Abstract
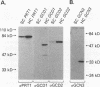
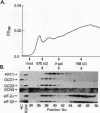
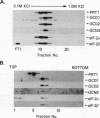
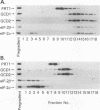
GCN4 is a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae whose expression is regulated by amino-acid availability at the translational level. GCD1 and GCD2 are negative regulators required for the repression of GCN4 translation under nonstarvation conditions that is mediated by upstream open reading frames (uORFs) in the leader of GCN4 mRNA. GCD factors are thought to be antagonized by the positive regulators GCN1, GCN2 and GCN3 in amino acid-starved cells to allow for increased GCN4 protein synthesis. Previous genetic studies suggested that GCD1, GCD2, and GCN3 have closely related functions in the regulation of GCN4 expression that involve translation initiation factor 2 (eIF-2). In agreement with these predictions, we show that GCD1, GCD2, and GCN3 are integral components of a high-molecular-weight complex of approximately 600,000 Da. The three proteins copurified through several biochemical fractionation steps and could be coimmunoprecipitated by using antibodies against GCD1 or GCD2. Interestingly, a portion of the eIF-2 present in cell extracts also cofractionated and coimmunoprecipitated with these regulatory proteins but was dissociated from the GCD1/GCD2/GCN3 complex by 0.5 M KCl. Incubation of a temperature-sensitive gcdl-101 mutant at the restrictive temperature led to a rapid reduction in the average size and quantity of polysomes, plus an accumulation of inactive 80S ribosomal couples; in addition, excess amounts of eIF-2 alpha, GCD1, GCD2, and GCN3 were found comigrating with free 40S ribosomal subunits. These results suggest that GCD1 is required for an essential function involving eIF-2 at a late step in the translation initiation cycle. We propose that lowering the function of this high-molecular-weight complex, or of eIF-2 itself, in amino acid-starved cells leads to reduced ribosomal recognition of the uORFs and increased translation initiation at the GCN4 start codon. Our results provide new insights into how general initiation factors can be regulated to affect gene-specific translational control.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Miller P. F., Jackson B. M., Hinnebusch A. G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol. 1991 Jan;11(1):486–496. doi: 10.1128/mcb.11.1.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad M. F., Nasrin N., Banerjee A. C., Gupta N. K. Purification and properties of eukaryotic initiation factor 2 and its ancillary protein factor (Co-eIF-2A) from yeast Saccharomyces cerevisiae. J Biol Chem. 1985 Jun 10;260(11):6955–6959. [PubMed] [Google Scholar]
- Anthony D. D., Jr, Kinzy T. G., Merrick W. C. Affinity labeling of eukaryotic initiation factor 2 and elongation factor 1 alpha beta gamma with GTP analogs. Arch Biochem Biophys. 1990 Aug 15;281(1):157–162. doi: 10.1016/0003-9861(90)90426-y. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Donahue T. F. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cigan A. M., Pabich E. K., Feng L., Donahue T. F. Yeast translation initiation suppressor sui2 encodes the alpha subunit of eukaryotic initiation factor 2 and shares sequence identity with the human alpha subunit. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2784–2788. doi: 10.1073/pnas.86.8.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti A., Baglioni C. Phosphorylation of initiation factor eIF-2 alpha, binding of mRNA to 48 S complexes, and its reutilization in initiation of protein synthesis. J Biol Chem. 1983 Dec 10;258(23):14556–14562. [PubMed] [Google Scholar]
- Dholakia J. N., Wahba A. J. Phosphorylation of the guanine nucleotide exchange factor from rabbit reticulocytes regulates its activity in polypeptide chain initiation. Proc Natl Acad Sci U S A. 1988 Jan;85(1):51–54. doi: 10.1073/pnas.85.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann C. L., Tzagoloff A. Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J Biol Chem. 1985 Feb 10;260(3):1513–1520. [PubMed] [Google Scholar]
- Donahue T. F., Cigan A. M., Pabich E. K., Valavicius B. C. Mutations at a Zn(II) finger motif in the yeast eIF-2 beta gene alter ribosomal start-site selection during the scanning process. Cell. 1988 Aug 26;54(5):621–632. doi: 10.1016/s0092-8674(88)80006-0. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feinberg B., McLaughlin C. S., Moldave K. Analysis of temperature-sensitive mutant ts 187 of Saccharomyces cerevisiae altered in a component required for the initiation of protein synthesis. J Biol Chem. 1982 Sep 25;257(18):10846–10851. [PubMed] [Google Scholar]
- Foiani M., Cigan A. M., Paddon C. J., Harashima S., Hinnebusch A. G. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Jun;11(6):3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M., Redman R., Kaplansky D. A. Evidence that the primary effect of phosphorylation of eukaryotic initiation factor 2(alpha) in rabbit reticulocyte lysate is inhibition of the release of eukaryotic initiation factor-2.GDP from 60 S ribosomal subunits. J Biol Chem. 1985 Aug 5;260(16):9491–9500. [PubMed] [Google Scholar]
- Gross M., Rubino M. S. Regulation of eukaryotic initiation factor-2B activity by polyamines and amino acid starvation in rabbit reticulocyte lysate. J Biol Chem. 1989 Dec 25;264(36):21879–21884. [PubMed] [Google Scholar]
- Gross M., Wing M., Rundquist C., Rubino M. S. Evidence that phosphorylation of eIF-2(alpha) prevents the eIF-2B-mediated dissociation of eIF-2 X GDP from the 60 S subunit of complete initiation complexes. J Biol Chem. 1987 May 15;262(14):6899–6907. [PubMed] [Google Scholar]
- Hanic-Joyce P. J., Singer R. A., Johnston G. C. Molecular characterization of the yeast PRT1 gene in which mutations affect translation initiation and regulation of cell proliferation. J Biol Chem. 1987 Feb 25;262(6):2845–2851. [PubMed] [Google Scholar]
- Hannig E. M., Hinnebusch A. G. Molecular analysis of GCN3, a translational activator of GCN4: evidence for posttranslational control of GCN3 regulatory function. Mol Cell Biol. 1988 Nov;8(11):4808–4820. doi: 10.1128/mcb.8.11.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E. M., Williams N. P., Wek R. C., Hinnebusch A. G. The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics. 1990 Nov;126(3):549–562. doi: 10.1093/genetics/126.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima S., Hannig E. M., Hinnebusch A. G. Interactions between positive and negative regulators of GCN4 controlling gene expression and entry into the yeast cell cycle. Genetics. 1987 Nov;117(3):409–419. doi: 10.1093/genetics/117.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima S., Hinnebusch A. G. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Nov;6(11):3990–3998. doi: 10.1128/mcb.6.11.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., McLaughlin C. S. A mutant of yeast apparently defective in the initiation of protein synthesis. Proc Natl Acad Sci U S A. 1969 Feb;62(2):468–474. doi: 10.1073/pnas.62.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. E., Struhl K. Molecular characterization of GCD1, a yeast gene required for general control of amino acid biosynthesis and cell-cycle initiation. Nucleic Acids Res. 1988 Oct 11;16(19):9253–9265. doi: 10.1093/nar/16.19.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A., Safer B. Purification of the eukaryotic initiation factor 2-eukaryotic initiation factor 2B complex and characterization of its guanine nucleotide exchange activity during protein synthesis initiation. J Biol Chem. 1983 Mar 10;258(5):3402–3408. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Martin T. E., Hartwell L. H. Resistance of active yeast ribosomes to dissociation by KCl. J Biol Chem. 1970 Mar 25;245(6):1504–1506. [PubMed] [Google Scholar]
- Matts R. L., Levin D. H., London I. M. Effect of phosphorylation of the alpha-subunit of eukaryotic initiation factor 2 on the function of reversing factor in the initiation of protein synthesis. Proc Natl Acad Sci U S A. 1983 May;80(9):2559–2563. doi: 10.1073/pnas.80.9.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldave K. Eukaryotic protein synthesis. Annu Rev Biochem. 1985;54:1109–1149. doi: 10.1146/annurev.bi.54.070185.005333. [DOI] [PubMed] [Google Scholar]
- Mueller P. P., Harashima S., Hinnebusch A. G. A segment of GCN4 mRNA containing the upstream AUG codons confers translational control upon a heterologous yeast transcript. Proc Natl Acad Sci U S A. 1987 May;84(9):2863–2867. doi: 10.1073/pnas.84.9.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Paddon C. J., Hannig E. M., Hinnebusch A. G. Amino acid sequence similarity between GCN3 and GCD2, positive and negative translational regulators of GCN4: evidence for antagonism by competition. Genetics. 1989 Jul;122(3):551–559. doi: 10.1093/genetics/122.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon C. J., Hinnebusch A. G. gcd12 mutations are gcn3-dependent alleles of GCD2, a negative regulator of GCN4 in the general amino acid control of Saccharomyces cerevisiae. Genetics. 1989 Jul;122(3):543–550. doi: 10.1093/genetics/122.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent S. A., Fenimore C. M., Bostian K. A. Vector systems for the expression, analysis and cloning of DNA sequences in S. cerevisiae. Yeast. 1985 Dec;1(2):83–138. doi: 10.1002/yea.320010202. [DOI] [PubMed] [Google Scholar]
- Peterson D. T., Merrick W. C., Safer B. Binding and release of radiolabeled eukaryotic initiation factors 2 and 3 during 80 S initiation complex formation. J Biol Chem. 1979 Apr 10;254(7):2509–2516. [PubMed] [Google Scholar]
- Rowlands A. G., Montine K. S., Henshaw E. C., Panniers R. Physiological stresses inhibit guanine-nucleotide-exchange factor in Ehrlich cells. Eur J Biochem. 1988 Jul 15;175(1):93–99. doi: 10.1111/j.1432-1033.1988.tb14170.x. [DOI] [PubMed] [Google Scholar]
- Salimans M., Goumans H., Amesz H., Benne R., Voorma H. O. Regulation of protein synthesis in eukaryotes. Mode of action of eRF, an eIF-2-recycling factor from rabbit reticulocytes involved in GDP/GTP exchange. Eur J Biochem. 1984 Nov 15;145(1):91–98. doi: 10.1111/j.1432-1033.1984.tb08526.x. [DOI] [PubMed] [Google Scholar]
- Siekierka J., Manne V., Ochoa S. Mechanism of translational control by partial phosphorylation of the alpha subunit of eukaryotic initiation factor 2. Proc Natl Acad Sci U S A. 1984 Jan;81(2):352–356. doi: 10.1073/pnas.81.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N. S., Matts R. L., Levin D. H., London I. M. The 60 S ribosomal subunit as a carrier of eukaryotic initiation factor 2 and the site of reversing factor activity during protein synthesis. J Biol Chem. 1985 Aug 15;260(17):9860–9866. [PubMed] [Google Scholar]
- Thomas N. S., Matts R. L., Petryshyn R., London I. M. Distribution of reversing factor in reticulocyte lysates during active protein synthesis and on inhibition by heme deprivation or double-stranded RNA. Proc Natl Acad Sci U S A. 1984 Nov;81(22):6998–7002. doi: 10.1073/pnas.81.22.6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D., Alexandraki D., Thireos G. Multiple cis-acting elements modulate the translational efficiency of GCN4 mRNA in yeast. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4849–4853. doi: 10.1073/pnas.83.13.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D., Roussou I., Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989 Jun 16;57(6):947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Warner J. R., Mitra G., Schwindinger W. F., Studeny M., Fried H. M. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol. 1985 Jun;5(6):1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Jackson B. M., Hinnebusch A. G. Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N. P., Hinnebusch A. G., Donahue T. F. Mutations in the structural genes for eukaryotic initiation factors 2 alpha and 2 beta of Saccharomyces cerevisiae disrupt translational control of GCN4 mRNA. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7515–7519. doi: 10.1073/pnas.86.19.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfner M., Yep D., Messenguy F., Fink G. R. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J Mol Biol. 1975 Aug 5;96(2):273–290. doi: 10.1016/0022-2836(75)90348-4. [DOI] [PubMed] [Google Scholar]