Abstract
Cancer cells must avoid succumbing to a variety of noxious conditions within their surroundings. Acidosis is one such prominent feature of the tumor microenvironment that surprisingly promotes tumor survival and progression. We recently reported that acidosis prevents apoptosis of starved or stressed lymphoma cells through regulation of several Bcl-2 family members (Ryder et al., JBC, 2012). Mechanistic studies in that work focused on the acid-mediated upregulation of anti-apoptotic Bcl-2 and Bcl-xL, while additionally showing inhibition of glutamine starvation-induced expression of pro-apoptotic PUMA by acidosis. Herein we report that amino acid (AA) starvation elevates PUMA, an effect that is blocked by extracellular acidity. Knockdown studies confirm that PUMA induction during AA starvation requires expression of both CHOP and c-Jun. Interestingly, acidosis strongly attenuates AA starvation-mediated c-Jun expression, which correlates with PUMA repression. As c-Jun exerts a tumor suppressive function in this and other contexts, its inhibition by acidosis has broader implications for survival of cancer cells in the acidic tumor milieu.
Keywords: Acidosis, Amino Acids, Apoptosis, PUMA, CHOP, c-Jun
Introduction
Tumor growth and progression breeds an increasingly inhospitable local environment, thereby imposing numerous obstacles to further expansion. In order to survive, cancer cells must overcome microenvironmental stresses such as hypoxia, nutrient limitation, and acidic stress [1]. Adaptations that facilitate malignant progression in the face of these harsh cell-extrinsic conditions are critical to the oncogenic process. In fact, deregulated responses to external stimuli figure prominently among the hallmarks of the disease [2].
One characteristic perturbation within the tumor micro-environment is the development of extracellular acidosis. Whereas most normal tissues exhibit a pHe ~7.4, numerous studies find intratumoral pHe measurements between 6.5–7.0 [3]. Despite this extracellular acidity, cancer cells maintain a slightly alkaline intracellular space. This pH gradient reversal is critical, as intracellular acidosis can activate nucleases and the apoptotic cascade [4,5]. Indeed, extracellular acidosis is toxic to some cell types [6,7]. In stark contrast, numerous reports show that acidosis promotes therapeutic resistance and invasive phenotypes [8,9,10]. Mechanisms continue to be elucidated for this surprising tumorigenic role for acidosis.
We recently reported that acidosis inhibits apoptosis of starved or stressed lymphoma cells through regulation of multiple members of the Bcl-2 family [11]. This group consists of over 20 proteins that share homology in at least one of four distinct Bcl-2 homology (BH) domains [12]. These proteins primarily control entry into the intrinsic apoptosis cascade, with some members promoting cell death and others having an inhibitory role. Our work revealed that induction of anti-apoptotic family members Bcl-2 and Bcl-xL by acidosis contributes significantly to its cytoprotective effect and that the elevation of these pro-survival proteins requires GPR65, an acid-sensing G protein-coupled receptor (GPCR). Additionally, we found acidification to strongly block starvation-induced elevation of pro-apoptotic PUMA (p53-upregulated mediator of apoptosis) at both the mRNA and protein level. Yet the mechanism for repression of PUMA by acidosis remains undetermined. Further inquiries in this direction stand to uncover pH-dependent regulatory factors that contribute to acidosis-mediated evasion of apoptosis.
Though originally discovered to be induced by p53, PUMA expression has since been shown to be controlled by numerous factors, primarily at the level of transcription (reviewed in [13]). Activation of this BH3-only protein is known to occur in response to diverse stimuli such as DNA damage, ER stress and growth factor withdrawal. Involved transcription factors include CCAAT/enhancer-binding protein (C/EBP) homologous protein (CHOP) and c-Jun, among others. PUMA exerts its pro-apoptotic function by directly activating Bax and Bak, leading to mitochondrial outer membrane permeabilization and apoptosis [14]. It is no surprise then that expression levels of this tumor suppressive gene are decreased in several tumor types, though genetic inactivation does not seem to be contributory [13]. Further understanding of PUMA repression in cancer remains an important area of investigation.
In this study we show that PUMA upregulation during amino acid (AA) starvation requires induction of both CHOP and c-Jun. Interestingly, we find that acidosis strongly represses starvation-induced c-Jun levels while not affecting CHOP expression. We propose that CHOP and c-Jun cooperate to elevate pro-apoptotic PUMA and that acidosis represses PUMA elevation by blocking c-Jun expression. These findings highlight a novel mechanism for the promotion of cancer cell survival mediated by tumor-associated acidity.
Materials and Methods
Cell culture
Maintenance and experimental conditions for wild type and Bcl-2-expressing WEHI7.2 cells were previously described [11]. Initial cell density of 3–6 × 105 cells/mL was used for all experiments. Control and acidic pH media were set to 7.55 ± 0.1 and 6.50 ± 0.1, respectively.
Immunoblot analysis
Protocol was described previously [11]. Antibodies used include: anti-PUMA, anti-Bim, anti-CHOP, anti-c-Jun, anti-cleaved caspase-3, and anti-PARP from Cell Signaling, and anti-Actin from Sigma. Protein expression was visualized with ECL reagent or ECL Prime (GE Healthcare).
RNA isolation and RT-PCR
RNA isolation and RT-PCR were performed as described previously [11]. All assays were created from Roche universal probe library. Primers are as follows (5′→ 3′): CHOP (Forward (F)-gcgacagagccagaataaca, Reverse (R)-gatgcacttccttctggaaca); c-Jun (F-ccagaagatggtgtggtgttt, R-ctgaccctctccccttgc); JunB (F-ccacggagggagagaaaatc, R-agttggcagctgtgcgtaa); c-Fos (F-gggacagcctttcctactacc, R-agatctgcgcaaaagtcctg).
RNA interference
For gene knockdown, 107 cells were electroporated with ON-TARGETplus SMARTpool siRNA (Dharmacon, Lafayette, CO), as described previously [11]. Cells were transiently transfected with either non-targeting control, CHOP-specific or c-Jun-specific siRNA.
Statistical analysis
All statistical analyses show the highest level of significance for repeated measures ANOVA with Tukey-Kramer post-test. Analyses were performed using GraphPad Prism software. Densitometric analysis was done using ImageJ software. Error bars represent ± standard error of the mean for at least three experiments.
Results
Starvation of different AAs causes similar acid-inhibitable increases in apoptosis and PUMA levels
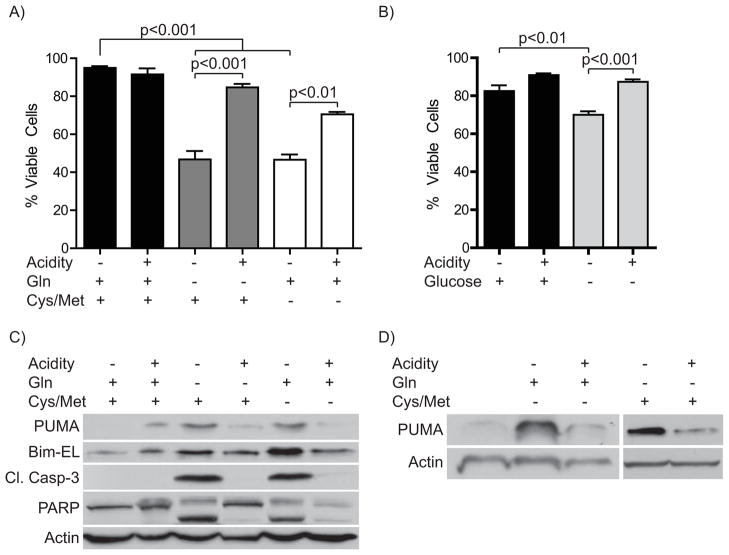
The goal of this study was to understand the mechanistic basis for the pH-dependent induction of PUMA during glutamine starvation of lymphoma cells. As a first step to address this question, we needed to understand the nature of the apoptotic stress. This knowledge would then inform our later investigation into the mediating factors. As glutamine is a vital fuel source for cancer cells in addition to its roles as a precursor for protein synthesis and transamination reactions, we investigated whether starvation of different AAs, namely the two sulfur-containing amino acids cysteine and methionine, would elicit a similar response and whether this cell death would also be inhibited by acidosis. Importantly, sulfur-containing AAs are among those decreased in the tumor microenvironment [15]. Therefore, we set up a direct comparison of glutamine versus cysteine/methionine starvation of WEHI7.2 murine lymphoma cells in the presence or absence of extracellular acidity. We found similar levels of cell death upon starvation of either AA(s) after 12 hours (Fig. 1a). Furthermore, acidosis inhibited the cell death in either starvation condition. In CEM-C7 human lymphoma cells, AA starvation caused minimal cell death before 72 hours (data not shown). As expected from our previous work we found the cell death to be apoptotic, as starvation markedly increased cleavage of caspase-3 and PARP (Fig. 1c). Acidosis strongly attenuated the appearance of these apoptotic markers. In contrast, the appearance of cell death upon glucose starvation only became detectable at 24 hours, when glutamine starved cells are nearly all dead (Fig. 1b and [11]) These data suggest that the apoptosis is an AA withdrawal response rather than a metabolic starvation.
Figure 1. Starvation of different AAs causes similar acid-inhibitable increases in apoptosis and PUMA levels.
A) WEHI7.2 cells were incubated in the presence or absence of either glutamine (Gln) or cysteine (Cys) and methionine (Met) with or without acidification of media for 12 hours. Cell viability was assessed by Trypan blue exclusion. B) Cells were starved of glucose for 24 hours with or without media acidification. Viability was determined as in A). C) Protein was isolated from cells treated as in A) for immunoblot analysis. D) Bcl-2-overexpressing WEHI7.2 cells were treated as in A) and harvested for Western blot analysis.
We next tested whether the two AA starvation protocols regulate the pro-apoptotic Bcl-2 family members PUMA and Bim similarly, as shown for glutamine withdrawal in our earlier studies [11]. In fact, robust elevation of both proteins occurred after starvation of either AA(s). In line with our previous findings acidosis strongly blocked PUMA induction, whereas the degree of Bim protein repression by acidity varied between experiments (Fig. 1c). Since acidosis elevates PUMA-interacting proteins Bcl-2 and Bcl-xL, we next examined the regulation of PUMA upon AA starvation of Bcl-2-overexpressing WEHI7.2 cells. This experiment tested whether the increase in anti-apoptotic Bcl-2 proteins by acidosis mediates changes in pro-apoptotic family members. AA starvation and acidosis had similar effects on PUMA levels in Bcl-2-expressing compared to wild type cells (Fig. 1c and d), suggesting that the control of PUMA levels occurs independently of expression changes for its inhibitory binding partners.
Starvation-induced CHOP mediates PUMA and Bim elevation
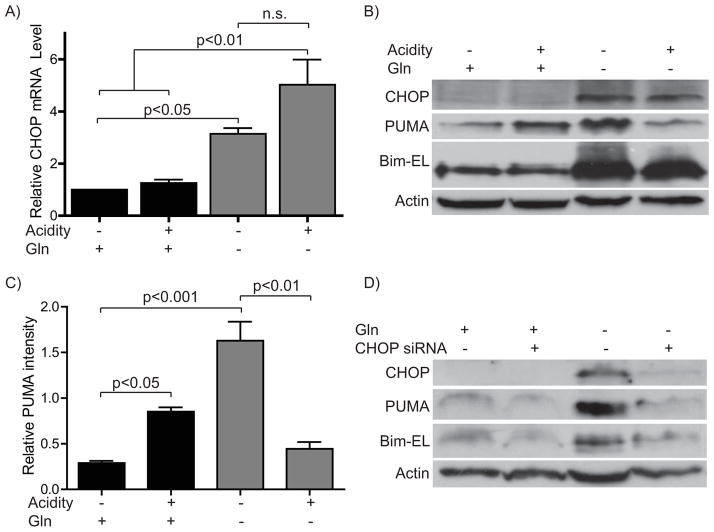
Because PUMA and Bim upregulation occurred in response to an AA starvation insult, we next focused on factors induced or activated by AA starvation that could, in turn, mediate increases in these BH3-only proteins. Among the downstream components of the AA response (AAR) is CHOP (C/EBPζ, CHOP10, DDIT3, GADD153) [16]. Importantly, CHOP has been shown to mediate expression of both PUMA and Bim in response to a variety of toxic stimuli [17,18]. To explore a role for CHOP in PUMA and Bim elevation during the AAR, we first examined CHOP expression upon AA starvation with or without acidosis. As expected, we found a robust induction of CHOP by AA limitation at both the mRNA and protein level (Fig. 2a and b). Elevated CHOP mRNA and protein were seen by 1–2 hours (data not shown). However, we saw no significant difference in CHOP levels when comparing normal and acidic pH in starved cells, whereas acidosis caused differential regulation of PUMA and Bim (Fig. 2b and c). Still, the strong and early induction of CHOP by AA starvation supported its possible contribution to the regulation of PUMA and Bim in this context. To test this hypothesis, we next sought to prevent CHOP elevation during AA starvation by transfection of CHOP-specific siRNA. This knockdown robustly inhibited starvation-induced increases in CHOP (Fig. 2d). Importantly, it also blocked the elevation of both PUMA and Bim during AA starvation. This finding confirmed that CHOP plays an essential role in the induction of these pro-apoptotic factors in response to AA starvation.
Figure 2. Starvation-induced CHOP mediates PUMA and Bim elevation.
A)–C) Cells were incubated in the presence or absence of glutamine with or without media acidification for 10–12 hours and harvested for A) RT-PCR or B) Western. C) Densitometry for PUMA protein relative to Actin for B). D) Cells were electroporated with siRNA either non-targeting or CHOP specific, allowed to recover overnight and treated as in A) then harvested for immunoblot analysis.
Other p53-regulated genes differ from PUMA in expression pattern during AA starvation and acidosis
The absence of an effect of acidosis on CHOP levels during AA starvation meant there must be an additional factor(s) that accounts for the pH-dependent regulation of PUMA. The most extensively studied PUMA regulatory factor is p53 [13]. To assess the possibility that p53 controls PUMA expression during AA starvation and acidosis, we investigated other p53-regulated genes under these conditions. None of the genes we examined (Apaf-1, Mdm2, Bax and p21) showed a pattern of regulation similar to that of PUMA, though some minor (less than two-fold) expression changes did occur (data not shown). Furthermore, we previously showed that another p53-regulated BH3-only protein, Noxa, was not altered by AA starvation and acidosis [11]. Thus, it is unlikely that p53 contributes significantly to the regulation of PUMA in this context.
AA starvation-induced c-Jun is blocked by acidosis and contributes to PUMA regulation
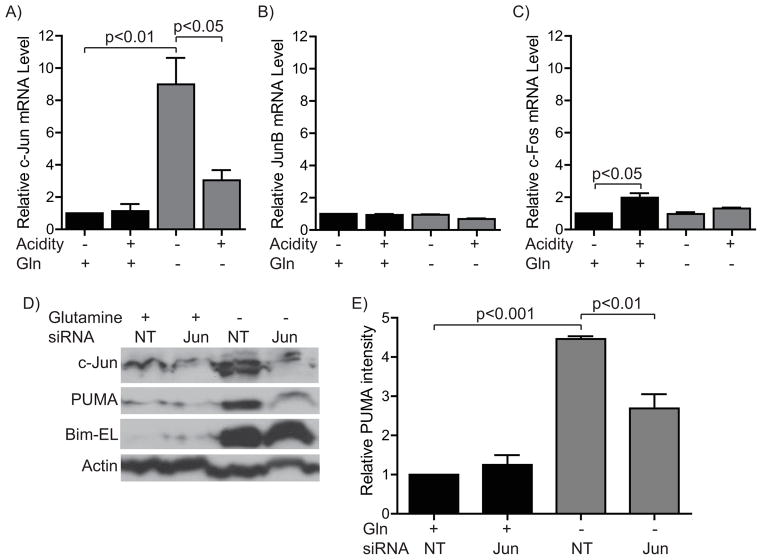
Among other factors that regulate PUMA gene expression, the AP-1 transcription factor c-Jun is known to cooperate with CHOP in this process [18]. Moreover, the AP-1 genes c-Jun, JunB and c-Fos have been shown previously to be induced during the AAR [19]. Therefore, we assayed for changes in mRNA levels of these genes during AA starvation with or without acidosis (Fig. 3a–c). Of these three genes, AA starvation strikingly elevated c-Jun alone. Impressively, acidosis completely blunted this induction. The other two AP-1 genes showed only slight variation in transcript levels in response to these conditions at the time points examined. The expression changes for c-Jun correlated well with PUMA regulation during AA starvation and acidosis, prompting us to investigate further the role of c-Jun in PUMA induction by inhibiting its expression during AA starvation with siRNA (Fig. 3d and e). Interestingly, c-Jun knockdown prevented induction of PUMA but had minimal effect on Bim elevation (Fig. 3d). These data indicate that CHOP and c-Jun cooperate in the elevation of PUMA during AA starvation. They additionally support a model wherein c-Jun expression changes contribute to the differential regulation of PUMA and Bim during acidosis in AA-starved cells.
Figure 3. AA starvation-induced c-Jun is blocked by acidosis and contributes to PUMA regulation.
A)–C) Cells were incubated in the presence or absence of glutamine with or without media acidification for 10–12 hours and harvested for RT-PCR for A) c-Jun, B) JunB, or C) c-Fos. D) Cells were electroporated with siRNA either non-targeting or c-Jun specific, allowed to recover overnight and treated as in A) then harvested for immunoblot analysis. Densitometric analysis for PUMA protein compared to Actin from D) is represented graphically in E).
Discussion
In this report we elucidate the role of the transcription factors CHOP and c-Jun in controlling the differential regulation of two pro-apoptotic BH3-only proteins during AA starvation and acidosis. We show that CHOP mediates the elevation of both PUMA and Bim during AA starvation. This bZIP (basic leucine zipper) transcription factor plays an essential role in the adaptive response to AA starvation. However, prolonged elevation of CHOP can also trigger apoptosis [20]. To our knowledge, this study is the first to link AA starvation to PUMA and Bim induction by CHOP. More interestingly, we demonstrate that the regulation of PUMA has an additional requirement for the AP-1 transcription factor c-Jun that correlates with the pH-dependence of PUMA induction by AA starvation. As acidosis is a common feature of the tumor microenvironment, this negative regulation of a pro-apoptotic activity of c-Jun has broader implications for the inappropriate survival of nutrient deprived cancer cells.
The c-Jun gene has been intensively studied for over 25 years [21], being implicated as both an oncogene and tumor suppressor and performing functions ranging from proliferation to apoptosis (reviewed in [22]). The outcome of c-Jun expression is determined by its available dimerization partners, leading to cell type and context dependent effects [23]. As a fellow bZIP transcription factor, c-Jun can dimerize with CHOP as well as itself and other related AP-1 factors [24,25]. Previous work in palmitate-treated hepatoma cells showed that cooperation between CHOP and c-Jun occurs at an AP-1 binding site in the proximal PUMA promoter [18]. Interestingly, Bim induction during ER stress requires a CHOP:C/EBPα binding site [17]. However, the PUMA gene lacks such a promoter element. As C/EBPα is another gene known to be elevated by AA limitation [26], the different transcriptional complexes (CHOP:c-Jun and CHOP:C/EBPα) are likely to mediate PUMA and Bim elevation, respectively.
Acidosis has been reported to have apparently contradictory effects on c-Jun and AP-1 activity across different model systems. Increased AP-1 levels and transcriptional activity has been shown to occur in the face of acidosis in several cell types [27,28]. However, a recent report showed that lactic acidosis blocked c-Jun phosphorylation in stimulated cytotoxic T lymphocytes [29]. The present study was performed in a T cell lymphoma cell line, raising the possibility that the effect of acidosis to inhibit c-Jun activity may be specific to lymphoid cells. Interestingly, mining of multiple microarray data sets with Oncomine showed that lymphoma cells had a greatly reduced c-Jun level compared to other cancer types [30,31,32]. This information raises two possibilities: First that the observed negative regulation of c-Jun is specific to lymphoid malignancies and secondly that lymphoma cells reside in an acidic microenvironment [33], accounting for the downregulation of c-Jun.
A critical question that remains is the identity of the upstream factors that inhibit AA starvation-induced c-Jun expression in response to acidosis. The cellular response to AA limitation is initiated by general control nonderepressed 2 (GCN2), which phosphorylates eukaryotic initiation factor 2 alpha (eIF-2α), thereby causing a stall of most protein translation [34]. However, some genes such as activating transcription factor 4 (ATF4) are then preferentially translated. Concurrently, AA starvation causes an activating phosphorylation of ATF2 [35]. Elevation of CHOP during AA deprivation requires both ATF4 upregulation and ATF2 phosphorylation [35]. However, since both CHOP and c-Jun induction during the AAR requires ATF2 activity [19] yet are differentially regulated by acidosis, it is unlikely that acidosis modulates this pathway. As another possibility, activating phosphorylation of c-Jun occurs via JNK (c-Jun N-terminal kinase) [36]. A recent report showed that phosphorylation of existing c-Jun facilitates its auto-regulation during AA limitation [19]. Interestingly, in that study upregulation of c-Jun during the AAR was inhibited by either JNK or MEK inhibitor treatment. Additionally, a requirement for JNK1 has been shown for an apoptotic pathway that culminates in CHOP- and AP-1-mediated PUMA expression [18,37]. Confusingly, acidosis has been reported to either positively or negatively regulate JNK activity, while others find no effect of acidosis [29,38,39]. In our hands, JNK inhibition fails to prevent AA starvation-induced PUMA elevation (data not shown). Yet potentially differing effects of JNK1 and JNK2 may confound inhibitor experiments [19].
Finally, the initial responder to extracellular acidosis represents an important target for investigation. As one possibility, the acid-sensing GPCRs GPR65 and GPR4 have been shown to be overexpressed in cancer and to function as oncogenes [40,41]. In normal immune cells, GPR65 also mediates inhibition of pro-inflammatory cytokine production during acidosis [42,43]. Importantly, c-Jun plays a role in induction of all the genes studied (IL-2, IL-6, TNF-α) [44,45,46]. Thus, the finding of c-Jun inhibition by acidosis may explain other related findings in normal immune cell biology. Future studies should address the potential link between c-Jun inhibition and upstream pH-responsive GPCRs.
Research Highlights.
Acidosis inhibits amino acid (AA) starvation-induced cell death of WEHI7.2 cells
AA starvation-mediated induction of PUMA and Bim requires CHOP
AA starvation-mediated induction of PUMA additionally requires c-Jun
Acidosis inhibits AA starvation-mediated c-Jun elevation
Acknowledgments
The authors would like to thank members of the Distelhorst lab as well as Dr. Maria Hatzoglou for their advice.
Abbreviations used
- AAR
amino acid response
- Apaf-1
apoptotic protease-activating factor-1
- ATF
activating transcription factor
- Bcl-2
B cell lymphoma-2
- Bax
Bcl-2-associated X protein
- Bim
Bcl-2-interacting mediator of cell death
- CHOP
CCAAT/enhancer-binding protein homologous protein
- eIF2α
eukaryotic initiation factor-2alpha
- GCN2
general control nonderepressed 2
- GPCR
G protein-coupled receptor
- Mdm2
Murine double minute 2
- PARP
poly-(ADP-ribose) polymerase
- pHe
extracellular pH
- PUMA
p53-upregulated mediator of apoptosis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 4.Matsuyama S, Llopis J, Deveraux QL, Tsien RY, Reed JC. Changes in intramitochondrial and cytosolic pH: early events that modulate caspase activation during apoptosis. Nat Cell Biol. 2000;2:318–325. doi: 10.1038/35014006. [DOI] [PubMed] [Google Scholar]
- 5.Famulski KS, Macdonald D, Paterson MC, Sikora E. Activation of a low pH-dependent nuclease by apoptotic agents. Cell Death Differ. 1999;6:281–289. doi: 10.1038/sj.cdd.4400495. [DOI] [PubMed] [Google Scholar]
- 6.Kubasiak LA, Hernandez OM, Bishopric NH, Webster KA. Hypoxia and acidosis activate cardiac myocyte death through the Bcl-2 family protein BNIP3. Proc Natl Acad Sci U S A. 2002;99:12825–12830. doi: 10.1073/pnas.202474099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding D, Moskowitz SI, Li R, Lee SB, Esteban M, Tomaselli K, Chan J, Bergold PJ. Acidosis induces necrosis and apoptosis of cultured hippocampal neurons. Exp Neurol. 2000;162:1–12. doi: 10.1006/exnr.2000.7226. [DOI] [PubMed] [Google Scholar]
- 8.Reichert M, Steinbach JP, Supra P, Weller M. Modulation of growth and radiochemosensitivity of human malignant glioma cells by acidosis. Cancer. 2002;95:1113–1119. doi: 10.1002/cncr.10767. [DOI] [PubMed] [Google Scholar]
- 9.Thews O, Gassner B, Kelleher DK, Schwerdt G, Gekle M. Impact of extracellular acidity on the activity of P-glycoprotein and the cytotoxicity of chemotherapeutic drugs. Neoplasia. 2006;8:143–152. doi: 10.1593/neo.05697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moellering RE, Black KC, Krishnamurty C, Baggett BK, Stafford P, Rain M, Gatenby RA, Gillies RJ. Acid treatment of melanoma cells selects for invasive phenotypes. Clin Exp Metastasis. 2008;25:411–425. doi: 10.1007/s10585-008-9145-7. [DOI] [PubMed] [Google Scholar]
- 11.Ryder C, McColl K, Zhong F, Distelhorst CW. Acidosis Promotes Bcl-2 Family-mediated Evasion of Apoptosis: INVOLVEMENT OF ACID-SENSING G PROTEIN-COUPLED RECEPTOR GPR65 SIGNALING TO MEK/ERK. J Biol Chem. 2012;287:27863–27875. doi: 10.1074/jbc.M112.384685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J, Zhang L. PUMA, a potent killer with or without p53. Oncogene. 2008;27(Suppl 1):S71–83. doi: 10.1038/onc.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrison SP, Phillips DC, Jeffers JR, Chipuk JE, Parsons MJ, Rehg JE, Opferman JT, Green DR, Zambetti GP. Genetically defining the mechanism of Puma- and Bim-induced apoptosis. Cell Death Differ. 2012;19:642–649. doi: 10.1038/cdd.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moyer GH, Pitot HC. Static and dynamic aspects of amino acid pools in rat liver and Morris hepatomas 9618A and 7800. Cancer Res. 1974;34:2647–2653. [PubMed] [Google Scholar]
- 16.Bruhat A, Jousse C, Wang XZ, Ron D, Ferrara M, Fafournoux P. Amino acid limitation induces expression of CHOP, a CCAAT/enhancer binding protein-related gene, at both transcriptional and post-transcriptional levels. J Biol Chem. 1997;272:17588–17593. doi: 10.1074/jbc.272.28.17588. [DOI] [PubMed] [Google Scholar]
- 17.Puthalakath H, O’Reilly LA, Gunn P, Lee L, Kelly PN, Huntington ND, Hughes PD, Michalak EM, McKimm-Breschkin J, Motoyama N, Gotoh T, Akira S, Bouillet P, Strasser A. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–1349. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 18.Cazanave SC, Elmi NA, Akazawa Y, Bronk SF, Mott JL, Gores GJ. CHOP and AP-1 cooperatively mediate PUMA expression during lipoapoptosis. Am J Physiol Gastrointest Liver Physiol. 2010;299:G236–243. doi: 10.1152/ajpgi.00091.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu L, Balasubramanian M, Shan J, Dudenhausen EE, Kilberg MS. Auto-activation of c-JUN gene by amino acid deprivation of hepatocellular carcinoma cells reveals a novel c-JUN-mediated signaling pathway. J Biol Chem. 2011;286:36724–36738. doi: 10.1074/jbc.M111.277673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–190. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maki Y, Bos TJ, Davis C, Starbuck M, Vogt PK. Avian sarcoma virus 17 carries the jun oncogene. Proc Natl Acad Sci U S A. 1987;84:2848–2852. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaulian E. AP-1--The Jun proteins: Oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22:894–899. doi: 10.1016/j.cellsig.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Meng Q, Xia Y. c-Jun, at the crossroad of the signaling network. Protein Cell. 2011;2:889–898. doi: 10.1007/s13238-011-1113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ubeda M, Vallejo M, Habener JF. CHOP enhancement of gene transcription by interactions with Jun/Fos AP-1 complex proteins. Mol Cell Biol. 1999;19:7589–7599. doi: 10.1128/mcb.19.11.7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vesely PW, Staber PB, Hoefler G, Kenner L. Translational regulation mechanisms of AP-1 proteins. Mutat Res. 2009;682:7–12. doi: 10.1016/j.mrrev.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Marten NW, Burke EJ, Hayden JM, Straus DS. Effect of amino acid limitation on the expression of 19 genes in rat hepatoma cells. FASEB J. 1994;8:538–544. doi: 10.1096/fasebj.8.8.8181673. [DOI] [PubMed] [Google Scholar]
- 27.Guan J, Wu X, Arons E, Christou H. The p38 mitogen-activated protein kinase pathway is involved in the regulation of heme oxygenase-1 by acidic extracellular pH in aortic smooth muscle cells. J Cell Biochem. 2008;105:1298–1306. doi: 10.1002/jcb.21930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimokawa N, Sugama S, Miura M. Extracellular H+ stimulates the expression of c-fos/c-jun mRNA through Ca2+/calmodulin in PC12 cells. Cell Signal. 1998;10:499–503. doi: 10.1016/s0898-6568(97)00176-9. [DOI] [PubMed] [Google Scholar]
- 29.Mendler AN, Hu B, Prinz PU, Kreutz M, Gottfried E, Noessner E. Tumor lactic acidosis suppresses CTL function by inhibition of p38 and JNK/c-Jun activation. Int J Cancer. 2012;131:633–640. doi: 10.1002/ijc.26410. [DOI] [PubMed] [Google Scholar]
- 30.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan J, Wei JS, Ringner M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci U S A. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallagher FA, Kettunen MI, Day SE, Hu DE, Ardenkjaer-Larsen JH, Zandt R, Jensen PR, Karlsson M, Golman K, Lerche MH, Brindle KM. Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature. 2008;453:940–943. doi: 10.1038/nature07017. [DOI] [PubMed] [Google Scholar]
- 34.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Averous J, Bruhat A, Jousse C, Carraro V, Thiel G, Fafournoux P. Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J Biol Chem. 2004;279:5288–5297. doi: 10.1074/jbc.M311862200. [DOI] [PubMed] [Google Scholar]
- 36.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 37.Cazanave SC, Mott JL, Elmi NA, Bronk SF, Werneburg NW, Akazawa Y, Kahraman A, Garrison SP, Zambetti GP, Charlton MR, Gores GJ. JNK1-dependent PUMA expression contributes to hepatocyte lipoapoptosis. J Biol Chem. 2009;284:26591–26602. doi: 10.1074/jbc.M109.022491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsuganezawa H, Sato S, Yamaji Y, Preisig PA, Moe OW, Alpern RJ. Role of c-SRC and ERK in acid-induced activation of NHE3. Kidney Int. 2002;62:41–50. doi: 10.1046/j.1523-1755.2002.00418.x. [DOI] [PubMed] [Google Scholar]
- 39.Martinez D, Vermeulen M, Trevani A, Ceballos A, Sabatte J, Gamberale R, Alvarez ME, Salamone G, Tanos T, Coso OA, Geffner J. Extracellular acidosis induces neutrophil activation by a mechanism dependent on activation of phosphatidylinositol 3-kinase/Akt and ERK pathways. J Immunol. 2006;176:1163–1171. doi: 10.4049/jimmunol.176.2.1163. [DOI] [PubMed] [Google Scholar]
- 40.Sin WC, Zhang Y, Zhong W, Adhikarakunnathu S, Powers S, Hoey T, An S, Yang J. G protein-coupled receptors GPR4 and TDAG8 are oncogenic and overexpressed in human cancers. Oncogene. 2004;23:6299–6303. doi: 10.1038/sj.onc.1207838. [DOI] [PubMed] [Google Scholar]
- 41.Ihara Y, Kihara Y, Hamano F, Yanagida K, Morishita Y, Kunita A, Yamori T, Fukayama M, Aburatani H, Shimizu T, Ishii S. The G protein-coupled receptor T-cell death-associated gene 8 (TDAG8) facilitates tumor development by serving as an extracellular pH sensor. Proc Natl Acad Sci U S A. 2010;107:17309–17314. doi: 10.1073/pnas.1001165107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mogi C, Tobo M, Tomura H, Murata N, He XD, Sato K, Kimura T, Ishizuka T, Sasaki T, Sato T, Kihara Y, Ishii S, Harada A, Okajima F. Involvement of proton-sensing TDAG8 in extracellular acidification-induced inhibition of proinflammatory cytokine production in peritoneal macrophages. J Immunol. 2009;182:3243–3251. doi: 10.4049/jimmunol.0803466. [DOI] [PubMed] [Google Scholar]
- 43.Onozawa Y, Fujita Y, Kuwabara H, Nagasaki M, Komai T, Oda T. Activation of T cell death-associated gene 8 regulates the cytokine production of T cells and macrophages in vitro. Eur J Pharmacol. 2012;683:325–331. doi: 10.1016/j.ejphar.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Yao J, Mackman N, Edgington TS, Fan ST. Lipopolysaccharide induction of the tumor necrosis factor-alpha promoter in human monocytic cells. Regulation by Egr-1, c-Jun, and NF-kappaB transcription factors. J Biol Chem. 1997;272:17795–17801. doi: 10.1074/jbc.272.28.17795. [DOI] [PubMed] [Google Scholar]
- 45.Granelli-Piperno A, Nolan P. Nuclear transcription factors that bind to elements of the IL-2 promoter. Induction requirements in primary human T cells. J Immunol. 1991;147:2734–2739. [PubMed] [Google Scholar]
- 46.Dendorfer U, Oettgen P, Libermann TA. Multiple regulatory elements in the interleukin-6 gene mediate induction by prostaglandins, cyclic AMP, and lipopolysaccharide. Mol Cell Biol. 1994;14:4443–4454. doi: 10.1128/mcb.14.7.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]