Salloum RM, Kasper M, Plumley DA, Dolson DJ, Hautamaki RD, Mendenhall WR, Bova FC, Bland KI, Copeland EM, 3rd, et al. Arch Surg. 1990;125:1040–1045. doi: 10.1001/archsurg.1990.01410200104017. [DOI] [PubMed] [Google Scholar]
- 36.Audette-Stuart M, Ferreri C, Festarini A, Carr J. Radiat Res. 178:173–181. doi: 10.1667/rr2820.1. [DOI] [PubMed] [Google Scholar]
- 37.Johnson CA, Balboa MA, Balsinde J, Dennis EA. J Biol Chem. 1999;274:27689–27693. doi: 10.1074/jbc.274.39.27689. [DOI] [PubMed] [Google Scholar]
- 38.Biniek K, Levi K, Dauskardt RH. Proc Natl Acad Sci U S A. doi: 10.1073/pnas.1206851109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klimovich MA, Kozlov MV, Shishkina LN. Radiats Biol Radioecol. 52:58–65. [PubMed] [Google Scholar]
- 40.Kulagina TP. Radiobiologiia. 1990;30:745–748. [PubMed] [Google Scholar]
- 41.Roh C, Yu DK, Kim I, Jo SK. Molecules. 2011;17:145–150. doi: 10.3390/molecules17010145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan TR, Jr, Dempsey DT, Milner R, Ritchie WP., Jr J Surg Res. 1994;56:112–116. doi: 10.1006/jsre.1994.1019. [DOI] [PubMed] [Google Scholar]
- 43.Lanz C, Ledermann M, Slavik J, Idle JR. Int J Radiat Biol. 2011;87:360–371. doi: 10.3109/09553002.2010.537432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lognonne JL, Ducousso R, Rocquet G, Kergonou JF. Biochimie. 1985;67:1015–1021. doi: 10.1016/s0300-9084(85)80296-0. [DOI] [PubMed] [Google Scholar]
- 45.Hahn GL, Menconi MJ, Cahill M, Polgar P. Prostaglandins. 1983;25:783–791. doi: 10.1016/0090-6980(83)90003-5. [DOI] [PubMed] [Google Scholar]
- 46.Dubois RN, Abramson SB, Crofford L, Gupta RA, Simon LS, Van De Putte LB, Lipsky PE. FASEB J. 1998;12:1063–1073. [PubMed] [Google Scholar]
- 47.Burger U, Wolf H, Bauer M. Infusionsther Klin Ernahr. 1978;5:254–260. [PubMed] [Google Scholar]
- 48.Maier K, Hofmann U, Reuss M, Mauch K. BMC Syst Biol. 2010;4:54. doi: 10.1186/1752-0509-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gougelet RM, Rea ME, Nicolalde RJ, Geiling JA, Swartz HM. Health Phys. 2010;98:118–127. doi: 10.1097/HP.0b013e3181a6de7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, Idle JR. Radiat Res. 2009;172:42–57. doi: 10.1667/RR1703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson CH, Patterson AD, Krausz KW, Kalinich JF, Tyburski JB, Kang DW, Luecke H, Gonzalez FJ, Blakely WF, Idle JR. Radiat Res. 2012 doi: 10.1667/rr2950.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu H, Wang Z, Zhang X, Qiao Y, Wu S, Dong F, Chen Y. Radiat Prot Dosimetry. 2012 doi: 10.1093/rpd/ncs138. [DOI] [PubMed] [Google Scholar]
- 53.Tyburski JB, Patterson AD, Krausz KW, Slavik J, Fornace AJ, Jr, Gonzalez FJ, Idle JR. Radiat Res. 2008;170:1–14. doi: 10.1667/RR1265.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fu Q, Berbee M, Boerma M, Wang J, Schmid HA, Hauer-Jensen M. Radiat Res. 2009;171:698–707. doi: 10.1667/RR1685.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramachandran R, Nair CKK. Iran J of Radiat Res. 2012;10:1–10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.