Abstract
The determination of final organ size is a highly coordinated and complex process that relies on the precise regulation of cell number and/or cell size. Perturbation of organ size control contributes to many human diseases, including hypertrophy, degenerative diseases, and cancer. Hippo and TOR are among the key signaling pathways involved in the regulation of organ size through their respective functions in the regulation of cell number and cell size. Here, we review the general mechanisms that regulate organ growth, describe how Hippo and TOR control key aspects of growth, and discuss recent findings that highlight a possible coordination between Hippo and TOR in organ size regulation.
Introduction
Precise control of organ size is a key feature of metazoans and a crucial process during animal development and regeneration. Classical organ transplantation studies provided the first clues that both intrinsic and extrinsic mechanisms operate in organ size control. For instance, transplantation of multiple fetal thymus glands into a developing mouse results in each thymus gland growing to its characteristic adult size, suggesting an organ-autonomous mechanism for size control [1]. Similarly, Drosophila imaginal discs grown outside their environment attain a normal size even if given additional time to grow, suggesting that growth determinants residing within the imaginal discs provide autonomous growth cues [2]. In contrast, transplantation of multiple spleens into a developing mouse results in the spleens collectively attaining the mass of one adult spleen, indicating a non-autonomous mechanism for organ size regulation [3].
Regeneration studies also revealed both intrinsic and extrinsic mechanisms for the regulation of organ size. For instance, Drosophila imaginal discs or the mammalian liver can regenerate to their original size following removal of part of their mass [4,5], implying that some form of memory is retained in these organs. In contrast, the intestine is incapable of recovering its length following resection, despite the remarkable self-renewal capacity of its stem cells [6]. These studies indicate that the ability to recover size and function following injury varies between organs.
In many cases, the regulation of organ size is achieved by systemic or ‘extrinsic’ factors, which can exert either positive or negative effects on size. In Drosophila and mammals, the rate of growth and final organ size of developing organs are dependent on nutritional status and are controlled by circulating factors, like growth hormone, insulin, and insulin-like growth factor (IGF). In the blood and central nervous system, final organ size is determined primarily by growth factors through the regulation of cell proliferation and apoptosis.
In some cases, progenitor cell number, independently of regulation by growth factors, is the critical determinant of organ size, as shown by studies using genetic methods for altering the number of organ-specific progenitor cells during early embryonic development. For instance, the final size of a pancreas from a primordium with a reduced number of progenitor cells is small whereas that of the liver is normal, suggesting that final pancreas size is controlled by an intrinsic program established early in development that is not subject to growth compensation, whereas final liver size is not limited by reductions in the progenitor cell number [7]. Thus, embryonic progenitor cells may represent a crucial and limiting determinant of some but not all organs.
In this review, we discuss the contributions of cell death, proliferation and growth to the regulation of organ size, and then focus on the roles of the highly conserved Hippo and target of rapamycin (TOR) signaling pathways in organ size control.
Regulation of Organ Size
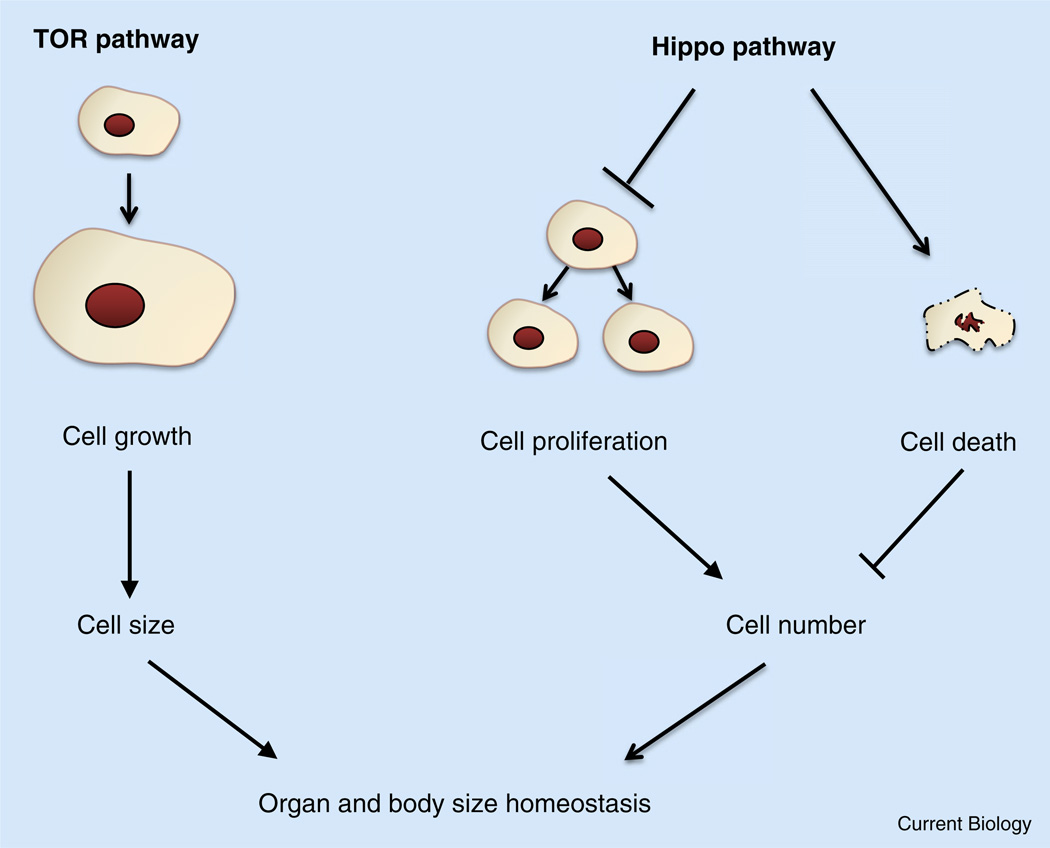
Organ growth is a consequence of increased cell number, cell size, or both(Figure 1).In general, cell number is dependent on the balance between cell proliferation and cell death, whereas cell size is dependent on cell growth.
Figure 1. The TOR and Hippo pathways in organ size control.
TOR regulates organ size by stimulating cell growth, thereby increasing cell size. Hippo controls organ size by restricting cell number via inhibition of proliferation and induction of apoptosis.
Role of Cell Proliferation
Cell proliferation is controlled by extracellular mitogens and inhibitory molecules to ensure that cell division takes place only when more cells are needed. Mitogenic signals, such as epidermal growth factor (EGF), activate intracellular signaling pathways to promote DNA replication and cell-cycle progression [8]. Conversely, inhibitory molecules activate intracellular signaling pathways to block cell-cycle progression and arrest cells in G1 [9]. A family of kinases called cyclin-dependent kinases (CDKs) are cyclically activated to trigger the different phases of the cell cycle, and various cell-cycle checkpoints exist to ensure proper progression [10]. Therefore, cell proliferation signals impinge on CDK activity to control cell division.
An important role of cell proliferation in organ size control is highlighted in transgenic mouse studies. One important regulator of the cell cycle is the p27 gene, which inhibits cyclin–CDK complexes and arrests cells at G1 in response to transforming growth factor β, cell-contact inhibition, and serum deprivation in epithelial cell lines [11]. Mice deficient in p27 have enlarged organs due to increased proliferation rather than decreased cell death or increased cell size [12–14], indicating a critical role of a cell-cycle regulator in proliferation and organ size control. However, it should be noted that overexpression or loss-of-function of most cell-cycle regulators has no effect on organ size, indicating that the cell-cycle machinery may not be the key determinant for organ size regulation.
A role of cell proliferation in organ size control is also demonstrated in transgenic mice in which stabilized β-catenin is overexpressed in neural precursors [15]. These mice have enlarged brains with an increased number of neural precursors. A detailed analysis of these cells reveals that they do not differentiate and are in a proliferative state, suggesting that prevention of cell-cycle exit and cell differentiation may serve as important mechanisms for the regulation of cell proliferation and organ growth. It should be noted that, while cell proliferation clearly plays an important role in organ size control, only a few of the many genes that regulate proliferation have critical functions in organ size control. Thus, the maintenance of organ size is a much more complex process that requires additional inputs and factors beyond cell proliferation.
Role of Cell Death
Apoptosis is a major form of cell death that controls cell number during animal development. Studies in Drosophila indicate that apoptosis is required to attain appropriate wing size during development [16]. This apoptosis-dependent regulation of wing size is achieved via induction of the pro-apoptotic gene hid by the growth regulator dMyc [16], suggesting that modulation of dMyc levels is a mechanism for the regulation of organ size during development. Studies in mammals also confirm a role of apoptosis in organ size control. For example, transgenic mice heterozygous for the gene Pax2 develop kidneys that are considerably smaller than wild-type mice, as a result of increased apoptosis of duct epithelial cells rather than decreased proliferation [17]. Similarly, mutations of the Drosophila homolog of Pax2 result in apoptosis of photoreceptors in the eyes and impair eye development [18], indicating a functional conservation of Pax2 in the regulation of apoptosis and organ size. Thus, in some cases apoptosis is a critical determinant of organ size.
Apoptosis is initiated in response to developmental cues, environmental insults, or a lack of survival factors. Survival factors induce the expression of genes that are important for the suppression of apoptosis, such as the inhibitor of apoptosis (IAP) and members of the Bcl-2 family of proteins. Overexpression of the anti-apoptotic gene bcl2 in mice has been shown to result in enlarged brains with increased numbers of neurons [19–21]. In contrast, overexpression of the pro-apoptotic gene p53 in mice results in smaller kidneys [22]. Many growth factors provide key survival signals for their respective target cells via activation of the phosphoinositide 3-kinase (PI3K) pathway, whereby the downstream target AKT suppresses activation of components of the cell death machinery.
Apoptosis is also induced by signals between cells. Genetic mosaic studies in Drosophila have uncovered a phenomenon called cell competition, in which cells that are otherwise viable get eliminated if their neighboring cells have a growth advantage, as a mechanism for organ size control [16]. For instance, slowly growing cells are eliminated when they are next to cells that grow at a normal rate [23]. Analysis of known signaling pathways has implicated the bone morphogenetic protein (BMP) family member Decapentaplegic (Dpp) and the Jun N-terminal kinase (JNK) pathway in cell competition. More recently, the Hippo pathway has been shown to play a role in cell competition through regulation of dMyc via Yorkie [24,25], with higher dMyc levels providing a competitive advantage.
Coordination of Cell Proliferation and Cell Death in Organ Size Control
Control of cell number is dependent on a balance between cell proliferation and cell death. As such, cell proliferation and cell death must be tightly regulated to maintain organ size. This coordination is particularly important during the process of regeneration, when both inducers of proliferation and inhibitors of apoptosis must be coordinately activated to support organ growth. Several tumor suppressor genes involved in organ size control through regulation of proliferation and apoptosis have been identified using genetic mosaic screens, including components of the recently discovered Hippo signaling pathway, which will be discussed in detail later in the review.
Role of Cell Growth
Cell growth is initiated in response to a myriad of signals, such as extracellular growth factors and nutrient sufficiency. Growth factors bind to cell surface receptors and activate intracellular signaling pathways that ultimately lead to increased protein synthesis and decreased protein degradation. Pioneering studies in Drosophila suggested that inhibition of the translation of a subset of mRNAs through mutation of the Drosophila ribosomal protein p70 S6 kinase(DS6K) alters growth rates, as well as cell and organ size [26]. Mutation of DS6K in larvae resulted in smaller flies that had smaller cells. However, the cell numbers are not significantly changed when compared with the wild-type flies, indicating that cell size, not cell number, accounted for the size change. Central to the regulation of cell size is the TOR kinase, which activates DS6K. In Drosophila, mutations in TOR inhibit larval growth, and this phenotype can be rescued by DS6K overexpression [27]. Another major regulator of growth is the Myc transcription factor [16,28]. Myc induces several genes involved in ribosome biogenesis and protein synthesis, including the eukaryotic translation initiation factors eIF4E and eIF2α [29,30], which are necessary for growth. In mammals overexpression of Myc results in increased liver size characterized by enlarged hepatocyte size [31].It should be noted that, although changes in cell size can have significant effects on organ size, changes in cell number more often account for organ size differences and that, in several cases, modulation of cell growth does not influence organ size (reviewed in [32]).
Coordination of Cell Number and Cell Size
Organ growth is often associated with increases in both cell number and cell size. This was recognized with the identification of the gene encoding Chico, a Drosophila insulin receptor substrate (IRS) protein. Flies mutant for Chico are small with reduced cell size and cell number [33], providing evidence for the role of the IGF pathway in imaginal disc growth control. Acting downstream of Chico, the Drosophila class 1A PI3K Dp110 and its adaptor p60 were also shown to regulate imaginal disc cell size, cell number, and organ size. Mutations of Dp110 and p60 in mitotic clones reduced cell size and cell number, while overexpression of Dp110 increased wing disc size and caused cells to accumulate in G2 phase [34]. Moreover, expression of Dp110 in one compartment of the wing imaginal disc increased not only the size of the compartment but also the size of the disc, indicating that Dp110 activation is sufficient for imaginal disc growth. Two prominent downstream targets of Dp110 are dAKT and dTOR. The PI3K pathway thus coordinately regulates cell number and cell size to promote organ growth. Coordination of cell number and cell size in organ size control is also supported by studies on the gene myostatin, wherein mice with myostatin deletion are larger than wild-type mice and have bigger muscles due to increases in both cell number and cell size [35].
The Hippo Pathway
An Overview
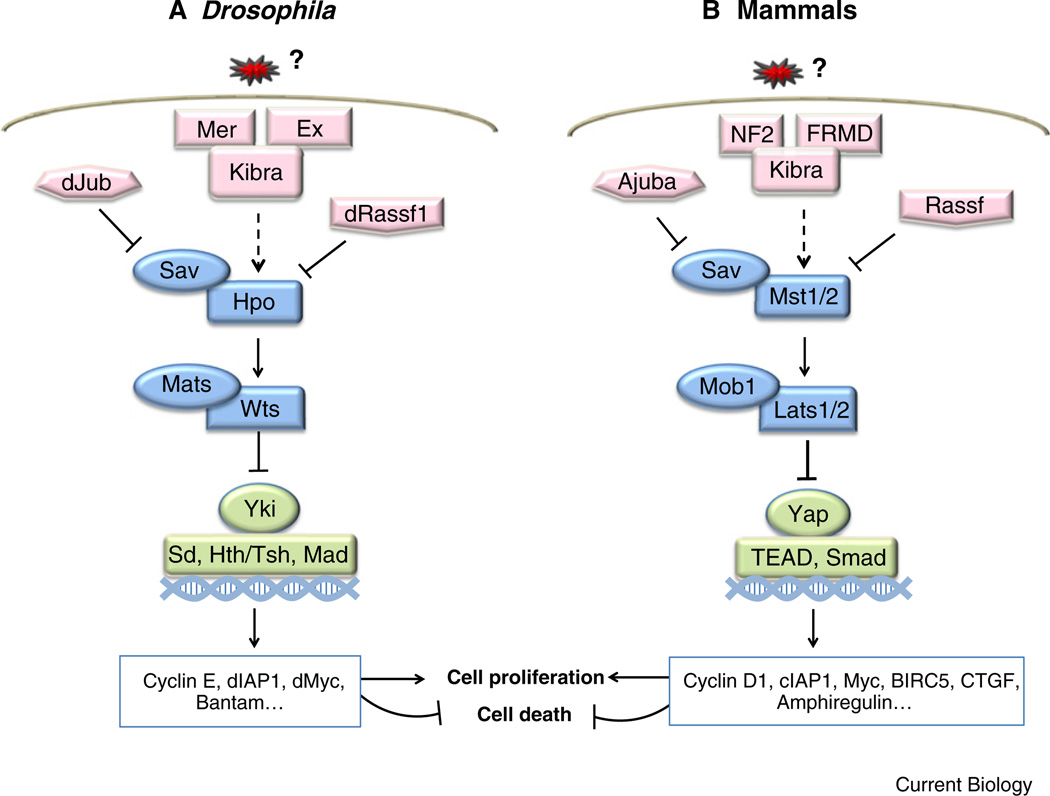
First elucidated in Drosophila, the Hippo pathway consists of the tumor suppressor genes Warts (wts), Salvador (sav), Hippo (hpo), and Mob as tumor suppressor (mats)(Figure 2), mutations of which lead to massive tissue overgrowth. wts encodes a member of the nuclear Dbf2-related (NDR) kinase family [36,37]. Loss of wts leads to robust cell-autonomous overgrowth in various epithelial structures such as the wings, the legs, and the eyes. sav encodes a WW-domain-containing protein. Mutation of sav results in a similar, although weaker, cell-autonomous overgrowth to that observed in wts mutant cones [38,39]. Additionally, loss of wts or sav results in increased proliferation and diminished apoptosis, indicating that wts and sav coordinately regulate both of these cellular processes to control cell number. hpo, from which the pathway name was derived, encodes a Ste20 family protein kinase. hpo exhibits a similar loss-of-function overgrowth phenotype to that reported for sav or wts. In vitro and cell culture-based studies reveal that Hpo phosphorylates and activates Wts, and that Sav potentiates this phosphorylation [40]. Mats, a protein of the Mob1 family, interacts with Wts and potentiates the intrinsic kinase activity of Wts 41] as well as Hippo-mediated growth [42]. Consistently, loss of mats function results in increased cell proliferation, reduced apoptosis, and induction of tissue overgrowth [41], similar to the phenotypes caused by loss of hpo, sav, or wts.
Figure 2. The Hippo pathway in Drosophila and mammals.
Corresponding genes in (A) Drosophila and (B) mammals are shown. Core components, upstream regulators, and downstream targets of the Hippo pathway are labeled in blue, pink, and green boxes, respectively. See text for details.
Loss of wts or sav results in an increase in the expression of the cell-cycle regulator cyclin E and the cell death inhibitor Diap1 [39] at the transcriptional level [40], suggesting an involvement of a transcriptional regulator acting downstream of the Hippo pathway. By yeast two-hybrid screening using Wts as bait, the transcription co-activator Yorkie (Yki) was identified as a major downstream target of Hippo signaling [43]. Overexpression of yki phenocopies the loss of Hippo pathway components and rescues the phenotypes of Hippo pathway activation. Biochemical studies indicate that Wts directly phosphorylates Yki at serine 168, creating a binding site for 14-3-3 proteins, which promote cytoplasmic translocation of Yki [44]. Hence, the Hippo pathway consists of a kinase cascade, in which Hpo interacts with Sav to directly phosphorylate and activate the complex formed by Wts and Sav. In turn, Wts phosphorylates and inactivates the Yki transcription co-activator.
The core components of the Drosophila Hippo pathway are highly conserved in mammals as Mst1/2 (ortholog of Hpo), Sav, Lats1/2 (ortholog of Wts), and Mob1 (ortholog of Mats) (Figure 2). The key downstream effectors of the mammalian Hippo pathway are the Yes-associated protein (Yap) and its paralog TAZ, which function as transcription co-activators. Biochemical studies in the mammalian Hippo pathway similarly establish a kinase cascade whereby Mst1/2 interacts with Sav to phosphorylate and activate the Lats1/2–Mob1 complex (reviewed in [45]). Moreover, Lats1/2 phosphorylates Yap and Taz, resulting in their cytoplasmic sequestration and inactivation [44,46,47]. Consistent with Drosophila studies, mutations of the components of the mammalian Hippo pathway generate tissue overgrowth phenotypes. For instance, loss of both Mst1 and Mst2 results in liver expansion that leads to hepatocellular carcinoma [48]. Transgenic mouse studies also show that activation of Yap in the liver promotes liver growth in an inducible and reversible manner [44,46]. Remarkably, expression of Yap, Lats1, Mst2, and Mob1 can rescue the phenotypes of their corresponding Drosophila mutants in vivo, highlighting a conserved role of the Hippo pathway in organ size control.
Upstream Regulators
Acting upstream of the Hippo pathway are two FERM-domain-containing cytoskeleton binding proteins, Merlin (Mer) and Expanded (Ex). A FERM-domain-binding region is present in Sav [49], suggesting a likely direct interaction between Mer/Ex and Sav. In support of this, Ex has been shown to co-immunoprecipitate with Hpo and Sav [49]. Loss of both Mer and Ex mimics some of the phenotypes of Hippo pathway mutations, such as extra interommatidial cells [50]. However, loss of either Mer or Ex only weakly resembles the extra interommatidial phenotype, indicating that Mer and Ex likely have independent contributions to overgrowth induced by the Hippo pathway. Consistently, Mer mutant clones exhibit defective apoptosis while Ex mutant clones show impaired cell-cycle exit [51].
In mammals the homolog of Mer is the neurofibromatosis 2 (NF2) gene, mutations of which cause an autosomal dominant disorder characterized by the development of benign tumors, such as schwannomas [52,53]. NF2 has been shown to function antagonistically with Yap to regulate liver development. Yap inactivation results in loss of hepatocytes and biliary epithelial cells, whereas NF2 inactivation leads to hepatocellular carcinoma and bile duct hamartoma [54]. Remarkably, the phenotypes induced by NF2 deficiency are suppressed by heterozygous deletion of Yap, thus establishing Yap as a major effector of NF2 in growth regulation.
Loss of a Mer/Ex-interacting protein, Kibra, leads to similar phenotypes as Hippo pathway mutations [49,55,56]. Epistatic analysis reveals that Kibra acts upstream of Hpo and Sav and that overexpression of Kibra results in increased phosphorylation of Wts and Yki [55,56]. Kibra has been shown to function together with Mer and Ex in a protein complex that localizes to the apical domain of epithelial cells, and this protein complex regulates the Hippo kinase cascade via direct binding to Hpo and Sav [49], suggesting the involvement of multiple protein–protein interactions in this regulation.
Recent findings have provided additional insight into the regulation of the Hippo pathway by Ex. A search for Ex-binding proteins using affinity chromatography and mass spectrometry identified Yki as a major Ex-binding protein in Drosophila S2 cells [57]. This interaction, mediated by the WW domain in Yki and the PPxY motif in Ex, results in the nuclear export of Yki independently of Yki S168 phosphorylation, which is critical for 14-3-3 binding. Thus, Ex regulates Yki via the core Hippo pathway components or via a direct interaction.
Another upstream regulator of the Hippo pathway is the Fat protocadherin, a cell surface molecule with multiple cadherin repeats [58–62]. Fat mutants exhibit a mild overgrowth phenotype similar to that of Ex mutants. Fat is proposed to activate the Hippo pathway by regulating the protein level and apical membrane localization of Ex [58,60–62]. The activity of Fat is enhanced upon binding to the protocadherin Dachsous (Ds) [63]. Fat is regulated by several other proteins, including the casein kinase Discs overgrown (Dco), the Golgi-resident kinase Four-joined (Fj), and the Fat/Ds-interacting protein Lowfat (Lft) [64–67], though the functional significance of these interactions remains to be investigated. Most recently, the Ste20-like kinase Tao-1 has been reported to regulate the Hippo pathway to control tissue growth [68]. Tao-1 activates the Hippo signaling through direct phosphorylation of Hpo/Mst [69], providing mechanistic insight into Hippo pathway activation.
Loss of apico-basal polarity is one of the crucial factors that drives epithelial tumor progression. Recent studies have suggested that proteins involved in cell polarity play important roles in the regulation of the Hippo pathway. These cell polarity determinants include the Scribble–Discs large–Lethal giant larvae (Scrib–Dlg–Lgl) protein complex, the atypical protein kinase C (aPKC), and Crumbs (Crb) [64–67]. In Drosophila, depletion of lgl in eye epithelial tissue, where polarity is maintained, results in Yki hyperactivation and, consequently, hyperproliferation and diminished apoptosis [65]. Further, lgl depletion or aPKC overexpression leads to mislocalization of hpo. In contrast, Crb overexpression leads to mislocalization of Ex away from the apical cortex [66,67]. Together, these observations implicate a role for cell polarity determinants in the regulation of tissue growth via the Hippo pathway.
α-Catenin, a component of the adherens junction, has recently been identified as a Yap-interacting protein that regulates Yap localization and activity [70,71]. It binds Yap in high-density human keratinocytes, and is a critical determinant of Yap nuclear activity. α-Catenin, 14-3-3 protein, and phosphorylated Yap can form a complex in the cytoplasm and disruption of this complex by loss of α-catenin results in Yap1 dephosphorylation by protein phosphatase 2A (PP2A) and subsequent nuclear translocation of Yap [71]. Interestingly, α-catenin does not affect the activation status of Mst1/2 and Lats1/2. Moreover, Yap1 is non-responsive to Mst1/2 and Lats1/2 depletion in epidermal cells, raising the intriguing question of the identity of the kinase that phosphorylates Yap in these cells. E-cadherin has also been shown to regulate Yap localization via catenins and the canonical Hippo pathway [72]. Moreover, Yap has been reported to interact with and be regulated by the tight junction protein angiomotin [73]. These findings suggest potential mechanisms for cell-contact-induced Yap inactivation.
Downstream Targets
As a transcription co-activator, Yki does not possess a DNA-binding domain and must interact with transcription factors to stimulate gene expression. One transcription factor that interacts with Yki is Scalloped (Sd), a critical regulator of proliferation and survival of wing imaginal disc cells [74]. On the other hand, Sd is largely dispensable for the normal growth of imaginal discs, a function for which Yki is critical, thus suggesting that other DNA-binding transcription factors regulate gene expression in response to basal levels of Yki. Transcription factors that interact with Yki and regulate Yki target gene expression have recently been reported, including Homothorax (Hth) and Smad proteins [75,76].
In Drosophila the Yki target genes cyclin E and Diap1 do not account for the overgrowth phenotype induced by Yki activation because cyclin E overexpression combined with inhibition of apoptosis does not result in the tissue overgrowth that is characteristic of Hpo inactivation or Yki activation [77]. A search for additional targets of Yki identified the bantam microRNA (miRNA) as a critical biological target of Yki. Two independent studies provide evidence to establish that bantam is an important transcriptional target of Yki. Bantam expression is increased by Yki overexpression, and loss of bantam partially suppresses Yki-induced overproliferation, while bantam overexpression partially rescues the growth defects caused by yki mutants [78,79]. The fact that bantam only partially rescues these defects is probably due to the contribution of other targets, such as cyclin E and Diap1. Consistently, simultaneous overexpression of bantam, cyclin E, and Diap1 results in synergetic tissue overgrowth[78]. Hth, which is important for cell survival and proliferation anterior to the morphogenetic furrow in the eye imaginal disc, mediates the induction of bantam expression by yki [75]. It should be noted that there is no mammalian homologue of the bantam miRNA. It remains to be investigated whether Yap also regulates a miRNA and, if so, whether the human homologue of Hth mediates this function.
In mammals, the TEAD1–4 transcription factors have been shown to interact with and mediate Yap-dependent gene expression [80]. The connective tissue growth factor (CTGF) has also been identified as a direct target of Yap/TEADs. CTGF plays an important role in Yap-induced proliferation and anchorage-independent growth [80]. However, CTGF alone does not account for the overgrowth phenotypes induced by Yap, indicating the existence of additional key targets of Yap in organ size regulation. Another transcriptional target of Yap is amphiregulin (AREG), a ligand for the epidermal growth factor receptor (EGFR). AREG induction has been shown to contribute to Yap-mediated cell proliferation and knockdown of AREG abrogates the effects of Yap overexpression [81]. Yap is also known to induce other genes, including survivin and cyclin D1, that may contribute to cell survival and proliferation [44].
Competition between different cell populations within a growing organ is proposed as a mechanism for organ size control. Studies have linked the Hippo pathway to cell competition by identifying dMyc, a potent inducer of ribosome biogenesis and cell growth, as a transcriptional target of Yki–Sd [24,25]. Interestingly, Yap also induces Myc expression in transgenic mouse liver, although the mechanism has not been reported [44]. In Drosophila local expression of dMyc induces cell competition and leads to death of nearby wild-type cells in developing wings [16]. Consistently, the Hippo pathway is implicated to play a role in cell competition and transcriptional induction of dMyc by Yki is required for the competitive behavior of yki-expressing cells. This finding is particularly important because it reveals a Yki target gene that is directly involved in cell growth.
The Hippo Pathway in Organ Size Control
A distinctive phenotype of Hippo pathway mutations is the dramatic overgrowth in the imaginal discs and in adult organs (reviewed in [82–84]). The adult heads of flies with Hippo pathway mutations appear larger compared with other structures, and the mutant cells proliferate faster to outcompete the normal wild-type cells. When compared with wild-type cells, the Hippo pathway mutants exhibit a dramatic increase in the numbers of interommatidial cells. Moreover, these mutant cells fail to stop proliferating even when imaginal tissues have reached their normal size, and are resistant to apoptosis. The effects of Hippo pathway mutants are observed in other structures, such as the wings, legs, and thorax [39,85,86], suggesting that the Hippo pathway is ubiquitously required for organ size regulation.
In mammals, activation of Yap in the liver promotes liver growth in an inducible and reversible manner [44,46]. Loss of both Mst1 and Mst2 and ablation of Mer or Sav in mice also result in liver expansion [48,54,87–89]. Remarkably, loss of one or both copies of Yap suppresses liver enlargement induced by Mer deficiency [54]. Most of the overgrowth phenotypes of Hippo pathway mutations are characterized by increased proliferation and diminished apoptosis. This role of the Hippo pathway in coordinating proliferation and apoptosis is crucial during regeneration. In mice, biliary ductal epithelial cells make a significant contribution to liver regeneration after injury. Interestingly, a tissue-specific knockout of Yap in the mouse liver causes a defect in bile duct development [54]. Yap expression is also induced during intestinal damage, and loss of Yap severely impairs intestinal regeneration induced by dextran sodium sulfate [90], highlighting an important function of Yap in growth control and regeneration.
Proliferation of tissue-specific stem cells is tightly regulated during development and regeneration to produce organs of predetermined size. Accumulating evidence supports a role for the Hippo pathway in regulating stem/ progenitor cell self-renewal and expansion. Yap activation is observed in induced pluripotent (iPS) cells and knockdown of Yap in mouse embryonic stem (ES) cells leads to loss of pluripotency [91]. In contrast, Yap is inactivated in differentiated mouse ES cells and ectopic expression of Yap prevents mouse ES cell differentiation [91]. In the intestine where endogenous Yap expression is restricted to the progenitor/stem cell compartment, activation of Yap leads to expansion of multipotent undifferentiated progenitor cells and these progenitor cells differentiate when Yap expression ceases [46]. These findings establish a function for Yap in inhibiting progenitor/stem cell differentiation.
A recent study also highlights a critical role of Yap in the regulation of epidermal stem cell proliferation and skin expansion. In a transgenic mouse model, activation of Yap in the skin causes epidermal thickening, characterized by expansion of basal epidermal progenitor cells, and leads to formation of squamous cell carcinoma-like tumors [71]. In contrast, knockout of Yap fails to expand basal epidermal progenitor cells. The hyperplasia in the skin induced by Yap is mediated by interaction with TEAD transcription factors. Consistently, knock-in of a Yap mutant defective in TEAD binding in the mouse skin results in reduced proliferation of epidermal basal cells and failure of skin expansion. The important function of the Hippo pathway in progenitor cell expansion is also demonstrated by studies on liver-specific NF2 deletion, as well as on Mst1/2 and Sav [87,89,92]. Most recently, Taz has been suggested to play a crucial role in cancer stem cell function. Taz activity, which correlates with metastasis, is required to sustain self-renewal and tumor-initiating properties of breast cancer cells [93].
The Hippo pathway has also been suggested to promote cell competition, which influences organ growth. Cell competition suggests that the properties of individual cells are monitored during development and that variant clones of progenitor cells can be favored or eliminated accordingly. It has been reported that cells carrying different doses of myc exhibit different behaviors [16]. Studies in Drosophila show that hypomorphic myc mutants, although viable, are outcompeted by wild-type cells in mosaics [28]. Recently, dMyc has been identified as a transcriptional target and mediator of Yki-induced cell competition [24,25]. Interestingly, dMyc in turn represses the expression of yki such that high levels of dMyc repress yki expression through transcriptional and posttranscriptional mechanisms [25]. This functional coordination of Yki and dMyc activities may serve as an important mechanism of organ size control.
The TOR Pathway
An Overview
Rapamycin is an immunosuppressant capable of initiating cell-cycle arrest in eukaryotic cells. The TOR kinase, mutations of which relieve the growth-suppressive effects of rapamycin, was first identified in yeast [94,95]. Rapamycin requires binding to a cellular cofactor, FK506 binding protein 12kDa (FKBP12), which is capable of directly binding to TOR causing potent inhibition of TOR activity [96]. Functionally, the TOR kinase acts as a central signaling hub, adjusting cellular metabolic output to match growth factor signaling as well as energy and nutrient availability. Hyperactivation of the TOR pathway results in increased cell growth and can cause some cells to enter cell cycle [97–99].
TOR is a large atypical serine-threonine protein kinase, which forms two complexes — TORC1 and TORC2 — in yeast and mammalian cells. Rapamycin specifically inhibits TORC1, whereas the TORC2 complex is resistant to short-term rapamycin treatment [100]. The mammalian TORC1 complex consists of mTOR, regulatory associated protein of mTOR (Raptor), proline rich AKT substrate 40kDa (PRAS40), mammalian lethal with Sec-13 protein 8 (mLST8 also known as GbL), and DEP-domain TOR-binding protein (DEPTOR) [101–105]. The Raptor subunit is essential for TORC1 activity and promotes the formation of the TORC1 complex and substrate binding [106–108]. PRAS40 binding to TORC1 in vitro inhibits TOR activation, and growth factor depletion represses TORC1 activity in part through PRAS40 [109,110]. mLST8 binds to mTOR and may be involved in the activation of mTOR in response to amino acids [107]. DEPTOR is capable of inhibiting both TORC1 and TORC2, and its degradationispromotedbyTORC1andTORC2 [105].
TORC2 shares common subunits with TORC1, including mTOR, DEPTOR, and mLST8. However, several unique TORC2 complex members have been described, including rapamycin-insensitive companion of mTOR (Rictor), Sin1, and protein binding Rictor (Protor) [105, 111–114]. The binding of Rictor and Raptor to mTOR are exclusive to the TORC2 and TORC1 complexes, respectively. Protor associates with TORC2 through direct binding to Rictor. However, the function of Protor is elusive as it is not required for TORC2 assembly or activity [113]. Sin1 has been described to promote Rictor–mTOR interaction and regulate the substrate specificity of TORC2 [115].
The majority of biological functions attributed to mTOR are a result of TORC1 activity due to the availability of rapamycin as a TORC1-specific inhibitor. TORC1 plays a central role in the regulation of cell growth by stimulating ribosome biogenesis and protein translation. TORC1 also plays an important function in autophagy, a catabolic process involving degradation of cellular components that is required to maintain essential cellular functions under periods of nutrient deprivation. Evidence suggests that TORC1 inhibits autophagic machinery through regulation of the mammalian ortholog of yeast ATG1 (ULK1/2), a serine threonine kinase that plays a key role in autophagy initiation [116–118]. By inhibiting ULK1, TORC1 suppresses autophagic degradation, therefore maintaining cell size.
Upstream Regulators
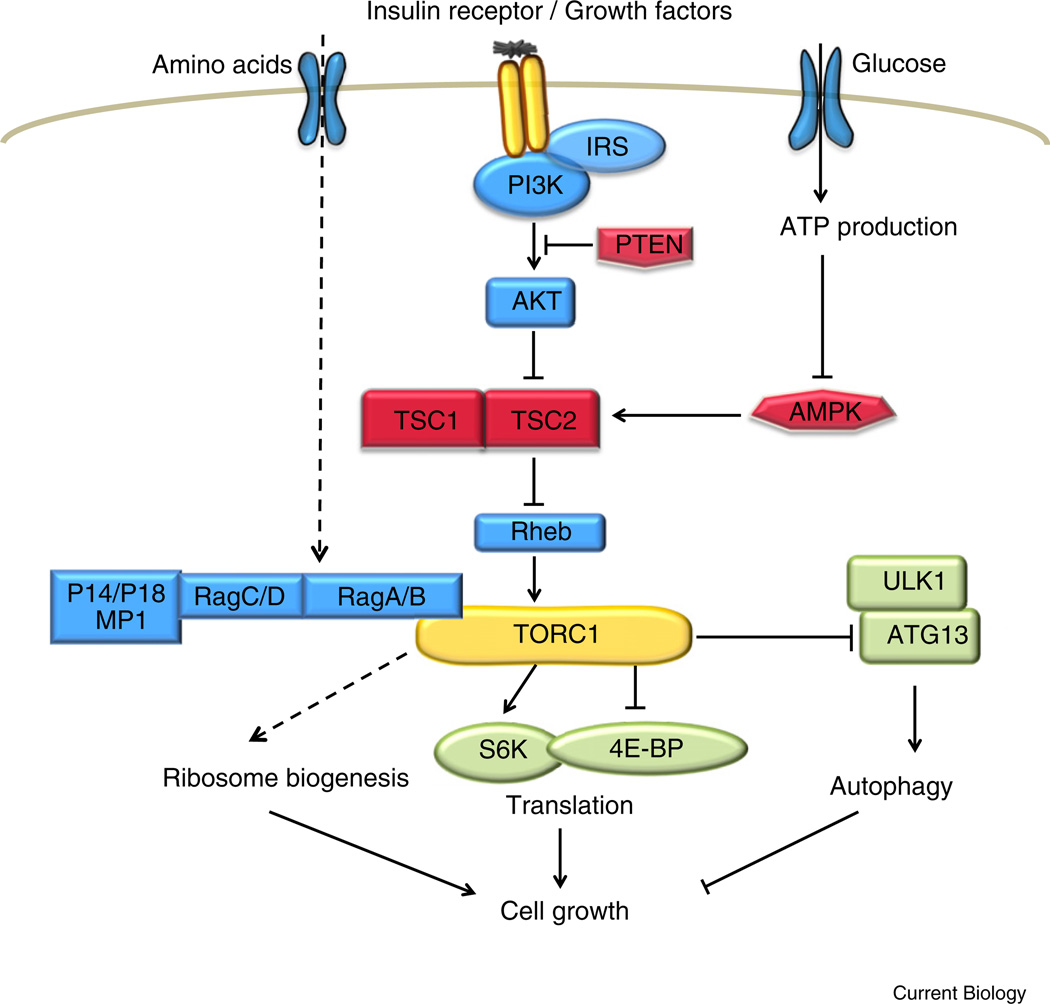
Upstream regulators of TORC1 signaling include growth factors, energy levels, nutrients, and oxygen (Figure 3). Collectively, these cellular cues are integrated by TORC1 to balance cellular energy consumption (ribosome biogenesis and translation) and cellular energy production (autophagic production of metabolites). Most of the signaling events, with the exception of growth factor signaling, are selective to TORC1 but do not affect TORC2.
Figure 3. The TOR pathway.
Genes that activate or inhibit TOR are labeled in blue and red boxes, respectively. Downstream targets of TOR are labeled in green boxes. See text for details.
The PI3K–AKT pathway is a key upstream activator of TORC1. Activation of AKT by PI3K promotes TORC1 activity by phosphorylating the tuberous sclerosis complex (TSC) protein TSC2. TSC is a target of AKT and is a potent repressor of TORC1 kinase activity in Drosophila and mammals [99, 119–121]. The TSC1–TSC2 complex functions as a GTPase-activating protein (GAP) for Rheb, a small GTPase that acts as a potent activator of TORC1 [122–124]. AKT activation also promotes TORC1 activity by phosphorylating PRAS40, relieving its inhibitory effect on TORC1 kinase activity [121,125,126].
Amino acids are required for cell growth and are potent inducers of TORC1 signaling. Evidence for the mechanistic link between TORC1 activity and amino-acid sufficiency comes from recent studies that identified and characterized the Rag GTPase pathway [127,128]. The Rag family is part of the Ras family of GTPases and consists of four family members (A, B, C, D). Activation of TORC1 by Rag GTPases requires the activity of Rheb, indicating that Rag proteins act in the same pathway as Rheb in the activation of TORC1 by amino acids. Other proteins have been reported to have a role in the regulation of TORC1 activity in response to amino acids, including VPS34 lipid kinase and the Ste20-related kinase MAP4K3 [129–133]. Interestingly, VPS34 is involved in endocytic vesicle transport and may affect the lysosomal activation of TORC1 by Rags. Additional studies are required to clarify the role of these genes in TORC1 activation.
The AMP-activated protein kinase (AMPK) plays an important role in cellular energy homeostasis. Under conditions of energy depletion, AMP levels are elevated and the corresponding response is activation of AMPK, which reduces energy-intensive processes, such as ribogenesis and translation [134]. AMPK inhibits the TORC1 complex by increasing TSC1–TSC2 complex activity through direct phosphorylation of TSC2, as well as inhibiting substrate recruitment to TORC1 via phosphorylation of Raptor [135,136]. Moreover, AMPK-mediated phosphorylation of TSC2 enhances the activation of TSC2by glycogen synthase kinase3 (GSK3) [137].
Downstream Targets
The two best characterized targets of TORC1 are the eIF4E-binding protein (4E-BP) and S6K, through which TORC1 regulates the assembly of the translation machinery on a subset of mRNA transcripts [138]. When conditions for growth are unfavorable, TORC1-mediated inhibition of 4E-BP is relieved, allowing 4E-BP to inhibit the pro-translation factor eIF4E and resulting in a decrease in protein translation [138,139]. TORC1 also targets S6K, a kinase that targets multiple substrates required for the progression of the ribosome towards the start codon of mRNA [138–140]. Importantly, TORC1 has also been shown to regulate ribosome biogenesis and tRNA production through increases in phosphorylation of transcriptional regulators of these processes [141,142]. TORC1 also regulates cellular catabolic process, such as inhibition of autophagy, by phosphorylating and inactivating the autophagy-initiating kinase ULK1 [143].
The TOR Pathway in Organ Size Control
Cell division is influenced by cell size. The ability of TORC1 to regulate cell size has been best described in Drosophila models. Disruption of dTORC1 activity results in a reduction of both cell size and cell proliferation, closely mimicking the effects of sustained nutrient deprivation [27]. Disruption of S6K or 4E-BP also results in decreased cell size [27,99,144,145]. Regulation of growth by TORC1 by cues from energy-sensing organs is also demonstrated in the Drosophila model. Deletion of slimfast, an amino-acid transporter, in the fat body of Drosophila results in a TORC1-dependent reduction of cell and organism size [146]. Similarly, rapamycin treatment reduces the size of mammalian cells in culture [140].
Given the central role of TORC1 in regulating growth, it is not surprising that disruption or activation of this pathway is linked to changes in organ size. For instance, deletion of dTOR leads to reduction in growth in larval development [27]. Moreover, dTOR deletion curbs the overgrowth phenotype caused by PTEN loss. Additionally, TSC1 or TSC2 deletion in Drosophila results in overgrowth of organs and body size [147]. Interestingly, lethality in flies harboring a loss-of-function mutation in the IGF-1 receptor can be rescued by deletion of a single TSC1 allele. Genetic studies have revealed that translational regulation by TORC1 is a key contributor to the changes observed in organ and tissue size. Deletion of the TORC1 target S6K decreases body size in Drosophila as a result of decreased cell size rather than decreased cell number [26]. This is in contrast to the dRheb deletion, which reduces both cell size and number, suggesting that TORC1 regulates organ size in a S6K-or translation-independent manner [148,149]. In mice, S6K1 deletion leads to a small body phenotype during embryogenesis that is partially compensated for by increases in S6K2 activity, indicating the conserved role of translational control by TORC1 in the regulation of cell and tissue size [150].
Disruption of insulin signaling to AKT results in retardation of organ and organism size in both Drosophila and mammalian models [151,152]. For example, mutation of IRS-1 in mice or Chico in Drosophila results in smaller body and organ size, highlighting the conservation of insulin signaling in the regulation and promotion of organ growth [153]. Conversely, promotion of insulin–AKT signaling by inactivation of PTEN results in overgrowth of organs and body size due to increased proliferation and cell size. The downstream AKT targets TORC1 and FoxO have both been implicated in the regulation of organ size and tissue overgrowth [27,154].
Interplay Between the Hippo and TOR Pathways
The roles of Hippo and TOR in organ size regulation are well established by their respective functions in controlling cell number and cell size. Emerging evidence suggests that components of the Hippo pathway play an important role in the regulation of TOR activity. For instance, deletion of NF2, the mammalian homologue of Mer, results in the activation of mTORC1 that is associated with meningioma and schwannoma growth [155,156]. Analysis of a panel of malignant mesothelioma cell lines further reveals a strong correlation between loss of Mer and activation of mTORC1 [156]. A recent study also indicates that Yap regulates IGF-1,aknown upstream activator of the mTOR pathway, to promote cardiomyocyte proliferation and embryonic heart size [157]. Similarly, loss of Mst1 or Mst2 leads to mTORC1 activation [48] and Mst1 affects mTORC2 downstream signaling [158]. Mst1 is suggested to promote oxidative-stress-induced cell death in primary mammalian neurons by directly activating Foxo transcription factors, substrates of AKT. Mst1 is shown to phosphorylate Foxo1 proteins at a conserved site within the forkhead domain that disrupts their interaction with 14-3-3 proteins, promotes Foxo nuclear translocation, and induces cell death in neurons [159].
Myc, a transcription factor important for cellular growth, has been identified as a transcriptional target of Yki. Importantly, Yki-induced upregulation of myc is required for the supercompetitive behavior of yki-expressing cells [24,25]. Among the transcriptional targets of Myc are translation initiation factors, which lie downstream of mTORC1 signaling [29,30]. Myc has also been shown to directly affect the transcription of TSC2 by binding to the TSC2 promoter [160]. These studies suggest an essential function of Yki in promoting cell growth via dMyc. It remains to be investigated whether Yap is directly involved in the regulation of mTOR activity.
Cell-cycle progression affects the rate of organ growth. Central to cell-cycle progression is the Retinoblastoma protein (pRB), which exerts its function as a tumor suppressor by promoting cell-cycle exit through control of the activity of the family of E2F transcription factors [161,162]. Overexpression of E2Fs results in cell-cycle advancement fromG1toSphase,and expression of pRB blocks this effect, providing a switch mechanism for the inhibition of cell proliferation [163]. A recent study has demonstrated that Lats2 cooperates with pRB to promote silencing of E2F target genes, resulting in cell cycle arrest [164]. The Lats2 effect does not seem to involve Yap but a novel substrate, DYRK1A. However, in Drosophila Yki itself plays a role in the induction of pRB phenotypes. The Yki–Sd complex has been reported to synergize with and require dE2F to induce a specific transcriptional program that is necessary to bypass the cell-cycle exit [165]. Yki–Sd and dE2F bind directly to the promoters of the Yki–Sd–dE2F shared target genes and activate the expression of these genes in a cooperative manner. Interestingly, E2F has been reported to regulate TORC1 by a transcriptional mechanism [166]. These studies imply that the TOR pathway is a downstream target of Hippo signaling.
Conclusions and Perspectives
As yet, there is no concrete link between the Hippo and TOR pathways that has been defined mechanistically. Hippo and TOR are well-established regulators of organ size, and their prominent roles in organ size control are highlighted by several studies demonstrating that genetic mutation in these pathways is sufficient to alter organ or body size through increases in cell number, cell size, or both. However, the individual roles of TOR and Hippo in organ size control still need to be precisely defined. In the case of TOR, targets beyond S6K, 4EBP1, and ULK1 in cell size control have not been extensively explored. TOR is a key integrator of multiple upstream signals and a regulator of diverse functions, including metabolism and aging. Given that translational control via S6K1 and 4E-BP1 only partially accounts for the role of TOR in maintaining whole-body metabolism, additional TOR targets must exist. The activity of autophagy certainly contributes to overall cellular contents and hence cell size. As for the Hippo pathway, several transcription factors interacting with Yap/Taz/Yki have been reported but the functional significance of most of these interactions has not been extensively studied. In mammals, Yap and Taz are known to induce genes involved in cell proliferation, including BIRC5 and cyclin D1, but it has not been investigated whether these are critical mediators of Yap-dependent functions. Moreover, it is likely that the Lats kinase has additional substrates that may play a role in organ size control.
The upstream regulators of the TOR pathway are well characterized, though in the case of Hippo the upstream signals are missing. Cell contact has been implicated to activate the Hippo pathway, but the key mediators of this regulation are unknown. Additionally, genes that are involved in cell contact, cell adhesion, and cell polarity have been described to affect Hippo pathway activity. However, how and under what conditions these genes activate the Hippo pathway are unclear. As cytoskeletal-associated proteins, the Hippo pathway components Mer and Ex might potentially be involved in relaying mechanical or morphological signals, or signals from as yet unidentified transmembrane receptors to activate the Hippo pathway. In addition, the cytoskeletal pathway involving the Rho GTPase has recently been shown to affect Yap activity, and Lats and Mob1 are suggested to interact with cytoskeletal proteins. However, the specific role of the cytoskeleton in the regulation of the Hippo pathway remains to be elucidated.
Existing evidence has established critical roles of the Hippo and TOR pathways in organ size regulation. Future studies should aim to delineate the mechanistic interactions between the two pathways and upstream signals for the Hippo pathway to establish a better understanding of organ size determination.
References
- 1.Metcalf D. The autonomous behaviour of normal thymus grafts. Aust. J. Exp. Biol. Med. Sci. Suppl. 1963;41:437–447. doi: 10.1038/icb.1963.64. [DOI] [PubMed] [Google Scholar]
- 2.Bryant PJ, Levinson P. Intrinsic growth control in the imaginal primordia of Drosophila, and the autonomous action of a lethal mutation causing overgrowth. Dev. Biol. 1985;107:355–363. doi: 10.1016/0012-1606(85)90317-3. [DOI] [PubMed] [Google Scholar]
- 3.Metcalf D. Restricted growth capacity of multiple spleen grafts. Transplantation. 1964;2:387–392. doi: 10.1097/00007890-196405000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Tsonis PA, Trombley MT, Rowland T, Chandraratna RA, del Rio-Tsonis K. Role of retinoic acid in lens regeneration. Dev. Dyn. 2000;219:588–593. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1082>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 5.Bryant PJ, Simpson P. Intrinsic and extrinsic control of growth in developing organs. Quart. Rev. Biol. 1984;59:387–415. doi: 10.1086/414040. [DOI] [PubMed] [Google Scholar]
- 6.Weale AR, Edwards AG, Bailey M, Lear PA. Intestinal adaptation after massive intestinal resection. Postgrad. Med. J. 2005;81:178–184. doi: 10.1136/pgmj.2004.023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445:886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- 8.Sherr CJ. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 9.Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 10.Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 11.Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, Koff A. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-beta and contact inhibition to cell cycle arrest. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 12.Kiyokawa H, Kineman RD, Manova-Todorova KO, Soares VC, Hoffman ES, Ono M, Khanam D, Hayday AC, Frohman LA, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh DY, Nakayama K. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
- 14.Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 15.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 16.de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- 17.Porteous S, Torban E, Cho NP, Cunliffe H, Chua L, McNoe L, Ward T, Souza C, Gus P, Giugliani R, et al. Primary renal hypoplasia in humans and mice with PAX2 mutations: evidence of increased apoptosis in fetal kidneys of Pax2(1Neu) +/− mutant mice. Hum. Mol. Genet. 2000;9:1–11. doi: 10.1093/hmg/9.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gehring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- 19.Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, Medema RH. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 2002;168:5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 20.Martinou JC, Dubois-Dauphin M, Staple JK, Rodriguez I, Frankowski H, Missotten M, Albertini P, Talabot D, Catsicas S, Pietra C, et al. Overexpression of BCL-2 in transgenic mice protects neurons from naturally occurring cell death and experimental ischemia. Neuron. 1994;13:1017–1030. doi: 10.1016/0896-6273(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 21.Kuida K, Haydar TF, Kuan CY, Gu Y, Taya C, Karasuyama H, Su MS, Rakic P, Flavell RA. Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell. 1998;94:325–337. doi: 10.1016/s0092-8674(00)81476-2. [DOI] [PubMed] [Google Scholar]
- 22.Godley LA, Kopp JB, Eckhaus M, Paglino JJ, Owens J, Varmus HE. Wild-type p53 transgenic mice exhibit altered differentiation of the ureteric bud and possess small kidneys. Genes Dev. 1996;10:836–850. doi: 10.1101/gad.10.7.836. [DOI] [PubMed] [Google Scholar]
- 23.Simpson P, Morata G. Differential mitotic rates and patterns of growth in compartments in the Drosophila wing. Dev. Biol. 1981;85:299–308. doi: 10.1016/0012-1606(81)90261-x. [DOI] [PubMed] [Google Scholar]
- 24.Ziosi M, Baena-Lopez LA, Grifoni D, Froldi F, Pession A, Garoia F, Trotta V, Bellosta P, Cavicchi S, Pession A. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001140. pii: e1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neto-Silva RM, de Beco S, Johnston LA. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev. Cell. 2010;19:507–520. doi: 10.1016/j.devcel.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 2000;14:2712–2724. doi: 10.1101/gad.835000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt EV. The role of c-myc in cellular growth control. Oncogene. 1999;18:2988–2996. doi: 10.1038/sj.onc.1202751. [DOI] [PubMed] [Google Scholar]
- 30.Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S, Li Q, Dang CV, Lee LA. Induction of ribosomal genes and hepatocyte hypertrophy by adenovirus-mediated expression of c-Myc in vivo. Proc. Natl. Acad. Sci. USA. 2000;97:11198–11202. doi: 10.1073/pnas.200372597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 33.Bohni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, Beckingham K, Hafen E. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. 1999;97:865–875. doi: 10.1016/s0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- 34.Weinkove D, Neufeld TP, Twardzik T, Waterfield MD, Leevers SJ. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr. Biol. 1999;9:1019–1029. doi: 10.1016/s0960-9822(99)80450-3. [DOI] [PubMed] [Google Scholar]
- 35.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 36.Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 37.Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 38.Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- 39.Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 40.Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 41.Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 42.Wei X, Shimizu T, Lai ZC. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J. 2007;26:1772–1781. doi: 10.1038/sj.emboj.7601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 47.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, Halder G. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 51.Pellock BJ, Buff E, White K, Hariharan IK. The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Dev. Biol. 2007;304:102–115. doi: 10.1016/j.ydbio.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McClatchey AI, Giovannini M. Membrane organization and tumorigenesis–the NF2 tumor suppressor, Merlin. Genes Dev. 2005;19:2265–2277. doi: 10.1101/gad.1335605. [DOI] [PubMed] [Google Scholar]
- 53.Okada T, You L, Giancotti FG. Shedding light on Merlin’s wizardry. Trends Cell Biol. 2007;17:222–229. doi: 10.1016/j.tcb.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 54.Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev. Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baumgartner R, Poernbacher I, Buser N, Hafen E, Stocker H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 56.Genevet A, Wehr MC, Brain R, Thompson BJ, Tapon N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le Bihan T, McNeill H. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 58.Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 2006;16:2101–2110. doi: 10.1016/j.cub.2006.09.045. [DOI] [PubMed] [Google Scholar]
- 59.Cho E, Feng Y, Rauskolb C, Maitra S, Fehon R, Irvine KD. Delineation of a Fat tumor suppressor pathway. Nat. Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 60.Silva E, Tsatskis Y, Gardano L, Tapon N, McNeill H. The tumor-suppressor gene fat controls tissue growth upstream of expanded in the hippo signaling pathway. Curr. Biol. 2006;16:2081–2089. doi: 10.1016/j.cub.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 61.Willecke M, Hamaratoglu F, Kango-Singh M, Udan R, Chen CL, Tao C, Zhang X, Halder G. The fat cadherin acts through the hippo tumor-suppressor pathway to regulate tissue size. Curr. Biol. 2006;16:2090–2100. doi: 10.1016/j.cub.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 62.Tyler DM, Baker NE. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev. Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matakatsu H, Blair SS. Separating the adhesive and signaling functions of the Fat and Dachsous protocadherins. Development. 2006;133:2315–2324. doi: 10.1242/dev.02401. [DOI] [PubMed] [Google Scholar]
- 64.Chen CL, Gajewski KM, Hamaratoglu F, Bossuyt W, Sansores-Garcia L, Tao C, Halder G. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc. Natl. Acad. Sci. USA. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 66.Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y, Wu S, Pan D. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc. Natl. Acad. Sci. USA. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr. Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poon CL, Lin JI, Zhang X, Harvey KF. The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador-Warts-Hippo pathway. Dev. Cell. 2011;21:896–906. doi: 10.1016/j.devcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 69.Boggiano JC, Vanderzalm PJ, Fehon RG. Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev. Cell. 2011;21:888–895. doi: 10.1016/j.devcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silvis MR, Kreger BT, Lien WH, Klezovitch O, Rudakova GM, Camargo FD, Lantz DM, Seykora JT, Vasioukhin V. alpha-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci. Signal. 2011;4:ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. USA. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- 75.Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oh H, Irvine KD. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev. Cell. 2011;20:109–122. doi: 10.1016/j.devcel.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- 78.Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 79.Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 80.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, Haber DA. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat. Cell Biol. 2009;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 83.Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- 84.Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- 85.Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 86.Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Benhamouche S, Curto M, Saotome I, Gladden AB, Liu CH, Giovannini M, McClatchey AI. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718–1730. doi: 10.1101/gad.1938710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc. Natl. Acad. Sci. USA. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shaw RL, Kohlmaier A, Polesello C, Veelken C, Edgar BA, Tapon N. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;137:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, Kim MC, Jeong WI, Calvisi DF, Kim JM, et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc. Natl. Acad. Sci. USA. 2010;107:8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 94.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 95.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 96.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 97.Soucek T, Pusch O, Wienecke R, DeClue JE, Hengstschlager M. Role of the tuberous sclerosis gene-2 product in cell cycle control. Loss of the tuberous sclerosis gene-2 induces quiescent cells to enter S phase. J. Biol. Chem. 1997;272:29301–29308. doi: 10.1074/jbc.272.46.29301. [DOI] [PubMed] [Google Scholar]
- 98.Soucek T, Rosner M, Miloloza A, Kubista M, Cheadle JP, Sampson JR, Hengstschlager M. Tuberous sclerosis causing mutants of the TSC2 gene product affect proliferation and p27 expression. Oncogene. 2001;20:4904–4909. doi: 10.1038/sj.onc.1204627. [DOI] [PubMed] [Google Scholar]
- 99.Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 2000;14:2689–2694. doi: 10.1101/gad.845700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 101.Takahara T, Hara K, Yonezawa K, Sorimachi H, Maeda T. Nutrient-dependent multimerization of the mammalian target of rapamycin through the N-terminal HEAT repeat region. J. Biol. Chem. 2006;281:28605–28614. doi: 10.1074/jbc.M606087200. [DOI] [PubMed] [Google Scholar]
- 102.Wang L, Rhodes CJ, Lawrence JC., Jr Activation of mammalian target of rapamycin (mTOR) by insulin is associated with stimulation of 4EBP1 binding to dimeric mTOR complex 1. J. Biol. Chem. 2006;281:24293–24303. doi: 10.1074/jbc.M603566200. [DOI] [PubMed] [Google Scholar]
- 103.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell. 2010;38:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Y, Billington CJ, Jr, Pan D, Neufeld TP. Drosophila target of rapamycin kinase functions as a multimer. Genetics. 2006;172:355–362. doi: 10.1534/genetics.105.051979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 107.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 108.Guertin DA, Guntur KV, Bell GW, Thoreen CC, Sabatini DM. Functional genomics identifies TOR-regulated genes that control growth and division. Curr. Biol. 2006;16:958–970. doi: 10.1016/j.cub.2006.03.084. [DOI] [PubMed] [Google Scholar]
- 109.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 2007;9:316–323. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 110.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 111.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 112.Yang Q, Inoki K, Ikenoue T, Guan KL. Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 2006;20:2820–2832. doi: 10.1101/gad.1461206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem. J. 2007;405:513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 115.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 116.Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 2002;4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- 120.Goncharova EA, Goncharov DA, Eszterhas A, Hunter DS, Glassberg MK, Yeung RS, Walker CL, Noonan D, Kwiatkowski DJ, Chou MM, et al. Tuberin regulates p70 S6 kinase activation and ribosomal protein S6 phosphorylation. A role for the TSC2 tumor suppressor gene in pulmonary lymphangioleiomyomatosis (LAM) J. Biol. Chem. 2002;277:30958–30967. doi: 10.1074/jbc.M202678200. [DOI] [PubMed] [Google Scholar]
- 121.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 122.Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- 123.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 124.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 126.Miron M, Lasko P, Sonenberg N. Signaling from Akt to FRAP/TOR targets both 4E-BP and S6K in Drosophila melanogaster. Mol. Cell Biol. 2003;23:9117–9126. doi: 10.1128/MCB.23.24.9117-9126.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Findlay GM, Yan L, Procter J, Mieulet V, Lamb RF. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem. J. 2007;403:13–20. doi: 10.1042/BJ20061881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yan L, Mieulet V, Burgess D, Findlay GM, Sully K, Procter J, Goris J, Janssens V, Morrice NA, Lamb RF. PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol. Cell. 2010;37:633–642. doi: 10.1016/j.molcel.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 131.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 2005;280:33076–33082. doi: 10.1074/jbc.M507201200. [DOI] [PubMed] [Google Scholar]
- 132.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J. Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 135.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 136.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 138.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 139.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 140.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kantidakis T, Ramsbottom BA, Birch JL, Dowding SN, White RJ. mTOR associates with TFIIIC, is found at tRNA and 5S rRNA genes, and targets their repressor Maf1. Proc. Natl. Acad. Sci. USA. 2010;107:11823–11828. doi: 10.1073/pnas.1005188107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mayer C, Zhao J, Yuan X, Grummt I. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 2004;18:423–434. doi: 10.1101/gad.285504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Radimerski T, Montagne J, Hemmings-Mieszczak M, Thomas G. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 2002;16:2627–2632. doi: 10.1101/gad.239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Radimerski T, Montagne J, Rintelen F, Stocker H, van der Kaay J, Downes CP, Hafen E, Thomas G. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat. Cell Biol. 2002;4:251–255. doi: 10.1038/ncb763. [DOI] [PubMed] [Google Scholar]
- 146.Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114:739–749. doi: 10.1016/s0092-8674(03)00713-x. [DOI] [PubMed] [Google Scholar]
- 147.Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- 148.Patel PH, Thapar N, Guo L, Martinez M, Maris J, Gau CL, Lengyel JA, Tamanoi F. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J. Cell Sci. 2003;116:3601–3610. doi: 10.1242/jcs.00661. [DOI] [PubMed] [Google Scholar]
- 149.Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 150.Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S, et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 152.Gao X, Pan D. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 2001;15:1383–1392. doi: 10.1101/gad.901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat. Cell Biol. 1999;1:500–506. doi: 10.1038/70293. [DOI] [PubMed] [Google Scholar]
- 154.Puig O, Tjian R. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 2005;19:2435–2446. doi: 10.1101/gad.1340505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.James MF, Han S, Polizzano C, Plotkin SR, Manning BD, Stemmer-Rachamimov AO, Gusella JF, Ramesh V. NF2/merlin is a novel negative regulator of mTOR complex 1, and activation of mTORC1 is associated with meningioma and schwannoma growth. Mol. Cell Biol. 2009;29:4250–4261. doi: 10.1128/MCB.01581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lopez-Lago MA, Okada T, Murillo MM, Socci N, Giancotti FG. Loss of the tumor suppressor gene NF2, encoding merlin, constitutively activates integrin-dependent mTORC1 signaling. Mol. Cell Biol. 2009;29:4235–4249. doi: 10.1128/MCB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, Bassel-Duby R, Olson EN. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 159.Yuan Z, Lehtinen MK, Merlo P, Villen J, Gygi S, Bonni A. Regulation of neuronal cell death by MST1-FOXO1 signaling. J. Biol. Chem. 2009;284:11285–11292. doi: 10.1074/jbc.M900461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ravitz MJ, Chen L, Lynch M, Schmidt EV. c-myc Repression of TSC2 contributes to control of translation initiation and Myc-induced transformation. Cancer Res. 2007;67:11209–11217. doi: 10.1158/0008-5472.CAN-06-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 162.Nevins JR. The Rb/E2F pathway and cancer. Hum. Mol. Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 163.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 164.Tschop K, Conery AR, Litovchick L, Decaprio JA, Settleman J, Harlow E, Dyson N. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 2011;25:814–830. doi: 10.1101/gad.2000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nicolay BN, Bayarmagnai B, Islam AB, Lopez-Bigas N, Frolov MV. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev. 2011;25:323–335. doi: 10.1101/gad.1999211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Real S, Meo-Evoli N, Espada L, Tauler A. E2F1 regulates cellular growth by mTORC1 signaling. PLoS One. 2011;6:e16163. doi: 10.1371/journal.pone.0016163. [DOI] [PMC free article] [PubMed] [Google Scholar]