Abstract
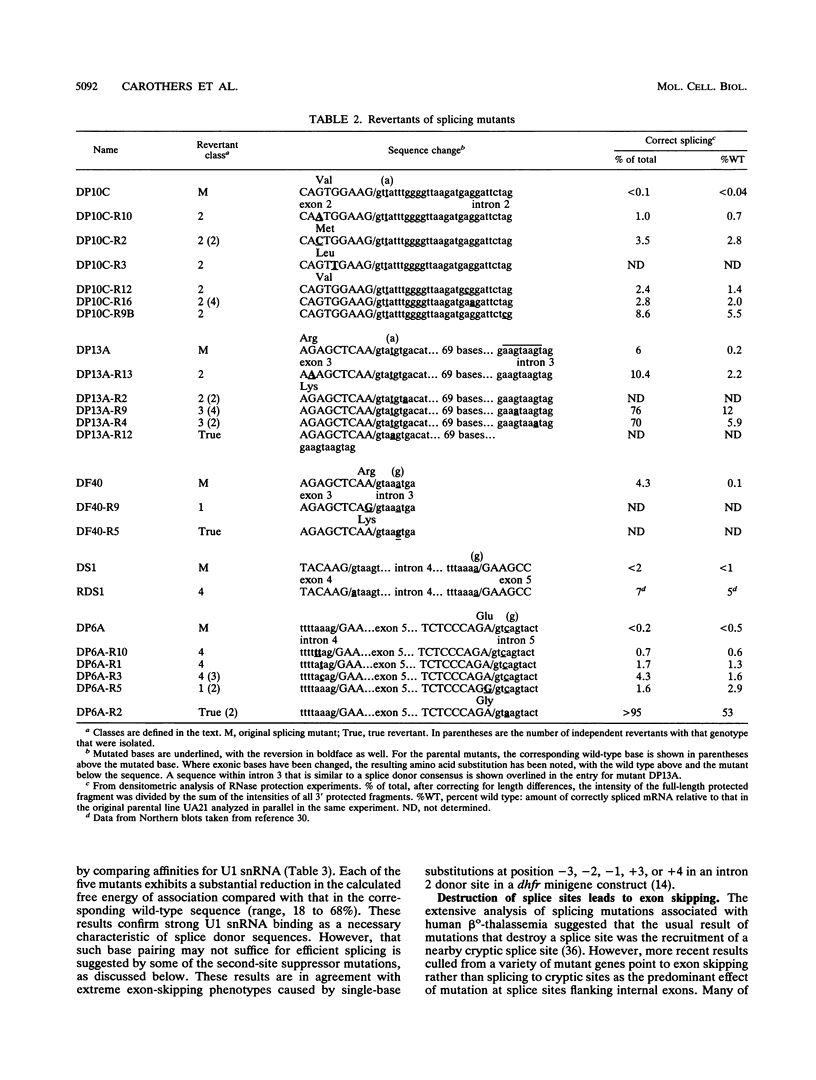
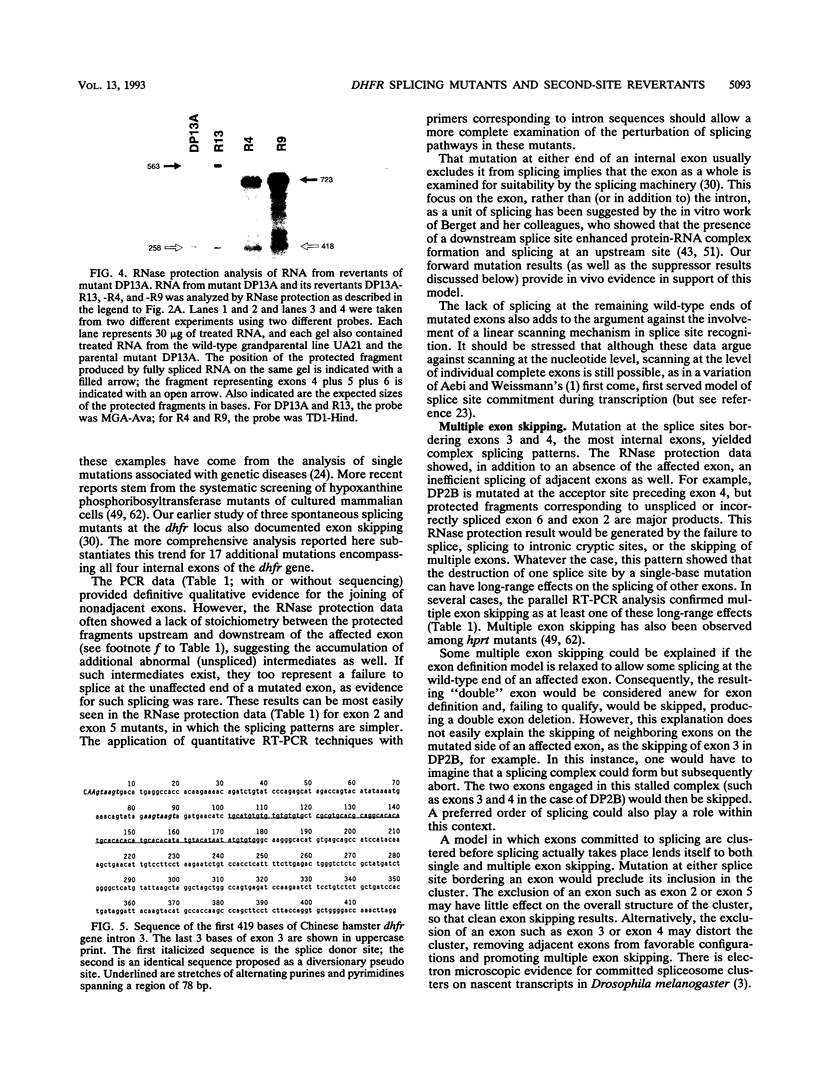
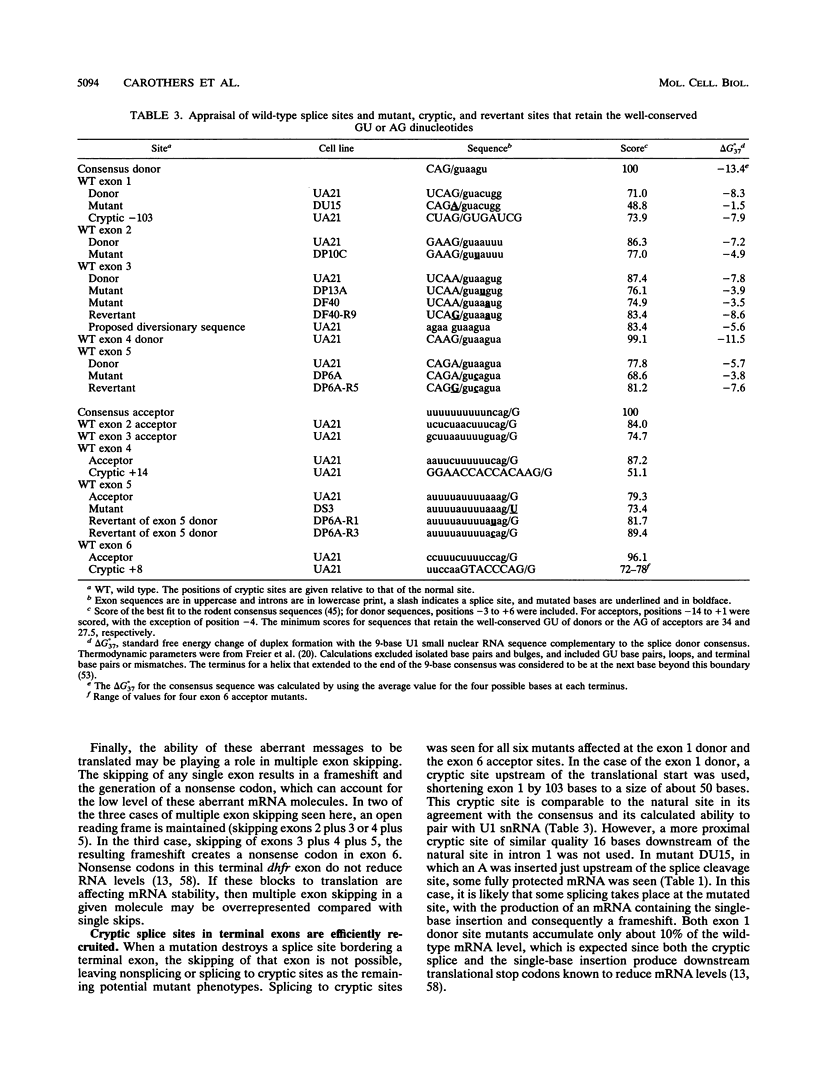
Point mutants induced with a variety of mutagens at the dihydrofolate reductase (dhfr) locus in Chinese hamster ovary (CHO) cells were screened for aberrantly spliced dhfr mRNA by RNase protection and/or reverse transcriptase coupled with cDNA amplification by the polymerase chain reaction (PCR). Of 115 mutants screened, 28 were found to be affected in splicing. All exhibited less than 1% correct splicing, probably because the selection procedure was stringent. All 26 unique mutations were located within the consensus splice sequences; changes were found at 9 of 10 possible sites in this 25-kb six-exon gene. Mutations at the sites flanking the first and last exons resulted in the efficient recruitment of a cryptic site within each exon. In contrast, mutations bordering internal exons caused predominantly exon skipping. In many cases, multiple exons were skipped, suggesting the clustering of adjacent exons prior to actual splicing. Six mutations fell outside the well-conserved GU and AG dinucleotides. All but one were donor site single-base substitutions that decreased the agreement with the consensus and resulted in little or no correct splicing. Starting with five of these donor site mutants, we isolated 31 DHFR+ revertants. Most revertants carried a single-base substitution at a site other than that of the original mutation, and most had only partially regained the ability to splice correctly. The second-site suppression occurred through a variety of mechanisms: (i) a second change within the consensus sequence that produced a better agreement with the consensus; (ii) a change close to but beyond the consensus boundaries, as far as 8 bases upstream in the exon or 28 bases downstream in the intron; (iii) mutations in an apparent pseudo 5' site in the intron, 84 and 88 bases downstream of a donor site; and (iv) mutations that improved the upstream acceptor site of the affected exon. Taken together, these second-site suppressor mutations extend the definition of a splice site beyond the consensus sequence.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashman C. R., Davidson R. L. Sequence analysis of spontaneous mutations in a shuttle vector gene integrated into mammalian chromosomal DNA. Proc Natl Acad Sci U S A. 1987 May;84(10):3354–3358. doi: 10.1073/pnas.84.10.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., Grunberger D. DNA base changes in benzo[a]pyrene diol epoxide-induced dihydrofolate reductase mutants of Chinese hamster ovary cells. Carcinogenesis. 1990 Jan;11(1):189–192. doi: 10.1093/carcin/11.1.189. [DOI] [PubMed] [Google Scholar]
- Carothers A. M., Mucha J., Grunberger D. DNA strand-specific mutations induced by (+/-)-3 alpha,4 beta-dihydroxy- 1 alpha,2 alpha-epoxy-1,2,3,4-tetrahydrobenzo[c]phenanthrene in the dihydrofolate reductase gene. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5749–5753. doi: 10.1073/pnas.88.13.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., Steigerwalt R. W., Urlaub G., Chasin L. A., Grunberger D. DNA base changes and RNA levels in N-acetoxy-2-acetylaminofluorene-induced dihydrofolate reductase mutants of Chinese hamster ovary cells. J Mol Biol. 1989 Aug 5;208(3):417–428. doi: 10.1016/0022-2836(89)90506-8. [DOI] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Ellis N., Chasin L. A. Structure of the dihydrofolate reductase gene in Chinese hamster ovary cells. Nucleic Acids Res. 1983 Apr 11;11(7):1997–2012. doi: 10.1093/nar/11.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Grunberger D., Chasin L. A. Mapping and characterization of mutations induced by benzo[a]pyrene diol epoxide at dihydrofolate reductase locus in CHO cells. Somat Cell Mol Genet. 1988 Mar;14(2):169–183. doi: 10.1007/BF01534402. [DOI] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Mucha J., Grunberger D., Chasin L. A. Point mutation analysis in a mammalian gene: rapid preparation of total RNA, PCR amplification of cDNA, and Taq sequencing by a novel method. Biotechniques. 1989 May;7(5):494-6, 498-9. [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Mucha J., Harvey R. G., Chasin L. A., Grunberger D. Splicing mutations in the CHO DHFR gene preferentially induced by (+/-)-3 alpha,4 beta-dihydroxy-1 alpha,2 alpha-epoxy-1,2,3,4- tetrahydrobenzo[c]phenanthrene. Proc Natl Acad Sci U S A. 1990 Jul;87(14):5464–5468. doi: 10.1073/pnas.87.14.5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carothers A. M., Zhen W., Mucha J., Zhang Y. J., Santella R. M., Grunberger D., Bohr V. A. DNA strand-specific repair of (+-)-3 alpha,4 beta-dihydroxy-1 alpha,2 alpha-epoxy-1,2,3,4-tetrahydrobenzo[c]phenanthrene adducts in the hamster dihydrofolate reductase gene. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11925–11929. doi: 10.1073/pnas.89.24.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasin L. A., Urlaub G., Mitchell P., Ciudad C., Barth J., Carothers A. M., Steigerwalt R., Grunberger D. RNA processing mutants at the dihydrofolate reductase locus in Chinese hamster ovary cells. Prog Clin Biol Res. 1990;340A:295–304. [PubMed] [Google Scholar]
- Chen I. T., Chasin L. A. Direct selection for mutations affecting specific splice sites in a hamster dihydrofolate reductase minigene. Mol Cell Biol. 1993 Jan;13(1):289–300. doi: 10.1128/mcb.13.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciudad C. J., Urlaub G., Chasin L. A. Deletion analysis of the Chinese hamster dihydrofolate reductase gene promoter. J Biol Chem. 1988 Nov 5;263(31):16274–16282. [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991 Jun 28;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Cohen J. B., Broz S. D., Levinson A. D. Expression of the H-ras proto-oncogene is controlled by alternative splicing. Cell. 1989 Aug 11;58(3):461–472. doi: 10.1016/0092-8674(89)90427-3. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., McEwan R. N., Pearson M. L. Expression and amplification of engineered mouse dihydrofolate reductase minigenes. Mol Cell Biol. 1983 Feb;3(2):257–266. doi: 10.1128/mcb.3.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshler J. O., Rossi J. J. Unexpected point mutations activate cryptic 3' splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 1991 Jul;5(7):1252–1263. doi: 10.1101/gad.5.7.1252. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall K. B., Konarska M. M. The 5' splice site consensus RNA oligonucleotide induces assembly of U2/U4/U5/U6 small nuclear ribonucleoprotein complexes. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10969–10973. doi: 10.1073/pnas.89.22.10969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman B. E., Grabowski P. J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3' splice site by a network of interactions spanning the exon. Genes Dev. 1992 Dec;6(12B):2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- Kedes D. H., Steitz J. A. Correct in vivo splicing of the mouse immunoglobulin kappa light-chain pre-mRNA is dependent on 5' splice-site position even in the absence of transcription. Genes Dev. 1988 Nov;2(11):1448–1459. doi: 10.1101/gad.2.11.1448. [DOI] [PubMed] [Google Scholar]
- Krawczak M., Reiss J., Cooper D. N. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992 Sep-Oct;90(1-2):41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Kühne T., Wieringa B., Reiser J., Weissmann C. Evidence against a scanning model of RNA splicing. EMBO J. 1983;2(5):727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K. M., Spritz R. A. RNA splice site selection: evidence for a 5' leads to 3' scanning model. Science. 1983 Jun 24;220(4604):1351–1355. doi: 10.1126/science.6304877. [DOI] [PubMed] [Google Scholar]
- Lear A. L., Eperon L. P., Wheatley I. M., Eperon I. C. Hierarchy for 5' splice site preference determined in vivo. J Mol Biol. 1990 Jan 5;211(1):103–115. doi: 10.1016/0022-2836(90)90014-D. [DOI] [PubMed] [Google Scholar]
- Libri D., Piseri A., Fiszman M. Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991 Jun 28;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Melera P. W., Davide J. P., Hession C. A., Scotto K. W. Phenotypic expression in Escherichia coli and nucleotide sequence of two Chinese hamster lung cell cDNAs encoding different dihydrofolate reductases. Mol Cell Biol. 1984 Jan;4(1):38–48. doi: 10.1128/mcb.4.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Urlaub G., Chasin L. Spontaneous splicing mutations at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol Cell Biol. 1986 Jun;6(6):1926–1935. doi: 10.1128/mcb.6.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasim F. H., Spears P. A., Hoffmann H. M., Kuo H. C., Grabowski P. J. A Sequential splicing mechanism promotes selection of an optimal exon by repositioning a downstream 5' splice site in preprotachykinin pre-mRNA. Genes Dev. 1990 Jul;4(7):1172–1184. doi: 10.1101/gad.4.7.1172. [DOI] [PubMed] [Google Scholar]
- Nelson K. K., Green M. R. Mechanism for cryptic splice site activation during pre-mRNA splicing. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6253–6257. doi: 10.1073/pnas.87.16.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. J., Norman C. U5 snRNA interacts with exon sequences at 5' and 3' splice sites. Cell. 1992 Feb 21;68(4):743–754. doi: 10.1016/0092-8674(92)90149-7. [DOI] [PubMed] [Google Scholar]
- Nigro J. M., Cho K. R., Fearon E. R., Kern S. E., Ruppert J. M., Oliner J. D., Kinzler K. W., Vogelstein B. Scrambled exons. Cell. 1991 Feb 8;64(3):607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- Orkin S. H., Kazazian H. H., Jr The mutation and polymorphism of the human beta-globin gene and its surrounding DNA. Annu Rev Genet. 1984;18:131–171. doi: 10.1146/annurev.ge.18.120184.001023. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Aebi M., Hornig H., Weissmann C., Sharp P. A. Nonconsensus branch-site sequences in the in vitro splicing of transcripts of mutant rabbit beta-globin genes. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8349–8353. doi: 10.1073/pnas.82.24.8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Siliciano P. G. Evidence for an essential non-Watson-Crick interaction between the first and last nucleotides of a nuclear pre-mRNA intron. Nature. 1993 Feb 18;361(6413):660–662. doi: 10.1038/361660a0. [DOI] [PubMed] [Google Scholar]
- Penotti F. E. Human pre-mRNA splicing signals. J Theor Biol. 1991 Jun 7;150(3):385–420. doi: 10.1016/s0022-5193(05)80436-9. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. The role of the mammalian branchpoint sequence in pre-mRNA splicing. Genes Dev. 1988 Oct;2(10):1268–1276. doi: 10.1101/gad.2.10.1268. [DOI] [PubMed] [Google Scholar]
- Reich C. I., VanHoy R. W., Porter G. L., Wise J. A. Mutations at the 3' splice site can be suppressed by compensatory base changes in U1 snRNA in fission yeast. Cell. 1992 Jun 26;69(7):1159–1169. doi: 10.1016/0092-8674(92)90637-r. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Greene J. M., Green M. R. Cryptic branch point activation allows accurate in vitro splicing of human beta-globin intron mutants. Cell. 1985 Jul;41(3):833–844. doi: 10.1016/s0092-8674(85)80064-7. [DOI] [PubMed] [Google Scholar]
- Senapathy P., Shapiro M. B., Harris N. L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- Siebel C. W., Fresco L. D., Rio D. C. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5' splice site control U1 snRNP binding. Genes Dev. 1992 Aug;6(8):1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Solnick D., Lee S. I. Amount of RNA secondary structure required to induce an alternative splice. Mol Cell Biol. 1987 Sep;7(9):3194–3198. doi: 10.1128/mcb.7.9.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsdottir H., Rowley G., Dorado G., Cole J., Lehmann A. R. Mutations which alter splicing in the human hypoxanthine-guanine phosphoribosyltransferase gene. Nucleic Acids Res. 1992 Mar 25;20(6):1201–1208. doi: 10.1093/nar/20.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A. Splicing takes a holliday. Science. 1992 Aug 14;257(5072):888–889. doi: 10.1126/science.1386941. [DOI] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992 Apr 10;256(5054):237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Chasin L. A. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4216–4220. doi: 10.1073/pnas.77.7.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Käs E., Carothers A. M., Chasin L. A. Deletion of the diploid dihydrofolate reductase locus from cultured mammalian cells. Cell. 1983 Jun;33(2):405–412. doi: 10.1016/0092-8674(83)90422-1. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Landzberg M., Chasin L. A. Selective killing of methotrexate-resistant cells carrying amplified dihydrofolate reductase genes. Cancer Res. 1981 May;41(5):1594–1601. [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Ciudad C. J., Chasin L. A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989 Jul;9(7):2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L., Urlaub G., Chasin L. A. Polyadenylation of Chinese hamster dihydrofolate reductase genomic genes and minigenes after gene transfer. Somat Cell Mol Genet. 1987 Sep;13(5):491–504. doi: 10.1007/BF01534491. [DOI] [PubMed] [Google Scholar]
- Winter E., Yamamoto F., Almoguera C., Perucho M. A method to detect and characterize point mutations in transcribed genes: amplification and overexpression of the mutant c-Ki-ras allele in human tumor cells. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7575–7579. doi: 10.1073/pnas.82.22.7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. H., Vrieling H., van Zeeland A. A., Jenssen D. Spectrum of spontaneously occurring mutations in the hprt gene of V79 Chinese hamster cells. J Mol Biol. 1992 Feb 5;223(3):627–635. doi: 10.1016/0022-2836(92)90979-t. [DOI] [PubMed] [Google Scholar]