Abstract
Mutations in genes necessary for survival in stationary phase were isolated to understand the ability of wild-type Saccharomyces cerevisiae to remain viable during prolonged periods of nutritional deprivation. Here we report results concerning one of these mutants, rvs167, which shows reduced viability and abnormal cell morphology upon carbon and nitrogen starvation. The mutant exhibits the same response when cells are grown in high salt concentrations and other unfavorable growth conditions. The RVS167 gene product displays significant homology with the Rvs161 protein and contains a SH3 domain at the C-terminal end. Abnormal actin distribution is associated with the mutant phenotype. In addition, while the budding pattern of haploid strains remains axial in standard growth conditions, the budding pattern of diploid mutant strains is random. The gene RVS167 therefore could be implicated in cytoskeletal reorganization in response to environmental stresses and could act in the budding site selection mechanism.
Full text
PDF



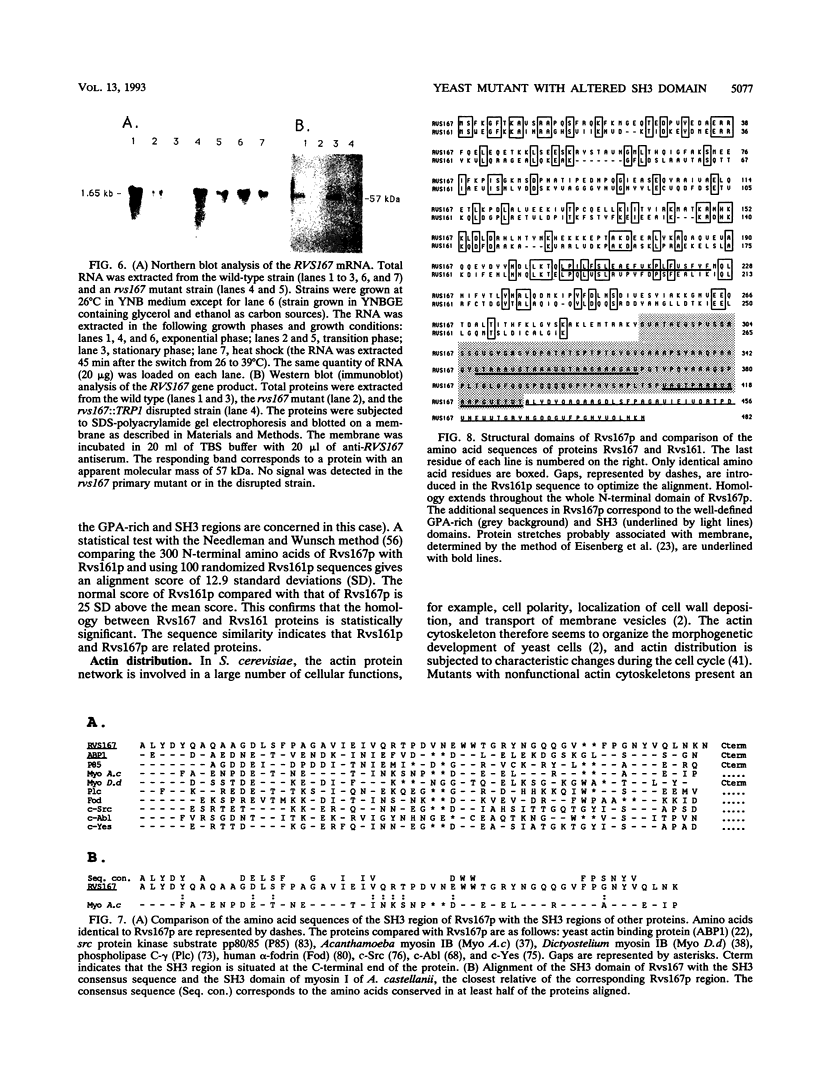



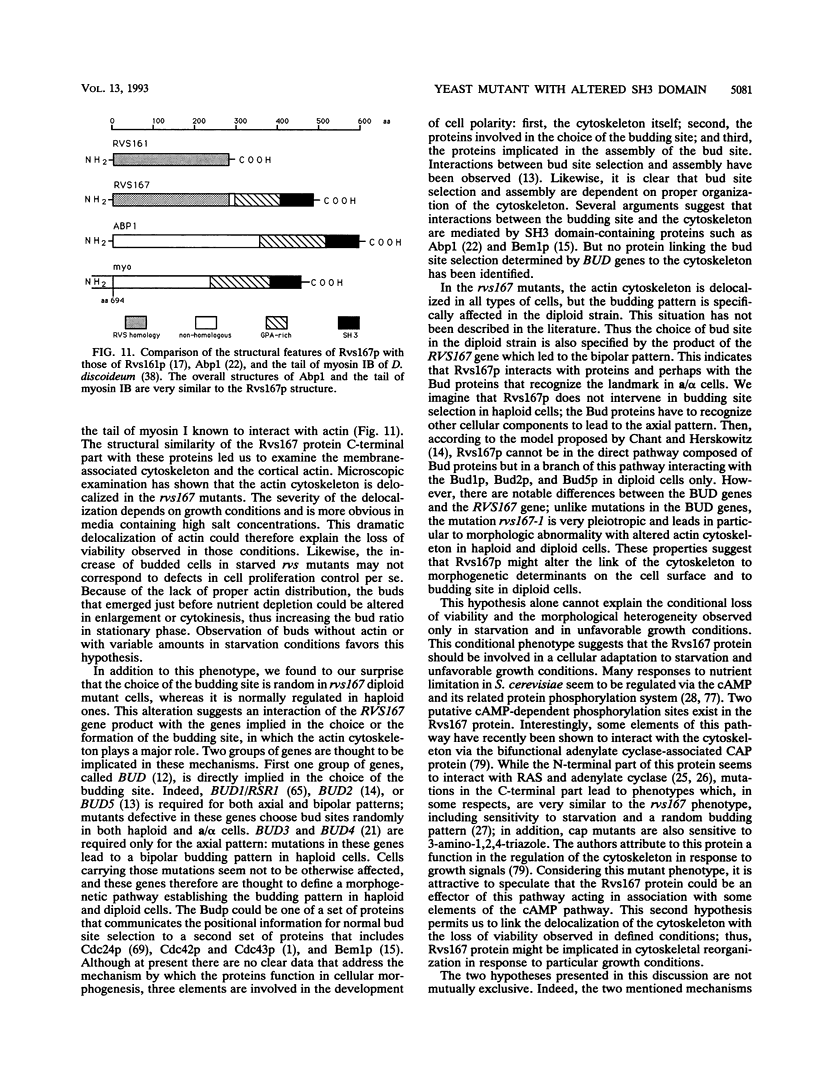



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A. E., Johnson D. I., Longnecker R. M., Sloat B. F., Pringle J. R. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990 Jul;111(1):131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A. E., Pringle J. R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984 Mar;98(3):934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R. J., Pollard T. D. Binding of myosin I to membrane lipids. Nature. 1989 Aug 17;340(6234):565–568. doi: 10.1038/340565a0. [DOI] [PubMed] [Google Scholar]
- Amatruda J. F., Cannon J. F., Tatchell K., Hug C., Cooper J. A. Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature. 1990 Mar 22;344(6264):352–354. doi: 10.1038/344352a0. [DOI] [PubMed] [Google Scholar]
- Amatruda J. F., Gattermeir D. J., Karpova T. S., Cooper J. A. Effects of null mutations and overexpression of capping protein on morphogenesis, actin distribution and polarized secretion in yeast. J Cell Biol. 1992 Dec;119(5):1151–1162. doi: 10.1083/jcb.119.5.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J. U. A method for glycogen determination in whole yeast cells. Anal Biochem. 1978 May;86(1):56–64. doi: 10.1016/0003-2697(78)90318-4. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneu M., Crouzet M., Urdaci M., Aigle M. Direct detection of yeast mutants with reduced viability on plates by erythrosine B staining. Anal Biochem. 1991 Mar 2;193(2):225–230. doi: 10.1016/0003-2697(91)90013-j. [DOI] [PubMed] [Google Scholar]
- Boucherie H. Protein synthesis during transition and stationary phases under glucose limitation in Saccharomyces cerevisiae. J Bacteriol. 1985 Jan;161(1):385–392. doi: 10.1128/jb.161.1.385-392.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D., Toda T., Michaeli T., Levin L., Birchmeier C., Zoller M., Powers S., Wigler M. The S. cerevisiae CDC25 gene product regulates the RAS/adenylate cyclase pathway. Cell. 1987 Mar 13;48(5):789–799. doi: 10.1016/0092-8674(87)90076-6. [DOI] [PubMed] [Google Scholar]
- Cameron S., Levin L., Zoller M., Wigler M. cAMP-independent control of sporulation, glycogen metabolism, and heat shock resistance in S. cerevisiae. Cell. 1988 May 20;53(4):555–566. doi: 10.1016/0092-8674(88)90572-7. [DOI] [PubMed] [Google Scholar]
- Camonis J. H., Kalékine M., Gondré B., Garreau H., Boy-Marcotte E., Jacquet M. Characterization, cloning and sequence analysis of the CDC25 gene which controls the cyclic AMP level of Saccharomyces cerevisiae. EMBO J. 1986 Feb;5(2):375–380. doi: 10.1002/j.1460-2075.1986.tb04222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant J., Corrado K., Pringle J. R., Herskowitz I. Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1. Cell. 1991 Jun 28;65(7):1213–1224. doi: 10.1016/0092-8674(91)90016-r. [DOI] [PubMed] [Google Scholar]
- Chant J., Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991 Jun 28;65(7):1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Chenevert J., Corrado K., Bender A., Pringle J., Herskowitz I. A yeast gene (BEM1) necessary for cell polarization whose product contains two SH3 domains. Nature. 1992 Mar 5;356(6364):77–79. doi: 10.1038/356077a0. [DOI] [PubMed] [Google Scholar]
- Cross F. R. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouzet M., Urdaci M., Dulau L., Aigle M. Yeast mutant affected for viability upon nutrient starvation: characterization and cloning of the RVS161 gene. Yeast. 1991 Oct;7(7):727–743. doi: 10.1002/yea.320070708. [DOI] [PubMed] [Google Scholar]
- Desfarges L., Durrens P., Juguelin H., Cassagne C., Bonneu M., Aigle M. Yeast mutants affected in viability upon starvation have a modified phospholipid composition. Yeast. 1993 Mar;9(3):267–277. doi: 10.1002/yea.320090306. [DOI] [PubMed] [Google Scholar]
- Drubin D. G. Development of cell polarity in budding yeast. Cell. 1991 Jun 28;65(7):1093–1096. doi: 10.1016/0092-8674(91)90001-f. [DOI] [PubMed] [Google Scholar]
- Drubin D. G., Mulholland J., Zhu Z. M., Botstein D. Homology of a yeast actin-binding protein to signal transduction proteins and myosin-I. Nature. 1990 Jan 18;343(6255):288–290. doi: 10.1038/343288a0. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Falco S. C., Li Y., Broach J. R., Botstein D. Genetic properties of chromosomally integrated 2 mu plasmid DNA in yeast. Cell. 1982 Jun;29(2):573–584. doi: 10.1016/0092-8674(82)90173-8. [DOI] [PubMed] [Google Scholar]
- Fedor-Chaiken M., Deschenes R. J., Broach J. R. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell. 1990 Apr 20;61(2):329–340. doi: 10.1016/0092-8674(90)90813-t. [DOI] [PubMed] [Google Scholar]
- Field J., Vojtek A., Ballester R., Bolger G., Colicelli J., Ferguson K., Gerst J., Kataoka T., Michaeli T., Powers S. Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell. 1990 Apr 20;61(2):319–327. doi: 10.1016/0092-8674(90)90812-s. [DOI] [PubMed] [Google Scholar]
- Gerst J. E., Ferguson K., Vojtek A., Wigler M., Field J. CAP is a bifunctional component of the Saccharomyces cerevisiae adenylyl cyclase complex. Mol Cell Biol. 1991 Mar;11(3):1248–1257. doi: 10.1128/mcb.11.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. B., Marshall M. S. The ras oncogene--an important regulatory element in lower eucaryotic organisms. Microbiol Rev. 1989 Jun;53(2):171–185. doi: 10.1128/mr.53.2.171-185.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992 Mar 20;68(6):1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Haarer B. K., Lillie S. H., Adams A. E., Magdolen V., Bandlow W., Brown S. S. Purification of profilin from Saccharomyces cerevisiae and analysis of profilin-deficient cells. J Cell Biol. 1990 Jan;110(1):105–114. doi: 10.1083/jcb.110.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Iida H., Yahara I. Durable synthesis of high molecular weight heat shock proteins in G0 cells of the yeast and other eucaryotes. J Cell Biol. 1984 Jul;99(1 Pt 1):199–207. doi: 10.1083/jcb.99.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida H., Yahara I. Specific early-G1 blocks accompanied with stringent response in Saccharomyces cerevisiae lead to growth arrest in resting state similar to the G0 of higher eucaryotes. J Cell Biol. 1984 Apr;98(4):1185–1193. doi: 10.1083/jcb.98.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet M., Camonis J. Contrôle du cycle de division cellulaire et de la sporulation chez Saccharomyces cerevisiae par le système de l'AMP cyclique. Biochimie. 1985 Jan;67(1):35–43. doi: 10.1016/s0300-9084(85)80228-5. [DOI] [PubMed] [Google Scholar]
- Johnston G. C., Pringle J. R., Hartwell L. H. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977 Mar 1;105(1):79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- Jung G., Korn E. D., Hammer J. A., 3rd The heavy chain of Acanthamoeba myosin IB is a fusion of myosin-like and non-myosin-like sequences. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6720–6724. doi: 10.1073/pnas.84.19.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G., Saxe C. L., 3rd, Kimmel A. R., Hammer J. A., 3rd Dictyostelium discoideum contains a gene encoding a myosin I heavy chain. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6186–6190. doi: 10.1073/pnas.86.16.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T., Powers S., McGill C., Fasano O., Strathern J., Broach J., Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984 Jun;37(2):437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Kieny M. P., Lathe R., Lecocq J. P. New versatile cloning and sequencing vectors based on bacteriophage M13. Gene. 1983 Dec;26(1):91–99. doi: 10.1016/0378-1119(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Kilmartin J. V., Adams A. E. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984 Mar;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C. A., Anderson D., Moran M. F., Ellis C., Pawson T. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science. 1991 May 3;252(5006):668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- Koerner T. J., Hill J. E., Myers A. M., Tzagoloff A. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- Kraft R., Tardiff J., Krauter K. S., Leinwand L. A. Using mini-prep plasmid DNA for sequencing double stranded templates with Sequenase. Biotechniques. 1988 Jun;6(6):544-6, 549. [PubMed] [Google Scholar]
- Kurjan J. Pheromone response in yeast. Annu Rev Biochem. 1992;61:1097–1129. doi: 10.1146/annurev.bi.61.070192.005313. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehto V. P., Wasenius V. M., Salvén P., Saraste M. Transforming and membrane proteins. Nature. 1988 Aug 4;334(6181):388–388. doi: 10.1038/334388a0. [DOI] [PubMed] [Google Scholar]
- Lillie S. H., Pringle J. R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980 Sep;143(3):1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. P., Bretscher A. Disruption of the single tropomyosin gene in yeast results in the disappearance of actin cables from the cytoskeleton. Cell. 1989 Apr 21;57(2):233–242. doi: 10.1016/0092-8674(89)90961-6. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Ishikawa T. Genetic analysis of the role of cAMP in yeast. Yeast. 1985 Sep;1(1):15–24. doi: 10.1002/yea.320010103. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Uno I., Oshima Y., Ishikawa T. Isolation and characterization of yeast mutants deficient in adenylate cyclase and cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2355–2359. doi: 10.1073/pnas.79.7.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh E. M., Haynes R. H. Sequence and expression of the dCMP deaminase gene (DCD1) of Saccharomyces cerevisiae. Mol Cell Biol. 1986 May;6(5):1711–1721. doi: 10.1128/mcb.6.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E. W., Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988 Mar;4(1):11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Novick P., Osmond B. C., Botstein D. Suppressors of yeast actin mutations. Genetics. 1989 Apr;121(4):659–674. doi: 10.1093/genetics/121.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okayama H., Berg P. High-efficiency cloning of full-length cDNA. Mol Cell Biol. 1982 Feb;2(2):161–170. doi: 10.1128/mcb.2.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesset J., Ludwig J. R., Cox B. S., McLaughlin C. S. Effect of cell cycle position on thermotolerance in Saccharomyces cerevisiae. J Bacteriol. 1987 Feb;169(2):779–784. doi: 10.1128/jb.169.2.779-784.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D., Doberstein S. K., Zot H. G. Myosin-I. Annu Rev Physiol. 1991;53:653–681. doi: 10.1146/annurev.ph.53.030191.003253. [DOI] [PubMed] [Google Scholar]
- Pringle J. R. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- Rodaway A. R., Sternberg M. J., Bentley D. L. Similarity in membrane proteins. Nature. 1989 Dec 7;342(6250):624–624. doi: 10.1038/342624a0. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Ruggieri R., Bender A., Matsui Y., Powers S., Takai Y., Pringle J. R., Matsumoto K. RSR1, a ras-like gene homologous to Krev-1 (smg21A/rap1A): role in the development of cell polarity and interactions with the Ras pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Feb;12(2):758–766. doi: 10.1128/mcb.12.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtivelman E., Lifshitz B., Gale R. P., Roe B. A., Canaani E. Alternative splicing of RNAs transcribed from the human abl gene and from the bcr-abl fused gene. Cell. 1986 Oct 24;47(2):277–284. doi: 10.1016/0092-8674(86)90450-2. [DOI] [PubMed] [Google Scholar]
- Sloat B. F., Adams A., Pringle J. R. Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1981 Jun;89(3):395–405. doi: 10.1083/jcb.89.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr Signal transduction in yeast mating: receptors, transcription factors, and the kinase connection. Trends Genet. 1991 Nov-Dec;7(11-12):393–398. [PubMed] [Google Scholar]
- Stahl M. L., Ferenz C. R., Kelleher K. L., Kriz R. W., Knopf J. L. Sequence similarity of phospholipase C with the non-catalytic region of src. Nature. 1988 Mar 17;332(6161):269–272. doi: 10.1038/332269a0. [DOI] [PubMed] [Google Scholar]
- Sudbery P. E., Goodey A. R., Carter B. L. Genes which control cell proliferation in the yeast Saccharomyces cerevisiae. Nature. 1980 Nov 27;288(5789):401–404. doi: 10.1038/288401a0. [DOI] [PubMed] [Google Scholar]
- Sukegawa J., Semba K., Yamanashi Y., Nishizawa M., Miyajima N., Yamamoto T., Toyoshima K. Characterization of cDNA clones for the human c-yes gene. Mol Cell Biol. 1987 Jan;7(1):41–47. doi: 10.1128/mcb.7.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J. M. Fermentable sugars and intracellular acidification as specific activators of the RAS-adenylate cyclase signalling pathway in yeast: the relationship to nutrient-induced cell cycle control. Mol Microbiol. 1991 Jun;5(6):1301–1307. doi: 10.1111/j.1365-2958.1991.tb00776.x. [DOI] [PubMed] [Google Scholar]
- Trueheart J., Boeke J. D., Fink G. R. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987 Jul;7(7):2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek A., Haarer B., Field J., Gerst J., Pollard T. D., Brown S., Wigler M. Evidence for a functional link between profilin and CAP in the yeast S. cerevisiae. Cell. 1991 Aug 9;66(3):497–505. doi: 10.1016/0092-8674(81)90013-1. [DOI] [PubMed] [Google Scholar]
- Wasenius V. M., Saraste M., Salvén P., Erämaa M., Holm L., Lehto V. P. Primary structure of the brain alpha-spectrin. J Cell Biol. 1989 Jan;108(1):79–93. doi: 10.1083/jcb.108.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]
- Wu H., Reynolds A. B., Kanner S. B., Vines R. R., Parsons J. T. Identification and characterization of a novel cytoskeleton-associated pp60src substrate. Mol Cell Biol. 1991 Oct;11(10):5113–5124. doi: 10.1128/mcb.11.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zot H. G., Doberstein S. K., Pollard T. D. Myosin-I moves actin filaments on a phospholipid substrate: implications for membrane targeting. J Cell Biol. 1992 Jan;116(2):367–376. doi: 10.1083/jcb.116.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]