Abstract
Flaviviruses are small spherical virus particles covered by a dense icosahedral array of envelope (E) proteins that mediate virus attachment to cells and the fusion of viral and cellular membranes. Our understanding of the mechanism by which flavivirus E proteins orchestrate entry into cells has been advanced by studies of E structure and arrangement on the virion at different steps of the virus entry/membrane fusion process. When combined with an increasingly clear (albeit still incomplete) view of the cell biology of virus entry, these advances suggest new antiviral strategies. Indeed, inhibitors that target cellular and viral processes involved in entry show promise as powerful tools to study this critical step of the viral lifecycle, and with luck, may ultimately lead to therapeutic advances.
Flaviviruses are a group of enveloped viruses vectored principally by arthropods, and include important human pathogens such as dengue (DENV), West Nile (WNV), and yellow fever viruses. These viruses cause a broad spectrum of disease in humans including fever, encephalitis, meningitis, and hemorrhage. Together the members of this genus are responsible each year for more than 50 million human infections worldwide [1]. Flaviviruses encapsidate an ~11kb positive-stranded RNA genome that is translated as a single polyprotein and subsequently cleaved into at least ten functionally distinct proteins; three of these (capsid, envelope (E), and premembrane (prM)) are incorporated into the virus particle [2]. Flavivirus entry into cells is coordinated by the activities of E proteins arrayed on the surface of the virion. These proteins orchestrate both the attachment of virus particles to cells, and the subsequent low pH-triggered fusion of viral and target cell membranes during endocytic entry. Significant mechanistic insight into these early steps in the virus lifecycle has arisen from structural and biochemical studies of the E protein of several different flaviviruses (reviewed in [3,4]). Interest in this process is enhanced by the fact that E proteins are the principle targets of neutralizing antibodies, and more recently, have been identified as targets for novel therapeutics [5,6]. Beyond their potential clinical utility, inhibitors that target flavivirus entry into cells have proven to be powerful tools for dissecting how viral envelope fusion proteins promote attachment to target cells and membrane fusion [7]. In this review, we will discuss recent progress towards understanding flavivirus entry and strategies being developed to block this critical phase of the virus lifecycle.
The envelope protein
The structure of the E protein ectodomain has been determined at high resolution for several flaviviruses (reviewed in [4])(Figure 1a). The E protein is an elongated predominantly β-stranded structure composed of three distinct domains connected by flexible linkers of one or more chains [8,9]. E protein domain II (E-DII) consists of two extended loops that contribute important dimerization contacts that coordinate the antiparallel E arrangement on mature virus particles [8,10,11](Figure 1b). A highly conserved glycine-rich fusion loop is located at the distal tip of E-DII [12]. Domain III (E-DIII) is an immunoglobulin-like structure thought to be the site of interactions with cellular receptors, although to date much of the evidence in support of this concept remains indirect [4]. Domain I (E-DI) is a central eight-stranded β-barrel structure that serves to connect E-DII and E-DIII. E proteins contain one or two asparagine-linked (N-linked) carbohydrates that may participate in stabilizing E protein dimers present on mature viruses [8,13] and can mediate interactions with cellular attachment factors during virus entry [14-17]. The E protein ectodomain is connected to the viral membrane by a helical region called the stem anchor [18] followed by two antiparallel transmembrane domains [19](Figure 3).
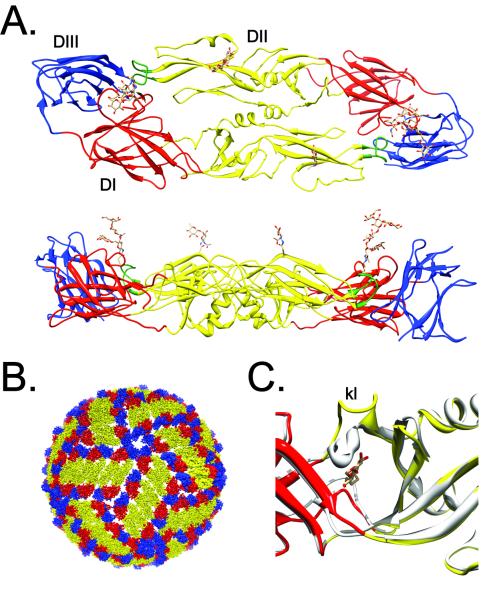
Figure 1. The structure and arrangement of the E protein on the mature virion.
(A.) Structure of the flavivirus E protein. E proteins form antiparallel dimers on the mature virus particle. The structure of the DENV3 E protein dimer (PDB 1UZG) is shown as a ribbon diagram from the top and side (top and bottom panels, respectively) [86]. Domains I, II, and III are colored in red, yellow, and blue, respectively, with the conserved fusion loop at the distal tip of E-DII in green. Asparagine linked carbohydrate modifications of E-DI and E-DII are shown as ball and stick representations. The E protein is linked to the protein’s transmembrane anchor by a helical stem (not depicted) at the carboxyl-terminus of E-DIII (B.) The mature flavivirus particle contains 90 E protein dimers arranged with pseudo-T=3 icosahedral symmetry. The domain structure of individual E proteins is colored as described above. Image constructed using PDB 1K4R. (C.) Structural details of the open and closed state of the β-OG pocket. The structure of the E protein of the DENV2 strain S1 was solved in the presence of the detergent n-octyl-β-D-glucoside [13]. The structure of E in the presence (colored ribbons; PDB 1OKE) or absence (grey ribbons; PDB 1OAN) of detergent are superimposed to illustrate changes in the orientation of the kl loop in the pocket “open” and “closed” state. The detergent molecule is shown as a ball and stick representation.
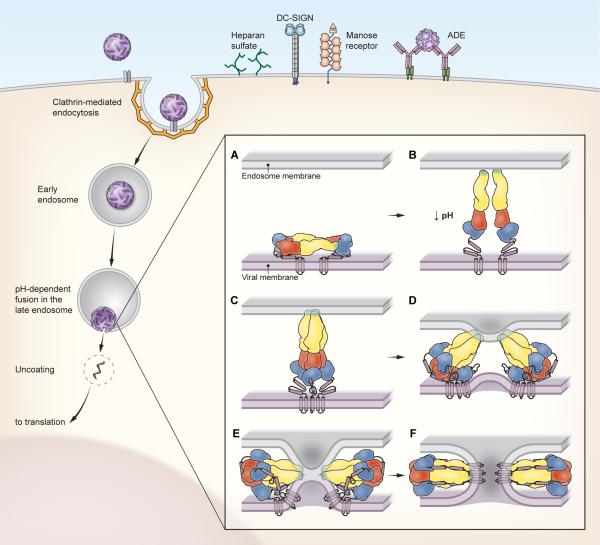
Figure 3. Entry and fusion of flaviviruses.
The cell biology of flavivirus entry into cells is incompletely understood. Viruses bind the cell surface via interactions with largely uncharacterized cellular receptors and/or attachment factors. Cellular factors shown to directly increase the efficiency of virus attachment are shown and include heparan sulfate, DC-SIGN, and the mannose receptor. In addition, antibodies may significantly increase the efficiency of virus attachment and entry via interactions with Fcγ-receptors (a process termed antibody-dependent enhancement of infection)[54]. Flaviviruses are internalized into cells via clathrin-mediated endocytosis and traffic into a late endosomal compartment in which fusion occurs (detailed in [60]). The current model for the fusion process is depicted in the inset. Upon exposure to an acidic pH, the E protein dimers present on mature virions (panel A) disassociate and project away from the cell surface. The extended intermediate shown in panel B was captured by the low pH structure of WNV bound to the neutralizing mAb E16 [72]. In this orientation, the fusion loop of the E protein is positioned to interact with the endosomal membrane coincident with the formation of the core trimer (panel C). E-DIII then folds back against the exterior surface of the core trimer (panel D). This rotation and the packing of the helical stem anchor into grooves on the exterior of the E protein core trimer (panel E) pulls the carboxyl-terminal viral membrane-anchored transmembrane domains towards the target cell membrane. This dramatic conformational change is thought to provide the free energy required for lipid mixing (hemifusion; panel E) and fusion (panel F).
The structure of the E ectodomain of DENV2 solved by Modis and Harrison revealed a hydrophobic pocket at the junction of E-DI and E-DII capable of accommodating an n-octyl-β-D-glucoside detergent molecule [13]. This structure will be referred to hereafter as the β-OG pocket (Figure 1c). Mutations around this pocket alter the pH required for E protein activation [20-23]. E protein structures solved in the presence or absence of this detergent suggest that a hairpin loop (the kl loop) regulates access to the pocket, rendering it in an “open” or “closed” state. In the closed state (captured by all other reported E protein structures) the kl loop lies over the top of the pocket. Because the β-OG pocket is located at an interface between E-DI and E-DII that rotates considerably during fusion (Figure 3), molecules that fit into the pocket may block this transition and thus have potential as therapeutics (Table 1).
Table 1.
Potential flavivirus entry inhibitors
| Step in Pathway | Target | Inhibitor examples |
|---|---|---|
| Endocytic entry | Receptor/attachment factor | Heparan |
| Endocytic uptake | Clathrin inhibitors | |
| Endosomal acidification | Chloroquine, bafilomycin | |
| Early fusion protein changes | E hinge movements | “Pocket” binding compounds |
| Fusion loop-membrane insertion | NA | |
| Trimerization | NA | |
| Late fusion protein changes | Outer layer packing | E-DIII |
| 1662G07 | ||
| E stem peptides | ||
| Lipid rearrangements during fusion | Membrane curvature | LJ001 |
| intermediates | Rigid amphiphiles |
Flavivirus virions
Flaviviruses are heterogeneous and dynamic structures (reviewed in [24-26]). Newly assembled flaviviruses bud into the lumen of the endoplasmic reticulum as immature non-infectious virions that incorporate sixty spikes, which are trimers of heterodimers of the transmembrane prM and E proteins [27,28]. As the virus particles transit the secretory pathway, the acidic environment of the exocytic pathway triggers a conformational rearrangement of the virus particle that allows cellular furin to process prM into pr peptide and transmembrane M [29-31]. The pr peptide remains bound to the virus particle until virus exit into the neutral pH of the extracellular space. prM processing and pr release are required for virion infectivity [32]. On mature virions, the E proteins are organized as 30 rafts of three anti-parallel dimers arranged with pseudo-icosahedral symmetry (Figure 1b) [10,19]. These dimers are oriented parallel to the viral membrane, resulting in the relatively smooth appearance of the mature virion. In this configuration, E proteins are located adjacent to a five-, three-, or two-fold symmetry axis. Whether E proteins located in each of these symmetry environments play distinct roles in virus entry or fusion (e.g. receptor binding) is unknown.
While cryo-electron microscopy studies (cryoEM) have provided valuable snapshots of the structure of flaviviruses [19,27,30,33], several recent studies suggest unresolved structural complexity may contribute significantly to the biology of flaviviruses. Under physiological conditions, flaviviruses likely exist as an ensemble of conformations at equilibrium; the “breathing” of flaviviruses has been suggested to markedly impact antibody recognition [25,26,34]. Whether the structural dynamics of the virion also regulate exposure of surfaces recognized by cellular proteins involved in virus attachment remains to be studied. Flaviviruses released from cells are also heterogeneous with respect to the extent of prM cleavage [35-39]. A recent investigation of the structure of such “partially mature” virions suggests these virus particles are a mosaic of regions of “spikey” immature character and those with a smooth structure consistent with mature viruses [40]. More than 90% of DENV virions retain at least some uncleaved prM [35]. Because of the very different orientation of the E proteins in these two regions (and the unresolved junction between regions), it is possible these portions of partially mature virions provide unique surfaces for interactions with cellular attachment factors and receptors.
Membrane fusion
Flavivirus membrane fusion involves a series of conformational and organizational changes in the E proteins triggered by exposure to the mildly acidic endosomal environment. Fusion is thought to proceed in several rapid steps (Figure 3). Exposure of the virion to low pH (~6.4 or below) results in the dissociation of the E homodimer and the rotation of E-DII away from E-DI and the viral membrane (Figure 2). The “hinge” region enabling this rotation includes the hydrophobic β-OG pocket. This rotation results in the exposure of the fusion loop, which inserts into the outer leaflet of the target cell membrane. E proteins in this extended state form a core trimer through interactions of E-DI and E-DII. E-DIII then moves towards the target cell membrane to pack against the core trimer, followed by packing of the stem anchor to produce the final post-fusion E structure, a “hairpin” in which the fusion loops and TM anchors are at the same end of the trimer. The well-studied fusion proteins of HIV-1 and influenza virus mediate fusion by refolding into a topologically similar (although structurally distinct) hairpin (reviewed by [41]). The number of E trimers required for flavivirus fusion is unknown, although inter-trimer interactions are observed in vitro and suggest possible cooperative effects [42].
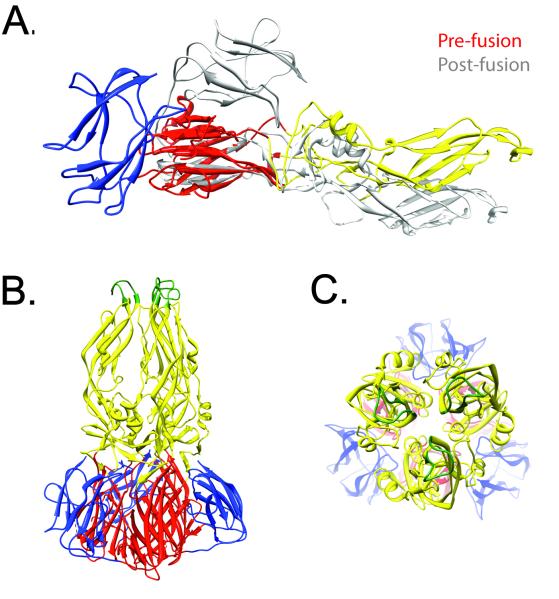
Figure 2. Structural transitions involved in E protein-mediated fusion.
Exposure of the E protein to an acidic environment triggers a conformational change in the E protein and a transition from antiparallel dimers (depicted in Figure 1) to a trimer that projects away from the surface of the virus particle. (A.) The structural changes that occur during this transition are shown. The structure of the DENV2 pre-fusion monomer (colored ribbons; PDB 1OAN) is shown superimposed on the post-fusion structure of E (grey ribbon; PDB 1OK8)[73,86]. This depiction illustrates the ~30° rotation that occurs between E-DI and E-DII, and the dramatic folding-back of E-DIII against the exterior surface of the trimer. (B.) Structure of the post-fusion trimer, showing the DI/DII core trimer packed by E-DIII. The stem region (not contained in this structure) is predicted to pack against the surface of the core trimer. (C.) Face-on view of the fusion loops at the tip of the E trimer from the perspective of the endosomal membrane.
The cell biology of flavivirus entry
Despite considerable interest, the identity and roles of cellular factors that participate in flavivirus entry into cells remain poorly understood. For many viruses, interactions between viral envelope proteins and cellular receptors play an essential role in determining cellular tropism and/or activating the fusion potential of virally-encoded fusion proteins (e.g. chemokine receptors and HIV-1). A required role for specific cellular factors as flavivirus “receptors” is not so clear. Flaviviruses are capable of fusing with synthetic membranes devoid of cellular proteins [43-45]. Thus, the cellular requirements for entry may be more general; several paths and/or cellular factors may contribute to delivering the virus into a low pH compartment in which membrane fusion can occur. The first (often quite inefficient) step in the entry of viruses into cells is the stable attachment of the virion to the cell surface [46]. Cellular factors that increase the efficiency of this process, but are not absolutely required for infection, are referred to as “attachment factors”. Flavivirus attachment to cells is mediated by the E protein [47], although interactions between cellular factors and prM may be sufficient for infectious entry in some contexts [36].
Two of the best characterized attachment structures for flaviviruses are heparan sulfate and the c-type lectin DC-SIGN. Heparan sulfates (HS) are a heterogeneous group of linear anionic polysaccharides that can decorate cell surface proteins and have been shown to interact with several flaviviruses [48-50]. HS binding is mediated by basic patches on the surface of the E protein [49,51], reflects an adaptation to propagation in tissue culture [51], and results in reduced virulence [52]. Thus, while HS binding can play a critical role in the entry of other viruses, the significance of HS interactions with flaviviruses is not yet clear. DC-SIGN (CD209) and DC-SIGNR (CD209L) bind DENV and WNV via interactions with carbohydrates present on the E and prM proteins [14-16,36]. While the expression of DC-SIGN on cells may dramatically increase their permissiveness to infection, experiments with internalization-deficient forms of DC-SIGN suggest that other cellular factors may actually mediate virus internalization [53]. It is also clear that antibody-bound flaviviruses can be internalized by the Fc-receptor in some cell types, a process that may enhance infection (reviewed by [54]).
Several lines of evidence indicate that flaviviruses enter mammalian and insect cells via clathrin-mediated endocytosis [46,55-60]. For example, the entry of DENV into living cells has been visualized using single virus particle tracking methodology [60]. This elegant study demonstrates that DENV binds cells and diffuses across the surface until it encounters a pre-formed clathrin coated pit. These structures are then internalized and deliver the virion to an early endosomal compartment (Rab5+) that subsequently matures into a late endosome with the acquisition of Rab7. Both early and late endosomal compartments have been implicated as the site of viral membrane fusion [55,60]. A recent study suggests the unique lipid composition of the late endosome may be required for DENV fusion [45].
Several recent studies suggest that under some circumstances flaviviruses may enter cells via routes that do not involve clathrin [61,62]. For example, the DENV2 strain NGC is markedly less sensitive to treatment of Vero cells by chlorpromazine, an inhibitor of clathrin-mediated endocytosis, than a DENV1 strain assayed in parallel. In contrast, DENV2 infection of A549 cells was reported to be dependent on clathrin [62]. The viral determinants that influence the pathway of virus internalization and its variation among cell types are not clear. Perhaps most importantly, does this matter? Does delivery of the virion to an acidic compartment via one endocytic pathway vs. another confer advantages to specific infectivity or virus yield?
Targeting virus entry: the potential for anti-viral therapies
Each step of the flavivirus entry and fusion pathway represents a potential druggable target (Figure 3 and Table 1). Cellular processes usurped by flaviviruses during virus entry, such as endocytosis and endosomal acidification, may be difficult to inhibit without significant cytotoxicity. Other steps involving specific flavivirus interactions with cells or the mechanics of E protein fusion represent promising targets for intervention. Targeting analogous steps during the virus entry pathway has led to clinically useful inhibitors of HIV-1 (reviewed by [63]). In contrast to HIV-1, flavivirus fusion is extremely rapid, being essentially complete within seconds of exposure to acidic conditions at 37°C [43-45]. Thus, the speed of fusion and its endosomal location are important challenges in the development of flavivirus fusion inhibitors.
Cell surface attachment
This step is critical for flavivirus infection, and its utility as a target is supported by the finding that a number of monoclonal antibodies (mAbs) neutralize infection by blocking initial attachment to cells (reviewed by [54]). In addition, highly sulfated forms of heparan can block infection by HS-utilizing flavivirus strains (reviewed in [6]). The identification of additional receptors and/or attachment factors may enable the development of more specific inhibitors in the future.
Endocytosis and acidification
Inhibitors of endocytosis or endosomal acidification have been useful in defining the entry pathway. Results from entry studies and genome-wide screens may identify host factors in these pathways that can be targeted without deleterious effects on cells. Translating these to clinical use will be complex, as shown by clinical trials with chloroquine, which efficiently inhibits acidification and DENV infection in vitro but does not have strong protective effects in vivo [64]. This may reflect the difficulties of raising endosomal pH above the relatively high DENV fusion threshold.
E protein early conformational changes
Identification of the β-OG pocket and its proposed role in the conformational changes that drive membrane fusion suggested its value as a target for antiviral drug discovery [13]. Numerous in silico screens for molecules capable of binding the β-OG pocket have identified lead compounds that inhibit flavivirus infection [65-68]. Several such compounds have subsequently been optimized for greater activity and reduced toxicity [69,70]. The most potent of these inhibitors block infection at high nanomolar/low micromolar concentrations in vitro. Time of addition experiments suggest an early step in the viral lifecycle is blocked by these molecules [67,68]. In support of this mechanism, several of these inhibitors have been shown more directly to inhibit fusion using cell-cell [65,68] and virus-liposome fusion assays [68]. A peptide targeting the hinge region of DENV E also inhibits virus infection, apparently inducing changes in the virus particle that block cell surface binding and/or E conformational changes [71]. The subsequent steps of fusion loop insertion into the target membrane and E core trimer formation, while clearly required for flavivirus fusion, have not yet been specifically targeted. The neutralizing mAb E16 binds E-DIII and blocks WNV fusion by interfering with initial rearrangements on the particle surface, trapping the particle in a radially expanded form thought to reflect the extension of the E protein stem away from the virion surface [72].
E protein refolding to final hairpin
Formation of the E homotrimer is completed by packing of the core trimer with an outer layer composed of E-DIII and the stem region [73,74]. This E refolding step and consequent virus fusion and infection can be blocked by the addition of soluble recombinant E-DIII molecules that contain the first helix of the stem [75,76]. The final zipping of the E protein stem against the core trimer can be inhibited by exogenous stem peptide fragments [77-79]. Peptides derived from the membrane proximal portion of the stem anchor (including the conserved spacer and helix two, Figure 3a) block infection and membrane fusion. Such stem peptides first interact hydrophobically with the viral membrane, and are thus carried into acidic endosomes where the E core trimer target is generated and inhibition of fusion occurs [77]. This endosomal delivery, which does not occur with DIII proteins, is a critical factor in stem peptide efficacy. The impact of virion maturation efficiency and structural dynamics on peptide access to the viral membrane and inhibition has not yet been investigated.
The interaction of a labeled stem peptide with a “stemless” E trimer was used as a screen for small molecule fusion inhibitors [80]. The inhibitor 1662G07 and derivatives thereof block stem-trimer binding, DENV infection, and membrane fusion. Rather than targeting the stem-trimer interaction directly, 1662G07 binds E-DI/DII and prefusion E dimers as well as the post-fusion trimer. These results suggest that 1662G07 inhibits fusion by trapping the trimer in a β-OG pocket-open state that is inactive in stem binding.
Inhibitors targeting the virus lipid bilayer
During fusion the virus lipid bilayer and target membrane form highly curved transient structures such as the hemifusion intermediate in which the outer leaflets of the two membranes have merged (Figure 3E). Such curvature is energetically unfavorable and can be inhibited by the presence of lipids or other amphiphiles whose shape favors the opposing curvature (reviewed by [81]). Two different approaches have identified compounds that intercalate into the virus membrane and block infection by flaviviruses and other enveloped viruses by interfering with late stages of lipid rearrangement during fusion [82,83]. Importantly, the lipid biosynthetic capacity of cells makes them relatively resistant to the inhibitors, and thus such antiviral compounds, while acting broadly to inhibit many enveloped viruses, have low cytotoxicity.
The pr-E interaction
Recent advances in understanding the biogenesis of the flavivirus fusion protein suggest an additional target for antiviral therapies. As discussed above, prM is cleaved during exocytic transit of the virion. Interaction of the pr peptide and the E protein is maintained in the mildly acidic pH of the exocytic pathway and serves to silence the fusion protein during virus exit [84,85]. pr-E binding has been reconstituted in vitro [85], suggesting a system to screen for small molecules that could block this interaction and thus allow premature triggering and inactivation of the fusion protein.
Summary
Flaviviruses enter cells via a multiple-step process orchestrated by E proteins on the virion. Insight into the mechanisms of attachment, entry, and fusion has informed the strategies for blocking this pathway. While at extremely early stages of development from a therapeutic perspective, these inhibitors will undoubtedly be of immediate use in studies to understand virus entry with greater precision and resolution.
Pierson and Kielian COIV highlights.
The flavivirus membrane contains a dense and organized array of E proteins that mediate receptor binding and membrane fusion.
Flaviviruses are heterogeneous and dynamic structures that may interact with cells via surfaces not predicted by existing models.
The functions of host proteins in virus binding and endocytic uptake remain largely undefined.
Recent advances in our understanding of virus fusion suggest potential antiviral targets.
Inhibitors that block distinct steps of virus fusion have been identified.
Acknowledgements
Work in the authors’ laboratories was supported by the intramural program of the National Institute of Allergy and Infectious Disease (TCP), and by National Institutes of Health grants GM057454, AI075647, and U54AI057158-Lipkin (MK). The authors wish to thank Dr. Heather Hickman and members of our laboratories for critical evaluation of this review. We would like to thank Ethan Tyler (NIH/OD) and Phong Lee (NIAID, NIH) for assistance with preparation of the figures. We acknowledge the important contributions of those researchers whose work was not fully cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gould EA, Solomon T. Pathogenic flaviviruses. Lancet. 2008;371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- 2.Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, editors. Fields Virology. edn 5th vol 1. Lippincott-Williams & Wilkins; 2007. pp. 1101–1152. [Google Scholar]
- 3.Sanchez-San Martin C, Liu CY, Kielian M. Dealing with low pH: entry and exit of alphaviruses and flaviviruses. Trends in microbiology. 2009;17:514–521. doi: 10.1016/j.tim.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 5.Diamond MS, Pierson TC, Roehrig JT. Antibody Therapeutics Against Flaviviruses. In: Shi PY, editor. Molecular Virology and Control of Flaviviruses. Caister Academic Press; 2012. [Google Scholar]
- 6.Perera R, Khaliq M, Kuhn RJ. Closing the door on flaviviruses: entry as a target for antiviral drug design. Antiviral research. 2008;80:11–22. doi: 10.1016/j.antiviral.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melikyan GB. Common principles and intermediates of viral protein-mediated fusion: the HIV-1 paradigm. Retrovirology. 2008;5:111. doi: 10.1186/1742-4690-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luca VC, AbiMansour J, Nelson CA, Fremont DH. Crystal structure of the Japanese encephalitis virus envelope protein. J Virol. 2012;86:2337–2346. doi: 10.1128/JVI.06072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J Virol. 2001;75:4268–4275. doi: 10.1128/JVI.75.9.4268-4275.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis CW, Mattei LM, Nguyen HY, Ansarah-Sobrinho C, Doms RW, Pierson TC. The location of asparagine-linked glycans on West Nile virions controls their interactions with CD209 (dendritic cell-specific ICAM-3 grabbing nonintegrin) J Biol Chem. 2006;281:37183–37194. doi: 10.1074/jbc.M605429200. [DOI] [PubMed] [Google Scholar]
- 15.Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL, et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J Exp Med. 2003;197:823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navarro-Sanchez E, Altmeyer R, Amara A, Schwartz O, Fieschi F, Virelizier JL, Arenzana-Seisdedos F, Despres P. Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 2003;4:723–728. doi: 10.1038/sj.embor.embor866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller JL, de Wet BJ, Martinez-Pomares L, Radcliffe CM, Dwek RA, Rudd PM, Gordon S. The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog. 2008;4:e17. doi: 10.1371/journal.ppat.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allison SL, Stiasny K, Stadler K, Mandl CW, Heinz FX. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J Virol. 1999;73:5605–5612. doi: 10.1128/jvi.73.7.5605-5612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn RJ. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beasley DW, Aaskov JG. Epitopes on the dengue 1 virus envelope protein recognized by neutralizing IgM monoclonal antibodies. Virology. 2001;279:447–458. doi: 10.1006/viro.2000.0721. [DOI] [PubMed] [Google Scholar]
- 21.Lee E, Weir RC, Dalgarno L. Changes in the dengue virus major envelope protein on passaging and their localization on the three-dimensional structure of the protein. Virology. 1997;232:281–290. doi: 10.1006/viro.1997.8570. [DOI] [PubMed] [Google Scholar]
- 22.Hurrelbrink RJ, McMinn PC. Attenuation of Murray Valley encephalitis virus by site-directed mutagenesis of the hinge and putative receptor-binding regions of the envelope protein. J Virol. 2001;75:7692–7702. doi: 10.1128/JVI.75.16.7692-7702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monath TP, Arroyo J, Levenbook I, Zhang ZX, Catalan J, Draper K, Guirakhoo F. Single mutation in the flavivirus envelope protein hinge region increases neurovirulence for mice and monkeys but decreases viscerotropism for monkeys: relevance to development and safety testing of live, attenuated vaccines. J Virol. 2002;76:1932–1943. doi: 10.1128/JVI.76.4.1932-1943.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierson TC, Diamond MS. Degrees of maturity: the complex structure and biology of flaviviruses. Curr Opin Virol. 2012;2:168–175. doi: 10.1016/j.coviro.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- 26.Dowd KA, Jost CA, Durbin AP, Whitehead SS, Pierson TC. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 2011;7:e1002111. doi: 10.1371/journal.ppat.1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG. Structures of immature flavivirus particles. EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG. Structure of immature West Nile virus. J Virol. 2007;81:6141–6145. doi: 10.1128/JVI.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadler K, Allison SL, Schalich J, Heinz FX. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71:8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu IM, Zhang W, Holdaway HA, Li L, Kostyuchenko VA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science. 2008;319:1834–1837. doi: 10.1126/science.1153264. [DOI] [PubMed] [Google Scholar]
- 31.Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 32.Elshuber S, Allison SL, Heinz FX, Mandl CW. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J Gen Virol. 2003;84:183–191. doi: 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- 33.Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile virus. Science. 2003;302:248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- 34.Sabo MC, Luca VC, Ray SC, Bukh J, Fremont DH, Diamond MS. Hepatitis C virus epitope exposure and neutralization by antibodies is affected by time and temperature. Virology. 2012;422:174–184. doi: 10.1016/j.virol.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Junjhon J, Edwards TJ, Utaipat U, Bowman VD, Holdaway HA, Zhang W, Keelapang P, Puttikhunt C, Perera R, Chipman PR, et al. Influence of pr-M cleavage on the heterogeneity of extracellular dengue virus particles. J Virol. 2010;84:8353–8358. doi: 10.1128/JVI.00696-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis CW, Nguyen HY, Hanna SL, Sanchez MD, Doms RW, Pierson TC. West Nile virus discriminates between DC-SIGN and DC-SIGNR for cellular attachment and infection. J Virol. 2006;80:1290–1301. doi: 10.1128/JVI.80.3.1290-1301.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson S, Jost CA, Xu Q, Ess J, Martin JE, Oliphant T, Whitehead SS, Durbin AP, Graham BS, Diamond MS, et al. Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog. 2008;4:e1000060. doi: 10.1371/journal.ppat.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang KJ, Yang YC, Lin YS, Huang JH, Liu HS, Yeh TM, Chen SH, Liu CC, Lei HY. The dual-specific binding of dengue virus and target cells for the antibody-dependent enhancement of dengue virus infection. J Immunol. 2006;176:2825–2832. doi: 10.4049/jimmunol.176.5.2825. [DOI] [PubMed] [Google Scholar]
- 39.Rodenhuis-Zybert IA, van der Schaar HM, da Silva Voorham JM, van der Ende-Metselaar H, Lei HY, Wilschut J, Smit JM. Immature dengue virus: a veiled pathogen? PLoS Pathog. 2010;6:e1000718. doi: 10.1371/journal.ppat.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plevka P, Battisti AJ, Junjhon J, Winkler DC, Holdaway HA, Keelapang P, Sittisombut N, Kuhn RJ, Steven AC, Rossmann MG. Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 2011;12:602–606. doi: 10.1038/embor.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stiasny K, Heinz FX. Effect of membrane curvature-modifying lipids on membrane fusion by tick-borne encephalitis virus. J Virol. 2004;78:8536–8542. doi: 10.1128/JVI.78.16.8536-8542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moesker B, Rodenhuis-Zybert IA, Meijerhof T, Wilschut J, Smit JM. Characterization of the functional requirements of West Nile virus membrane fusion. J Gen Virol. 2010;91:389–393. doi: 10.1099/vir.0.015255-0. [DOI] [PubMed] [Google Scholar]
- 44.Gollins SW, Porterfield JS. pH-dependent fusion between the flavivirus West Nile and liposomal model membranes. J Gen Virol. 1986;67(Pt 1):157–166. doi: 10.1099/0022-1317-67-1-157. [DOI] [PubMed] [Google Scholar]
- 45.Zaitseva E, Yang ST, Melikov K, Pourmal S, Chernomordik LV. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog. 2010;6:e1001131. doi: 10.1371/journal.ppat.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gollins SW, Porterfield JS. Flavivirus infection enhancement in macrophages: an electron microscopic study of viral cellular entry. J Gen Virol. 1985;66(Pt 9):1969–1982. doi: 10.1099/0022-1317-66-9-1969. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y, Maguire T, Marks RM. Demonstration of binding of dengue virus envelope protein to target cells. J Virol. 1996;70:8765–8772. doi: 10.1128/jvi.70.12.8765-8772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee E, Lobigs M. Substitutions at the putative receptor-binding site of an encephalitic flavivirus alter virulence and host cell tropism and reveal a role for glycosaminoglycans in entry. J Virol. 2000;74:8867–8875. doi: 10.1128/jvi.74.19.8867-8875.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 50.Kroschewski H, Allison SL, Heinz FX, Mandl CW. Role of heparan sulfate for attachment and entry of tick-borne encephalitis virus. Virology. 2003;308:92–100. doi: 10.1016/s0042-6822(02)00097-1. [DOI] [PubMed] [Google Scholar]
- 51.Mandl CW, Kroschewski H, Allison SL, Kofler R, Holzmann H, Meixner T, Heinz FX. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J Virol. 2001;75:5627–5637. doi: 10.1128/JVI.75.12.5627-5637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee E, Lobigs M. Mechanism of virulence attenuation of glycosaminoglycan-binding variants of Japanese encephalitis virus and Murray Valley encephalitis virus. J Virol. 2002;76:4901–4911. doi: 10.1128/JVI.76.10.4901-4911.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lozach PY, Burleigh L, Staropoli I, Navarro-Sanchez E, Harriague J, Virelizier JL, Rey FA, Despres P, Arenzana-Seisdedos F, Amara A. Dendritic cell-specific intercellular adhesion molecule 3-grabbing non-integrin (DC-SIGN)-mediated enhancement of dengue virus infection is independent of DC-SIGN internalization signals. J Biol Chem. 2005;280:23698–23708. doi: 10.1074/jbc.M504337200. [DOI] [PubMed] [Google Scholar]
- 54.Dowd KA, Pierson TC. Antibody-mediated neutralization of flaviviruses: a reductionist view. Virology. 2011;411:306–315. doi: 10.1016/j.virol.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krishnan MN, Sukumaran B, Pal U, Agaisse H, Murray JL, Hodge TW, Fikrig E. Rab 5 is required for the cellular entry of dengue and West Nile viruses. J Virol. 2007;81:4881–4885. doi: 10.1128/JVI.02210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chu JJ, Ng ML. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J Virol. 2004;78:10543–10555. doi: 10.1128/JVI.78.19.10543-10555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu JJ, Leong PW, Ng ML. Analysis of the endocytic pathway mediating the infectious entry of mosquito-borne flavivirus West Nile into Aedes albopictus mosquito (C6/36) cells. Virology. 2006;349:463–475. doi: 10.1016/j.virol.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 58.Nawa M, Takasaki T, Yamada K, Kurane I, Akatsuka T. Interference in Japanese encephalitis virus infection of Vero cells by a cationic amphiphilic drug, chlorpromazine. J Gen Virol. 2003;84:1737–1741. doi: 10.1099/vir.0.18883-0. [DOI] [PubMed] [Google Scholar]
- 59.Acosta EG, Castilla V, Damonte EB. Functional entry of dengue virus into Aedes albopictus mosquito cells is dependent on clathrin-mediated endocytosis. J Gen Virol. 2008;89:474–484. doi: 10.1099/vir.0.83357-0. [DOI] [PubMed] [Google Scholar]
- 60.van der Schaar HM, Rust MJ, Chen C, van der Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 2008;4:e1000244. doi: 10.1371/journal.ppat.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suksanpaisan L, Susantad T, Smith DR. Characterization of dengue virus entry into HepG2 cells. J Biomed Sci. 2009;16:17. doi: 10.1186/1423-0127-16-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Acosta EG, Castilla V, Damonte EB. Alternative infectious entry pathways for dengue virus serotypes into mammalian cells. Cell Microbiol. 2009;11:1533–1549. doi: 10.1111/j.1462-5822.2009.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Didigu CA, Doms RW. Novel approaches to inhibit HIV entry. Viruses. 2012;4:309–324. doi: 10.3390/v4020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tricou V, Minh NN, Van TP, Lee SJ, Farrar J, Wills B, Tran HT, Simmons CP. A randomized controlled trial of chloroquine for the treatment of dengue in Vietnamese adults. PLoS Negl Trop Dis. 2010;4:e785. doi: 10.1371/journal.pntd.0000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kampmann T, Yennamalli R, Campbell P, Stoermer MJ, Fairlie DP, Kobe B, Young PR. In silico screening of small molecule libraries using the dengue virus envelope E protein has identified compounds with antiviral activity against multiple flaviviruses. Antiviral research. 2009;84:234–241. doi: 10.1016/j.antiviral.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 66.Zhou Z, Khaliq M, Suk JE, Patkar C, Li L, Kuhn RJ, Post CB. Antiviral compounds discovered by virtual screening of small-molecule libraries against dengue virus E protein. ACS Chem Biol. 2008;3:765–775. doi: 10.1021/cb800176t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang QY, Patel SJ, Vangrevelinghe E, Xu HY, Rao R, Jaber D, Schul W, Gu F, Heudi O, Ma NL, et al. A small-molecule dengue virus entry inhibitor. Antimicrob Agents Chemother. 2009;53:1823–1831. doi: 10.1128/AAC.01148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poh MK, Yip A, Zhang S, Priestle JP, Ma NL, Smit JM, Wilschut J, Shi PY, Wenk MR, Schul W. A small molecule fusion inhibitor of dengue virus. Antiviral research. 2009;84:260–266. doi: 10.1016/j.antiviral.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 69.Mayhoub AS, Khaliq M, Botting C, Li Z, Kuhn RJ, Cushman M. An investigation of phenylthiazole antiflaviviral agents. Bioorg Med Chem. 2011;19:3845–3854. doi: 10.1016/j.bmc.2011.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Z, Khaliq M, Zhou Z, Post CB, Kuhn RJ, Cushman M. Design, synthesis, and biological evaluation of antiviral agents targeting flavivirus envelope proteins. J Med Chem. 2008;51:4660–4671. doi: 10.1021/jm800412d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Costin JM, Jenwitheesuk E, Lok SM, Hunsperger E, Conrads KA, Fontaine KA, Rees CR, Rossmann MG, Isern S, Samudrala R, et al. Structural optimization and de novo design of dengue virus entry inhibitory peptides. PLoS Negl Trop Dis. 2010;4:e721. doi: 10.1371/journal.pntd.0000721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaufmann B, Chipman PR, Holdaway HA, Johnson S, Fremont DH, Kuhn RJ, Diamond MS, Rossmann MG. Capturing a flavivirus pre-fusion intermediate. PLoS Pathog. 2009;5:e1000672. doi: 10.1371/journal.ppat.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 74.Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 2004;23:728–738. doi: 10.1038/sj.emboj.7600064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liao M, Kielian M. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus membrane fusion. J Cell Biol. 2005;171:111–120. doi: 10.1083/jcb.200507075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liao M, Sanchez-San Martin C, Zheng A, Kielian M. In vitro reconstitution reveals key intermediate states of trimer formation by the dengue virus membrane fusion protein. J Virol. 2010;84:5730–5740. doi: 10.1128/JVI.00170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of flavivirus entry derived from the E protein stem. J Virol. 2010;84:12549–12554. doi: 10.1128/JVI.01440-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmidt AG, Yang PL, Harrison SC. Peptide inhibitors of dengue-virus entry target a late-stage fusion intermediate. PLoS Pathog. 2010;6:e1000851. doi: 10.1371/journal.ppat.1000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hrobowski YM, Garry RF, Michael SF. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol J. 2005;2:49. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schmidt AG, Lee K, Yang PL, Harrison SC. Small-molecule inhibitors of dengue-virus entry. PLoS Pathog. 2012;8:e1002627. doi: 10.1371/journal.ppat.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Melikyan GB. Driving a wedge between viral lipids blocks infection. Proc Natl Acad Sci U S A. 2010;107:17069–17070. doi: 10.1073/pnas.1012748107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolf MC, Freiberg AN, Zhang T, Akyol-Ataman Z, Grock A, Hong PW, Li J, Watson NF, Fang AQ, Aguilar HC, et al. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci U S A. 2010;107:3157–3162. doi: 10.1073/pnas.0909587107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.St Vincent MR, Colpitts CC, Ustinov AV, Muqadas M, Joyce MA, Barsby NL, Epand RF, Epand RM, Khramyshev SA, Valueva OA, et al. Rigid amphipathic fusion inhibitors, small molecule antiviral compounds against enveloped viruses. Proc Natl Acad Sci U S A. 2010;107:17339–17344. doi: 10.1073/pnas.1010026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu IM, Holdaway HA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J Virol. 2009;83:12101–12107. doi: 10.1128/JVI.01637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zheng A, Umashankar M, Kielian M. In vitro and in vivo studies identify important features of dengue virus pr-E protein interactions. PLoS Pathog. 2010;6:e1001157. doi: 10.1371/journal.ppat.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol. 2005;79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
References of significant interest
- *.Kaufmann B, Chipman PR, Holdaway HA, Johnson S, Fremont DH, Kuhn RJ, Diamond MS, Rossmann MG. Capturing a flavivirus pre-fusion intermediate. PLoS Pathog. 2009;5:e1000672. doi: 10.1371/journal.ppat.1000672. The mAb E16 neutralizes WNV infection by inhibiting viral membrane fusion in the endosome. This manuscript describes the structure of WNV bound to E16 at low pH, revealing a previous unidentified extended conformation of the E protein stem region occurring at an early stage of the fusion process.
- *.Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol. 2008;15:312–317. doi: 10.1038/nsmb.1382. In this study, the authors solve the structure of the mAb 1A1D-2 bound to the DENV E protein and identify how it interacts with the intact virion using cyro-electron microscopy. MAb 1A1D-2 binds the virion in a temperature-dependent manner and traps it in a previously unidentified structure thought to arise through the dynamic motion, or “breathing”, of the virus particle. These studies suggest DENV exists as an ensemble of conformations at physiological temperatures.
- *.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci U S A. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. This manuscript describes the structure of the ectodomain of DENV serotype 2 in the presence or absence of a n-octyl-β-D-glucoside detergent molecule. The detergent molecule in this structure was found in a hydrophobic pocket at the junction between domain I and domain II shown previously to play a role in the fusion process. The existence of a pocket into which small molecules may insert identified a novel therapeutic target that has since been exploited for drug development.
- **.Plevka P, Battisti AJ, Junjhon J, Winkler DC, Holdaway HA, Keelapang P, Sittisombut N, Kuhn RJ, Steven AC, Rossmann MG. Maturation of flaviviruses starts from one or more icosahedrally independent nucleation centres. EMBO Rep. 2011;12:602–606. doi: 10.1038/embor.2011.75. In this study, the authors employ cryo-electron tomographic methods to determine the structure of partially mature dengue viruses that retain uncleaved prM protein. The results presented within suggest the E proteins of partially mature virions are organized as distinct patches of dimers or trimers similar to those found on the mature or immature virions, respectively.
- **.Schmidt AG, Lee K, Yang PL, Harrison SC. Small-molecule inhibitors of dengue-virus entry. PLoS Pathog. 2012;8:e1002627. doi: 10.1371/journal.ppat.1002627. This manuscript describes a small molecule DENV inhibitor that inhibits stem peptide binding to post-fuson E protein trimers though interactions with the β-OG pocket.
- **.St Vincent MR, Colpitts CC, Ustinov AV, Muqadas M, Joyce MA, Barsby NL, Epand RF, Epand RM, Khramyshev SA, Valueva OA, et al. Rigid amphipathic fusion inhibitors, small molecule antiviral compounds against enveloped viruses. Proc Natl Acad Sci U S A. 2010;107:17339–17344. doi: 10.1073/pnas.1010026107. Viral membrane fusion is driven by conformational changes in envelope proteins incorporated into the virion that induce membrane curvature. This manuscript (and the study by Wolf et al.) identifies a novel and general method for inhibiting viral infection through the insertion of molecules in the membrane that oppose the curvature required for fusion.
- *.van der Schaar HM, Rust MJ, Chen C, van der Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 2008;4:e1000244. doi: 10.1371/journal.ppat.1000244. In this manuscript, the authors employ cutting-edge confocal microscopic techniques to visualize the entry of single DENV particles into cells. This pioneering study not only defines the features of the endocytic pathway utilized by flaviviruses, but also the kinetics of each step in the virus entry process.
- **.Wolf MC, Freiberg AN, Zhang T, Akyol-Ataman Z, Grock A, Hong PW, Li J, Watson NF, Fang AQ, Aguilar HC, et al. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc Natl Acad Sci U S A. 2010;107:3157–3162. doi: 10.1073/pnas.0909587107. Viral membrane fusion is driven by conformational changes in envelope proteins incorporated into the virion that induce membrane curvature. This manuscript (and the study by St. Vincent et al.) identifies a novel and general method for inhibiting viral infection through the insertion of molecules in the membrane that oppose the curvature required for fusion.
- *.Zaitseva E, Yang ST, Melikov K, Pourmal S, Chernomordik LV. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog. 2010;6:e1001131. doi: 10.1371/journal.ppat.1001131. This study identifies a requirement for the unique lipid composition of the late endosome for membrane fusion of DENV.