Abstract
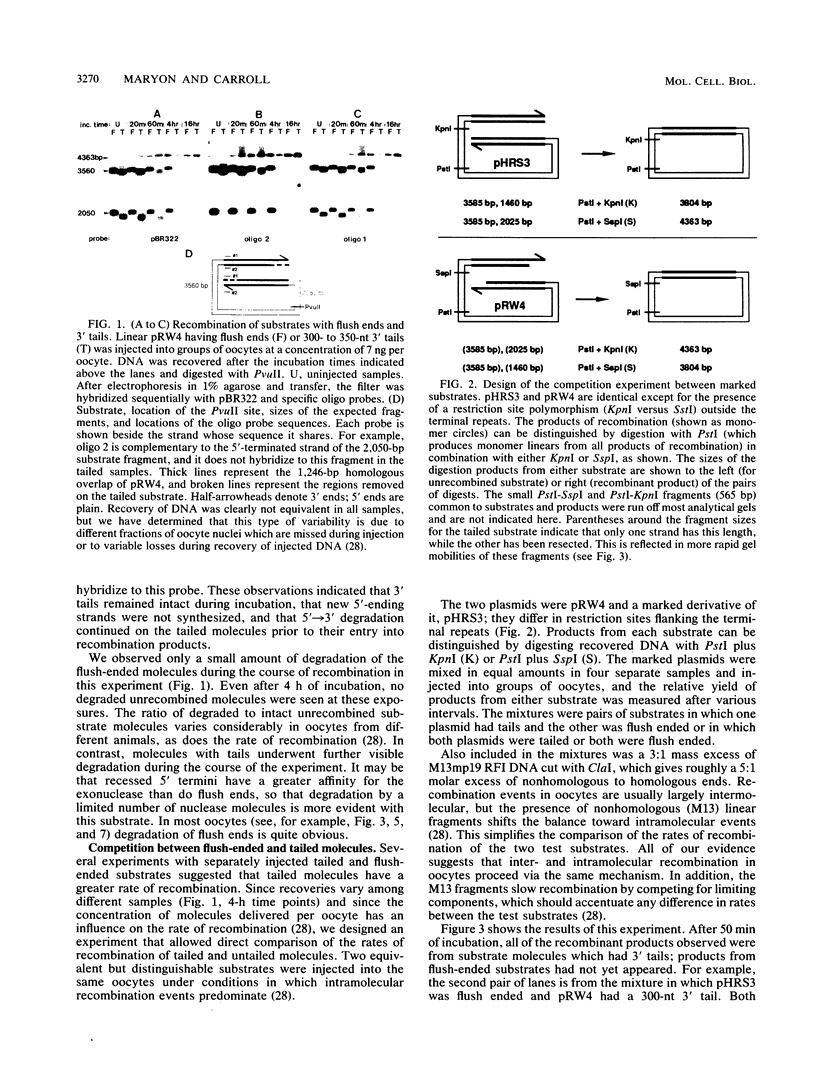
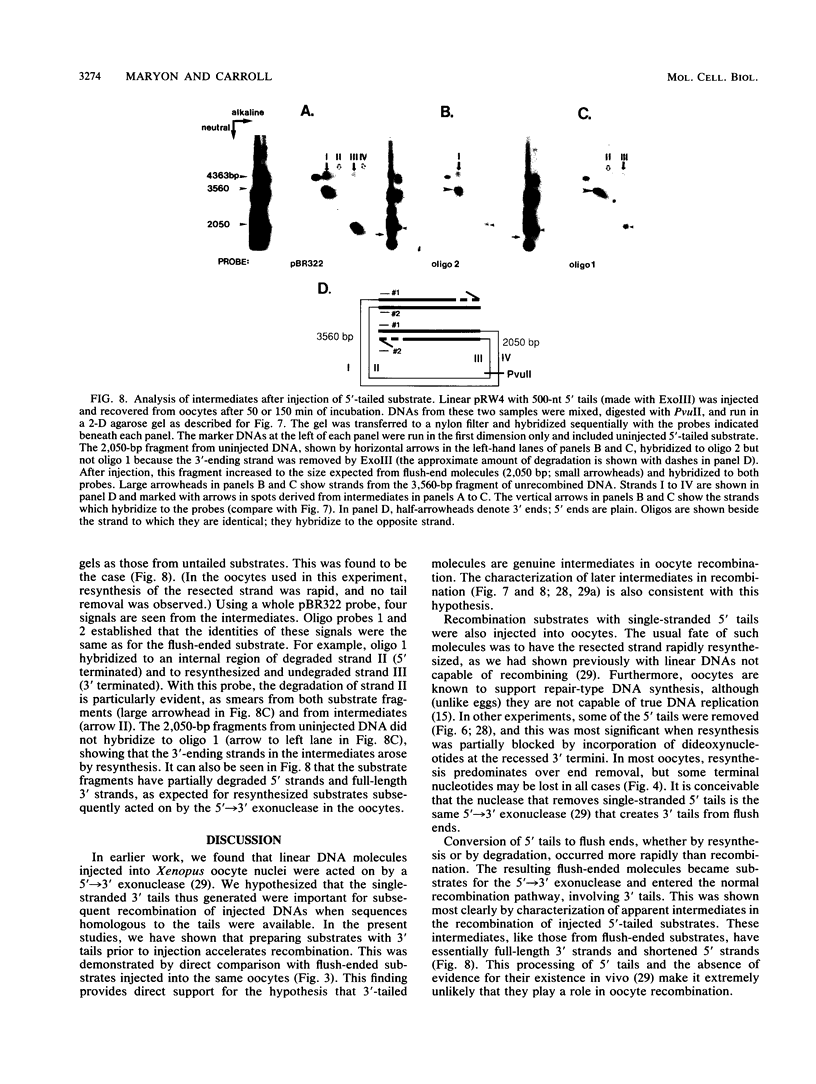
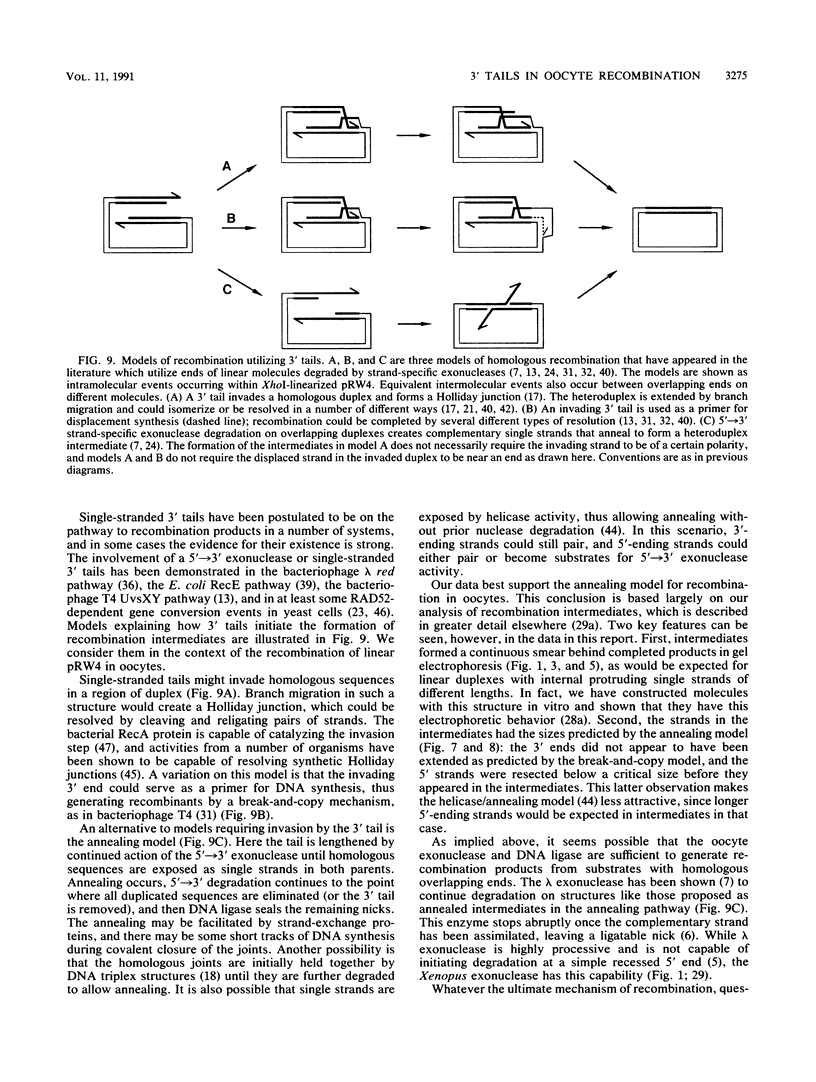
Homologous recombination of DNA molecules injected into Xenopus laevis oocyte nuclei is extremely efficient when those molecules are linear and have overlapping homologous ends. It was previously shown that a 5'----3' exonuclease activity in oocytes attacks injected linear DNAs and leaves them with single-stranded 3' tails. We tested the hypothesis that such tailed molecules are early intermediates on the pathway to recombination products. Substrates with 3' tails were made in vitro and injected into oocytes, where they recombined rapidly and efficiently. In experiments with mixed substrates, molecules with 3' tails entered recombination intermediates and products more rapidly than did molecules with flush ends. Molecules endowed in vitro with 5' tails also recombined efficiently in oocytes, but their rate was not faster than for flush-ended substrates. In most cases, the 5' tails served as templates for resynthesis of the 3' strands, regenerating duplex ends which then entered the normal recombination pathway. In oocytes from one animal, some of the 5' tails were removed, and this was exacerbated when resynthesis was partially blocked. Analysis by two-dimensional gel electrophoresis of recombination intermediates from 5'-tailed substrates confirmed that they had acquired 3' tails as a result of the action of the 5'----3' exonuclease. These results demonstrate that homologous recombination in oocytes proceeds via a pathway that involves single-stranded 3' tails. Molecular models incorporating this feature are discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. A., Eliason S. L. Recombination of homologous DNA fragments transfected into mammalian cells occurs predominantly by terminal pairing. Mol Cell Biol. 1986 Sep;6(9):3246–3252. doi: 10.1128/mcb.6.9.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur M., Potrykus I., Paszkowski J. Intermolecular homologous recombination in plants. Mol Cell Biol. 1990 Feb;10(2):492–500. doi: 10.1128/mcb.10.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990 Jun 15;61(6):1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Carroll D., Wright S. H., Wolff R. K., Grzesiuk E., Maryon E. B. Efficient homologous recombination of linear DNA substrates after injection into Xenopus laevis oocytes. Mol Cell Biol. 1986 Jun;6(6):2053–2061. doi: 10.1128/mcb.6.6.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D. M., Radding C. M. The role of exonuclease and beta protein of phage lambda in genetic recombination. II. Substrate specificity and the mode of action of lambda exonuclease. J Biol Chem. 1971 Apr 25;246(8):2502–2512. [PubMed] [Google Scholar]
- Cassuto E., Lash T., Sriprakash K. S., Radding C. M. Role of exonuclease and beta protein of phage lambda in genetic recombination. V. Recombination of lambda DNA in vitro. Proc Natl Acad Sci U S A. 1971 Jul;68(7):1639–1643. doi: 10.1073/pnas.68.7.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto E., Radding C. M. Mechanism for the action of lambda exonuclease in genetic recombination. Nat New Biol. 1971 Jan 6;229(1):13–16. doi: 10.1038/newbio229013a0. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. Enzymes of general recombination. Annu Rev Biochem. 1987;56:229–262. doi: 10.1146/annurev.bi.56.070187.001305. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Mechanism of transposition of bacteriophage Mu: structure of a transposition intermediate. Cell. 1985 Jul;41(3):867–876. doi: 10.1016/s0092-8674(85)80067-2. [DOI] [PubMed] [Google Scholar]
- Doherty M. J., Morrison P. T., Kolodner R. Genetic recombination of bacterial plasmid DNA. Physical and genetic analysis of the products of plasmid recombination in Escherichia coli. J Mol Biol. 1983 Jul 5;167(3):539–560. doi: 10.1016/s0022-2836(83)80097-7. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Formosa T., Alberts B. M. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986 Dec 5;47(5):793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- Gurdon J. B., Wickens M. P. The use of Xenopus oocytes for the expression of cloned genes. Methods Enzymol. 1983;101:370–386. doi: 10.1016/0076-6879(83)01028-9. [DOI] [PubMed] [Google Scholar]
- Haber J. E., Borts R. H., Connolly B., Lichten M., Rudin N., White C. I. Physical monitoring of meiotic and mitotic recombination in yeast. Prog Nucleic Acid Res Mol Biol. 1988;35:209–259. [PubMed] [Google Scholar]
- Hsieh P., Camerini-Otero R. D. Formation of joint DNA molecules by two eukaryotic strand exchange proteins does not require melting of a DNA duplex. J Biol Chem. 1989 Mar 25;264(9):5089–5097. [PubMed] [Google Scholar]
- Klar A. J., Miglio L. M. Initiation of meiotic recombination by double-strand DNA breaks in S. pombe. Cell. 1986 Aug 29;46(5):725–731. doi: 10.1016/0092-8674(86)90348-x. [DOI] [PubMed] [Google Scholar]
- Kolodkin A. L., Klar A. J., Stahl F. W. Double-strand breaks can initiate meiotic recombination in S. cerevisiae. Cell. 1986 Aug 29;46(5):733–740. doi: 10.1016/0092-8674(86)90349-1. [DOI] [PubMed] [Google Scholar]
- Legerski R. J., Penkala J. E., Peterson C. A., Wright D. A. Removal of 2',3'-dideoxynucleotide residues from injected DNA in Xenopus laevis oocytes. Mutat Res. 1990 Jul;236(1):1–7. doi: 10.1016/0921-8777(90)90026-2. [DOI] [PubMed] [Google Scholar]
- Lichten M., Goyon C., Schultes N. P., Treco D., Szostak J. W., Haber J. E., Nicolas A. Detection of heteroduplex DNA molecules among the products of Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7653–7657. doi: 10.1073/pnas.87.19.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T., Gally J. A., Edelman G. M. Deoxyribonuclease IV: a new exonuclease from mammalian tissues. Proc Natl Acad Sci U S A. 1969 Feb;62(2):597–603. doi: 10.1073/pnas.62.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T. The action pattern of mammalian deoxyribonuclease IV. Eur J Biochem. 1971 Feb 1;18(3):415–421. doi: 10.1111/j.1432-1033.1971.tb01258.x. [DOI] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: evidence for a nonconservative mechanism of homologous recombination. Mol Cell Biol. 1991 Jun;11(6):3278–3287. doi: 10.1128/mcb.11.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Degradation of linear DNA by a strand-specific exonuclease activity in Xenopus laevis oocytes. Mol Cell Biol. 1989 Nov;9(11):4862–4871. doi: 10.1128/mcb.9.11.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosig G. The essential role of recombination in phage T4 growth. Annu Rev Genet. 1987;21:347–371. doi: 10.1146/annurev.ge.21.120187.002023. [DOI] [PubMed] [Google Scholar]
- Resnick M. A. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976 Jun;59(1):97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Rudin N., Sugarman E., Haber J. E. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989 Jul;122(3):519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman M. M. Intermolecular homologous recombination between transfected sequences in mammalian cells is primarily nonconservative. Mol Cell Biol. 1987 Oct;7(10):3561–3565. doi: 10.1128/mcb.7.10.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R., Amundsen S. K., Chaudhury A. M., Cheng K. C., Ponticelli A. S., Roberts C. M., Schultz D. W., Taylor A. F. Roles of RecBC enzyme and chi sites in homologous recombination. Cold Spring Harb Symp Quant Biol. 1984;49:485–495. doi: 10.1101/sqb.1984.049.01.055. [DOI] [PubMed] [Google Scholar]
- Stahl F. W. Roles of double-strand breaks in generalized genetic recombination. Prog Nucleic Acid Res Mol Biol. 1986;33:169–194. doi: 10.1016/s0079-6603(08)60023-9. [DOI] [PubMed] [Google Scholar]
- Sun H., Treco D., Schultes N. P., Szostak J. W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989 Mar 2;338(6210):87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Morrison P., Kolodner R. Intramolecular recombination of linear DNA catalyzed by the Escherichia coli RecE recombination system. J Mol Biol. 1985 Dec 5;186(3):515–525. doi: 10.1016/0022-2836(85)90126-3. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Taylor A., Smith G. R. Unwinding and rewinding of DNA by the RecBC enzyme. Cell. 1980 Nov;22(2 Pt 2):447–457. doi: 10.1016/0092-8674(80)90355-4. [DOI] [PubMed] [Google Scholar]
- Thaler D. S., Stahl F. W. DNA double-chain breaks in recombination of phage lambda and of yeast. Annu Rev Genet. 1988;22:169–197. doi: 10.1146/annurev.ge.22.120188.001125. [DOI] [PubMed] [Google Scholar]
- Thaler D. S., Stahl M. M., Stahl F. W. Double-chain-cut sites are recombination hotspots in the Red pathway of phage lambda. J Mol Biol. 1987 May 5;195(1):75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C. Processing of recombination intermediates in vitro. Bioessays. 1990 Apr;12(4):151–154. doi: 10.1002/bies.950120402. [DOI] [PubMed] [Google Scholar]
- White C. I., Haber J. E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990 Mar;9(3):663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A. M., Kahn R., DasGupta C., Radding C. M. Formation of nascent heteroduplex structures by RecA protein and DNA. Cell. 1982 Aug;30(1):37–44. doi: 10.1016/0092-8674(82)90009-5. [DOI] [PubMed] [Google Scholar]