Abstract
Several new taxa belonging to the genus Francisella have been described recently. The present study describes the prevalence of Francisella tularensis and Francisella-like endosymbionts (FLE) in ticks collected from Hungary from 2007 to 2009 and characterizes the genetic variability of FLEs. A total of 5402 Ixodid ticks (Ixodes ricinus, I. acuminatus, Dermacentor marginatus, D. reticulatus, Haemaphysalis inermis, H. concinna, H. punctata) were collected from vegetation and animal hosts and tested with conventional PCR, detecting the 16S rRNA and tul4 genes. F. tularensis ssp. holarctica was found in 2 pools of H. concinna and 1 pool of D. reticulatus, both representing minimum prevalence (calculated with 1 infected tick per pool) of 0.27% whereas the sequences of a FLE were detected in 11 pools of D. reticulatus showing a minimum prevalence of 3%. Although the tul4 gene sequence of this FLE was identical to all Hungarian and Portuguese FLEs found earlier, and the 16S rRNA sequence was also identical to the sequence of the endosymbiont of D. reticulatus described in Bulgaria, these 16S rRNA gene coding sequences differed in 2 nucleotides from the one found earlier in this tick species in Hungary. This divergence may appear to be a minor difference between the sequences, potentially even resulting from a technical failure, but it could also indicate a significant difference stemming from the conservative genetic character of Francisellaceae. Thus, it raises a question about the number of FLE variants circulating in D. reticulatus in Europe and indicates the need for further data about the FLEs described in other parts of the continent and new FLE genotyping markers.
Key Words: Francisella-like endosymbiont, Francisella tularensis, Tick, Zoonosis
Introduction
Francisella tularensis is the causative agent of tularemia, a highly infectious zoonotic disease. This bacterium has a worldwide distribution and a broad host spectrum. People can become infected in several ways, such as by direct contact, ingestion or inhalation, via conjunctiva, or through wounds or small cuts. F. tularensis can be transmitted by the infected animals' tissues or fluids, by contaminated water, food, or aerosols, or by the bites of infected arthropods like ticks, which serve both as reservoirs and vectors (Keim et al. 2007). Studies in Central Europe (Austria, Czech Republic, Slovakia) demonstrated a 0.1–2.8% prevalence of F. tularensis in Ixodid ticks (Guryčová et al. 1995, Hubálek et al. 1997).
In the past few years, researchers have explored the genus Francisella, describing new pathogenic taxa from fish and humans and several Francisella variants from ticks and environmental samples with unknown pathogenicity (Keim et al. 2007). Initial attempts at the detection of tularemia based on conventional PCR amplification have led to the misidentification of Francisella-like endosymbionts (FLE) and F. tularensis. However, newer molecular methods (e.g., real-time PCR methods) are more appropriate for the differentiation of these species; otherwise the sequencing of DNA products is essential for the confirmation of the PCR results (Kugeler et al. 2005, Escudero et al. 2008). FLEs were detected in several countries throughout Europe by amplifying the sequences of the 16S rRNA and/or tul4 genes. Endosymbionts of Dermacentor reticulatus were identified in Bulgaria (Ivanov et al. 2011), in Hungary (Sréter-Lancz et al. 2009), in Portugal (de Carvalho et al. 2011), and in Serbia (HM629448, HM629449). Another two new FLEs were described in Bulgaria, 1 from a Hyalomma aegyptium and 1 from Hy. marginatum and Rhipicephalus sanguineus ticks (Ivanov et al. 2011). The aim of this study was to investigate the prevalence of F. tularensis and FLEs and the genetic variability of FLEs in ticks in Hungary.
The DNA of 5402 ticks was available in 552 pools for this examination, from which 5024 were questing ticks (nymphs and adults; 3222 Ixodes ricinus, 369 Dermacentor marginatus, 361 D. reticulatus, 315 Haemaphysalis inermis, 735 H. concinna, 22 H. punctata), collected from 39 different sites in 15 out of the 19 counties in Hungary. The dragging-flagging method was used at fringes of pastures on bushy hillsides, fringes of meadows, and wide paths in mountain forests and at lowland areas, from March until October in 2007 and 2009. A further 374 samples of Ixodes acuminatus were collected from common hamsters (Cricetus cricetus) and 4 D. reticulatus nymphs from dogs during this period.
After identification of the ticks by miscroscopy on the basis of their morphological features, DNA was extracted in pools, using the MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche Diagnostics, Rotkreuz, Switzerland). The pools contained 10 or fewer (when less than 10 remained) ticks, grouped by sampling location, collection date, species, developmental stage, and gender. The samples were tested with the 16S rRNA gene based Francisella-specific conventional PCR, using the primer pairs Fr153/Fr1281 (Barns et al. 2005). Positive samples were further tested with conventional PCR based on the tul4 gene specific for the genus Francisella, using the primer pair FT393/FT642 (Long et al. 1993). The subspecies of the detected F. tularensis strains were confirmed with canonical single-nucleotide polymorphism (canSNP) assays according to Vogler et al. (2009). PCR products were extracted from agarose gel (QIAquick Gel Extraction Kit, Qiagen, Inc., Valencia, CA), and direct cycle sequencing was performed with the primers used for amplification on an ABI PRISM 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA). Nucleic acid databases were searched using the BLASTN program in GenBank. The reading errors of the chromatograms were corrected and alignments (16S rRNA, 1021 bp; tul4, 248 bp) of the obtained DNA sequences were performed with programs of the Lasergene package (DNASTAR Inc., Madison, WI). JModeltest was used to identify nucleotide substitution models best fitting for both groups of sequences (Posada 2008). Based on Akaike information criterion (AIC) the Tamura–Nei 1993 model was chosen for further analysis from a range of models that possessed a 100% confidence interval, built on the models' cumulative weight gained during the calculations (Posada 2008). Phylogenetic analysis was conducted with the neighbor-joining method using the maximum composite likelihood model (equivalent with Tamura–Nei 1993 model) and 1000 bootstraps in MEGA5 software (Tamura et al. 2011).
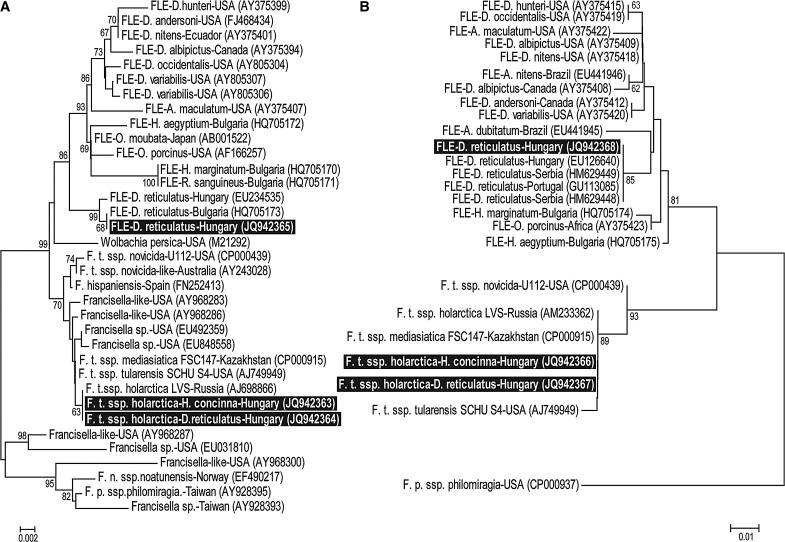
From the 552 tick pools examined, F. tularensis ssp. holarctica was detected in 1 nymph and 1 female H. concinna pool collected from a meadow in southeastern Hungary in 2009, and in 1 D. reticulatus pool of females collected from the environment in southwestern Hungary in 2007. In the case of these positive samples, each representing a minimum prevalence (calculating with only 1 infected tick per pool) of 0.27%, both 16S rRNA and tul4 gene coding regions were sequenced (Fig. 1A, B).
FIG. 1.
Neighbor-joining phylogenetic tree showing relationships of 16S rRNA gene (A) and tul4 gene (B) sequences obtained from Francisella species and Francisella-like endosymbionts (FLEs) with highlights of the Francisella isolates from this study. Bootstrap values of neighbor-joining (1000 replicates) of >60 are shown.
FLEs were found in 11 pools of D. reticulatus ticks collected in 2007 (2 pools of nymphs, 1 pool of males, and 3 pools of females originating from the environment in the northeastern part of Hungary, and 5 pools of females collected from the vegetation in southern counties), showing a minimum prevalence of 3%. Both 16S rRNA and tul4 gene coding sequences were identical in all 11 FLEs (Fig. 1A, B). Comparing obtained sequences with those deposited in GenBank, we found that the 16S rRNA gene sequence was identical to the FLE of a D. reticulatus from Bulgaria (HQ705173; Ivanov et al. 2011) and differed in 2 nucleotides from the endosymbiont found earlier in 3 D. reticulatus samples in Hungary (EU234535; Sréter-Lancz et al. 2009) (Fig. 1A). However, the tul4 gene coding sequences of the present FLEs proved to be identical to the endosymbiont found earlier in Portugal (GU113085; de Carvalho et al. 2011) and in Hungary (EU126640; Sréter-Lancz et al. 2009) (Fig. 1B).
Tularemia is known to be endemic in Hungary (Gyuranecz et al. 2010), and the results of this study corroborate that ticks carrying this pathogen could pose a threat to public health. The prevalence of F. tularensis in the Hungarian tick population was within the range found in the neighboring countries (Guryčová et al. 1995, Hubálek et al. 1997). This relatively low value could be explained with the moderate activity of the disease (seropositivity in the European brown hare [Lepus europaeus] population, 0.66–1.1%; annual number of human cases 20–25, Gyuranecz et al. 2010) during the tick collection period. Francisellaceae have a very conservative genetic character (Keim et al. 2007). The 16S rRNA sequence divergence (2 nucleotides) between the FLEs from D. reticulatus detected earlier in Hungary (Sréter-Lancz et al. 2009) and those described in the present study and earlier in Bulgaria (Ivanov et al. 2011) is equivalent in magnitude to the difference between the type strain (Schu S4, accession number: AJ749949) of the highly virulent F. tularensis ssp. tularensis, found in North America and type strain (LVS, accession number, AM233362) of the moderately virulent F. tularensis ssp. holarctica, found in the Northern Hemisphere. Thus, this is a notable difference that could lead to the hypothesis that there may be 2 distinct FLEs circulating in D. reticulatus populations in Hungary and therefore in Europe. However, the fact that the tul4 gene sequences of D. reticulatus endosymbionts from Hungary were identical in a previous study (Sréter-Lancz et al. 2009) and the present study, and that the samples were collected from the same geographical regions within a relatively short time, suggest that a technical error during sequencing can not be ruled out. Therefore, more information is needed about the tul4 gene coding sequence of the Bulgarian FLE (Ivanov et al. 2011) and the 16S rRNA gene of the Portuguese (de Carvalho et al. 2011) and Serbian FLEs; the identification of novel FLE genetic markers would also be required to decide whether there is only 1 or at least 2 distinct FLEs present in the D. reticulatus population in Europe.
Our knowledge on the genetic diversity in the Francisella genus is expanding rapidly, and this will necessitate progressive adaptation of molecular tools allowing the accurate identification of FLE species in the near future. If only 1 FLE species is harbored by D. reticulatus in Europe and this FLE infects D. reticulatus exclusively, then a host adaptation and a host species–linked evolution of this FLE species could be assumed. This would be quite an interesting finding because the genetically closely related and pathogenic F. tularensis is one of the bacterial agents with the broadest host range (Keim et al. 2007). The GenBank accession numbers of the sequences obtained in this study are JQ942363, JQ942364, JQ942365, JQ942366, JQ942367, and JQ942368.
Acknowledgments
This study was supported by the Lendület program of the Hungarian Academy of Sciences and by the OTKA-78139 grant of the Hungarian Scientific Research Fund. The laboratory work was in part performed with logistic support from the Center for Clinical Studies at the Vetsuisse Faculty of the University of Zurich. S.H. and K.E. are Bolyai János Research Fellows of the Hungarian Academy of Sciences.
Author Disclosure Statement
No competing financial interests exist.
References
- Barns SM. Grow CC. Okinaka RT. Keim P, et al. Detection of diverse new Francisella-like bacteria in environmental samples. Appl Environ Microbiol. 2005;71:5494–5500. doi: 10.1128/AEM.71.9.5494-5500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho IL. Santos N. Soares T. Zé-zé L, et al. Francisella-like endosymbiont in Dermacentor reticulatus collected in Portugal. Vector Borne Zoonot Dis. 2011;11:185–188. doi: 10.1089/vbz.2010.0014. [DOI] [PubMed] [Google Scholar]
- Escudero R. Toledo A. Gil H. Kovácsová K, et al. Molecular method for discrimination between Francisella tularensis and Francisella-like endosymbionts. J Clin Microbiol. 2008;46:3139–3143. doi: 10.1128/JCM.00275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryčová D. Kocianová E. Výrosteková V. Řeháček J. Prevalence of ticks infected with Francisella tularensis in natural foci of tularemia in western Slovakia. Eur J Epidemiol. 1995;11:469–474. doi: 10.1007/BF01721235. [DOI] [PubMed] [Google Scholar]
- Gyuranecz M. Fodor L. Makrai L. Monse L, et al. Epidemiology of tularemia, with special regard to the infection of European brown hare (Lepus europaeus) Magy Állatorv Lapja. 2010;132:39–45. [Google Scholar]
- Hubálek Z. Sixl V. Halouzka J. Mikulášková M. Prevalence of Francisella tularensis in Dermacentor reticulatus ticks collected in adjacent areas of the Czech and Austrian Republics. Centr Eur J Publ Hlth. 1997;5:199–201. [PubMed] [Google Scholar]
- Ivanov IN. Mitkova N. Reye AL. Hübschen JM, et al. Detection of new Francisella-like tick endosymbionts in Hyalomma ssp. and Rhipicephalus spp. (Acari:Ixodidae) from Bulgaria. Appl Environ Microbiol. 2011;77:5562–5565. doi: 10.1128/AEM.02934-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P. Johansson A. Wagner DM. Molecular epidemiology, evolution and ecology of Francisella. Ann NY Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- Kugeler KJ. Gurfield N. Creek JG. Mahoney KS, et al. Discrimination between Francisella tularensis and Francisella-like endosymbionts when screening ticks by PCR. Appl Environ Microbiol. 2005;71:7594–7597. doi: 10.1128/AEM.71.11.7594-7597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long GW. Oprandy JJ. Narayanan RB. Fortier AH, et al. Detection of Francisella tularensis in blood by polymerase chain reaction. J Clin Microbiol. 1993;31:152–154. doi: 10.1128/jcm.31.1.152-154.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Sréter-Lancz Zs. Széll L. Sréter T. Márialigeti K. Detection of a novel Francisella in Dermacentor reticulatus: A need for careful evaluation of PCR-based identification of Francisella tularensis in Eurasian ticks. Vector Borne Zoonot Dis. 2009;9:123–126. doi: 10.1089/vbz.2008.0010. [DOI] [PubMed] [Google Scholar]
- Tamura K. Peterson D. Peterson N. Stecher G, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler AJ. Birdsell D. Price LB. Bowers JR, et al. Phylogeography of Francisella tularensis: global expansion of a highly fit clone. J Bacteriol. 2009;191:2474–2484. doi: 10.1128/JB.01786-08. [DOI] [PMC free article] [PubMed] [Google Scholar]