Abstract
Micro-RNAs (miRNAs) are short non-coding RNA species, thought to act primarily through downregulation of target mRNA species with subsequent decrease in encoded proteins. Recent studies revealed that miRNAs play pivotal roles in physiology and disease, and therapeutic targeting has started being investigated. Generally, the up-regulation of miRNAs is achieved through administration of synthetic miRNAs or administration of miRNA expressing vectors. The down-regulation of miRNAs is achieved through administration of anti-sense nucleotides, often chemically modified to ensure stability and specificity. There are multiple potential limitations associated with the development and testing of miRNAs-based therapeutics. These issues include, but are not limited to, off-target effect, avoidance from internal nucleases, and toxicity for miRNA therapy. In this review, we will discuss recent advances in miRNA based therapeutic strategies.
Keywords: microRNA, miRNA, Therapy, Mimic, Inhibitor, Antagomir, Off-target effect, Cancer, OncomiRNAs, Tumor suppressor miRNAs, TSmiRNAs, Fibrosis, Hepatocellular carcinoma, Spinal and bulbar muscular atrophy, SBMA, Pathobiology
Introduction
MicroRNAs (miRNAs) are short (approximately 22 nucleotides), single stranded, non-coding RNA species that were first reported to have a function in 1993 [1]. It is believed that miRNAs exert their function mainly through binding of 6–8 nucleotide sequences (miRNA seed) to complementary sequences in target messenger RNA (mRNA), resulting in transcript degradation or translation inhibition, with the net effect of decreasing target protein amount [2]. While some miRNA species are transcribed independently, others originate in introns or exons of genes and are transcribed in concert with mRNA species [3]. The transcribed RNA (pri-miRNA) contains the mature miRNA sequence (guide strand) as well as the complementary sequence (passenger strand). Within the nucleus, the enzyme Drosha cuts the pri-miRNA to pre-miRNA [4], which is then transferred out of nucleus by a carrier protein, Exportin-5 [5]. Within the cytoplasm, pre-miRNA is processed by the enzyme Dicer to 20–25 base mature miRNAs [6]. Mature miRNA is then incorporated into the RNA-induced silencing complex (RISC), and interacts with target messenger RNA species [7]. Following initial reports of miRNA function, a plethora of studies identified salient roles for miRNAs in regulating a variety of physiologic and pathologic states. As the mechanism and role of miRNAs in human disease is gradually unraveled, recent studies also started exploring the role of miRNAs as therapeutics.
MiRNA-based therapeutic strategies – design, pitfalls and challenges
The process of developing miRNA-based therapeutics is somewhat similar to conventional drug discovery and testing protocols. An overview of miRNA-based therapeutic strategies is shown in Figure 1. From a historical perspective, the development of conventional drugs generally starts with large screening tests, without preexisting hypotheses regarding mechanism of action. In contrast, most efforts to date to select miRNAs as targets for therapy were based on prior knowledge regarding disease specific miRNA dysregulation [8]. Although hypothesis-free screening of miRNAs as therapeutics can hypothetically be employed, the strategies used so far are likely based on the fact that miRNAs are intrinsic regulatory molecules, with defined, physiological functions. For example, miRNA-34 was found to have tumor suppressive roles in a variety of human tumors, and later became the focus of miRNA-based therapeutics [9, 10]. Following identification of miRNA species dysregulated in cancer, the next step usually entails functional characterization of the candidate miRNA species in vitro and in vivo through gain or loss of function studies. Gain of function can be accomplished through introduction of synthetic miRNA (either transfection or viral transduction). Loss of function can be accomplished by employing complementary antisense oligonucleotides, usually modified chemically to ensure stability and specificity.
Figure 1. Typical algorithm for identification and testing of miRNA species as therapeutics.
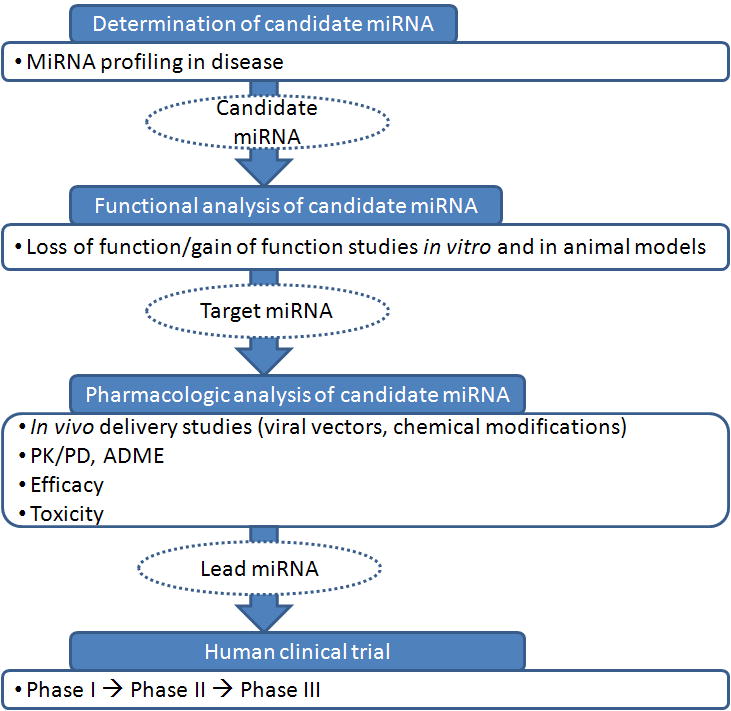
The typical first step is identification of miRNA species that are differentially expressed between the diseased tissue of interest and normal tissue. Next, the miRNA is tested for function, as the screening alone cannot rule out that a miRNA is differentially expressed, but plays no etiologic role. Next, the miRNA species is optimized in terms of in vivo delivery, pharmacokinetics (PK), pharmacodynamics (PD), and absorption, distribution, metabolism, and excretion (ADME). Animal studies are then performed, followed by clinical trials.
MiRNA mimic-based therapeutics - technology and challenges
RNA molecules are anionic, hydrophilic and are generally not uptaken by cells via passive effusion [11]. In addition, miRNAs may also be unstable in vivo due to rapid degradation by endogenous nucleases as well as rapid elimination through hepatic and renal metabolism and excretion [12]. One method to circumvent some of these limitations is local, rather than systemic administration of RNA based therapeutics [13]. Systemic administration is more challenging and efforts are underway to alter miRNA sequences through chemical modifications [12]. Atelocollagen coupling to miRNA molecules is such a modification, and offers several advantages: increased cellular uptake and nuclease resistance, without evoking interferon-based immune responses [14, 15]. MiRNA can also be delivered as lipid-based nanoparticles [16, 17]. Lipid nanoparticles can reduce the negative electrical charge of RNA nucleotides and promote their cellular uptake [18]. Another strategy for efficient systemic delivery of miRNA therapeutics is through viral vectors [19]. Adeno-associated virus vectors [20] or lentiviral vectors [21] can be used to transfer miRNAs to cells. Specificity of viral-mediated delivery continues to be a challenge, and current approaches employ viral serotypes demonstrated to more selectively transduce the cell type of interest, as well as using specific promoters [22, 23].
MiRNA inhibitor-based therapeutics – technology and challenges
To reduce endogenous miRNA levels, anti-sense oligonucleotides [24], called AMO (anti-miRNA antisense oligomer), also called antagomiR [25], are usually employed. Various methods have been tested to render AMO constructs more stable in vivo and to ensure adequate tissue availability and specificity. Two′-O-methyl (2′-OMe) modified RNA is chemically more stable than unmodified RNA, and more resistant to nucleases due to the methylated hydroxyl base [26–28]. Two′-O-methoxyethyl (2′-MOE) phosphorothioate and 2′-fluoro/2′-methoxyethyl (2′-F/MOE) modifications have also been employed to increase the efficiency of the synthetic oligonucleotide against the target miRNA [29–32]. In addition, conjugation with cholesterol appears to improve activity of the nucleotide sequence [27, 33, 34]. Locked nucleic acid (LNA) is another method to modify antisense oligonucleotides, where the 2′ oxygen and 4′ carbon of the nucleotide is bridged with methylene and form a cyclic structure. Because LNA is locked by this bridge, it does not transition between conformations, in contrast to the naked nucleotide sequence. LNA is more resistant to the endogenous nucleases, has stronger affinity to the target nucleotide [35, 36], and displays lower toxicity [37]. Last, peptide nucleic acid (PNA) is an artificially synthesized polymer that has a peptide structure in the main chain and similarity to DNA and RNA [38]. Because it does not have an organophosphate moiety, these constructs bind to target nucleotide sequences more tightly than nucleotide/nucleotide binding. In addition, PNA appears to be relatively stable and can be administered systemically with relatively low toxicity [39].
MiRNA sponge and miRNA masking: an indirect method to reduce endogenous miRNA species
MiRNA sponges and masking are relatively new strategies employed to down-regulate miRNAs. MiRNA sponges contain multiple target sites complementary to a target miRNA that is deemed to be of potential therapeutic relevance [40, 41]. MiRNA masking entails the design and administration of sequences complementary to a miRNA binding site in the 3′ UTR of the target mRNA. Through the competitive inhibition of the interaction between the miRNA and mRNA species, the miRNA mask reduces the activity of endogenous miRNA [42].
MiRNA off-target effects
Decades of conventional drug discovery and design suggest that drug off target effects are a salient aspect of the development process. Off target effects challenges are applicable, to a certain extent, to developing miRNA based therapeutics [43]. Small RNA species, of the miRNA, as well as siRNA type, can bind not only to fully complementary nucleotide sequences, but also to similar sequences [44]. Off-target effect may cause unexpected consequences, sometimes completely negating the utility of a particular small RNA species as therapeutics. The probability of off-target effect can be predicted for small interfering (si) RNA [45, 46], however, the true magnitude of off target effects cannot be fully known until in vivo studies are performed. Off-target studies with miRNAs are, in comparison with siRNA studies, not as well developed. In addition, although not proven on a large scale, we believe that miRNA species may escape the “off target effects curse”, at least to a certain extent. The main advantage of miRNA species when compared to synthetic siRNA species is the fact that miRNAs are intrinsic molecules. If we hypothesize that their modus operandi is through effects of small amplitude exerted on multiple targets simultaneously, then, by definition, their effects cannot be considered off-target. While in vitro and some preliminary in vivo studies suggest that this hypothesis is true, further in vivo studies are clearly needed [47].
Specific miRNA-based therapeutic advances
Cancer
Dysregulated miRNA species have been reported to be involved in carcinogenesis, cancer progression, and drug resistance [48, 49]. Theoretically, miRNA species upregulated in cancer (oncomiRNAs) as well as downregulated in cancer (tumor suppressor miRNAs, or TSmiRNAs) can be targeted for miRNA based therapeutics. Table 1 contains a list of the more studied oncomiRNAs as well as TSmiRNAs.
Table 1.
Representative oncomiRNA and TS miRNA species
| Cancer | OncomiRNA | TSmiRNA |
|---|---|---|
| Breast cancer | mir-21 [9] | mir-34b [78] |
| Colorectal cancer | mir-211 [79] | mir-33a [80] |
| Gastric cancer | mir-192 [81], mir-215 [81] | mir-125a [82] |
| Glioma | mir-21 [83] | mir-34a [10] |
| Hepatocellular carcinoma | mir-21 [50] | mir-26a [22] |
| Leukemia | mir-100 [84] | mir-33a [80] |
| Lung cancer | mir-31 [85] | mir-101 [86] |
| Ovarian cancer | mir-21 [87] | let-7 [88] |
| Pancreatic cancer | mir-21 [89] | mir-150 [90] |
| Prostate cancer | mir-32 [91] | mir-34a [92] |
Therapeutic miRNA-21 blockade in cancer
MiRNA-21, located in a coding gene, transmembrane protein 49, is one of the most studied miRNA species in human cancer. The collective knowledge from several studies indicates that miRNA-21 regulates a multitude of target genes, most of which are known to exert tumor suppressor effects. The targets of miRNA-21 include phosphatase and tensin homolog (PTEN), sprouty homolog 1 (SPRY1), B-cell CLL/lymphoma 2 (BCL2), and others [50]. MiRNA-21 was associated with a variety of cancers, including leukemia [51], lung [52], liver [50], stomach [53], pancreas [54], and brain [55]. Several in vitro as well as in vivo studies demonstrated that reducing miRNA-21 levels with anti-sense nucleotides results in delayed cancer growth in gastric cancer [53], gliomas [56] and other cancers.
Therapeutic miRNA-26 upregulation in hepatocellular carcinoma
MiRNA-26 was first implicated in liver cancers in a profiling study performed on a large cohort of human specimens [57]. Additional mRNA profiling also identified a possible involvement of miRNA-26 in nuclear factor Kappa B and Interleukin-6 pathways, further suggesting an etiologic role for this miRNA in liver cancer genesis or homeostasis. Building on prior data regarding the downregulation of miRNA-26a in liver cancers, another group investigated the usage of miRNA-26a as liver cancer therapeutics in vivo [22]. This is the first study to successfully use adeno-associated viruses (AAVs) to deliver miRNA in vivo to a mouse model of hepatocellular carcinoma, and resurrected the interest in AAVs as delivery methods for miRNAs. This study, however, also raises an interesting point regarding organ and tissue specific expression of miRNAs. Specifically, miRNA-26 does not appear to be underexpressed in all human tumors, suggesting that miRNA expression is highly tissue-dependent. This finding advocates caution in using miRNAs for therapeutics, in particular if the specificity of tissue delivery is not finely tuned. For example, miRNA-26 was found to be up-regulated in glioblastoma and over-expression of miRNA-26 increases tumor growth and proliferation [58]. Although it is currently not known if upregulation of miRNA-26 in normal cells can, under certain conditions, promote de novo cancer genesis, these findings are of unquestionable concern. It is conceivable that liver-specific delivery of miRNA-26 may be possible, however, hepatocellular cancer cell, or hepatocyte specific deliver may be significantly more difficult to accomplish. The need for hepatocyte-specific delivery was further suggested by a recent study, that found overexpression of miRNA-26a in human cholangiocarcinoma (CCA) [59]. These findings are more so concerning since the same study reported an etiologic role for miRNA-26 in CCA growth. These recent data raise a serious concern regarding miRNA-26 supplementation in hepatocellular cancers and further emphasize the need for tissue and cell type specific targeting of miRNA based therapies.
Therapeutic Let-7 upregulation in cancers
Let-7 family is one of the first identified miRNAs, and it is highly conserved in a wide range of species, suggestive of its functional role [60]. Further studies identified a tumor suppressive role for this miRNA family in a variety of cancers [61–63]. Mechanistic studies found that Let-7 directly binds to the 3′UTR of v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) resulting in its downregulation [64]. In addition, let-7 has been reported to downregulate other oncogenes, such as high-mobility group AT-hook 2 (HMGA2) and v-myc myelocytomatosis viral oncogene homolog (MYC) [64]. Gain of function studies demonstrated that induction of let-7 inhibits DNA replication and regulates apoptosis [64]. These data suggested that let-7 is an attractive miRNA for treatment for cancer. In vivo upregulation of let-7 in various cancers, such as prostate cancer [65], and lung cancer by nasal administration [66], by inducible vector [67], or exogenous delivery [23], induced decreased cell proliferation. Further studies are needed to accurately assess the utility of let-7 based miRNA therapeutics in vivo.
Hepatitis C infection
Therapeutic miRNA-122 blockade in Hepatitis C
miRNA-122 is one of the most studied miRNAs as in vivo therapeutics. Several studies demonstrated that miRNA-122 plays a pivotal role in a variety of liver-centric physiologic and pathologic processes. In vivo antisense blockade of miRNA-122 in liver results in significantly decreased plasma cholesterol levels in mice, as well as in non-human primates [29, 35]. Interestingly, these studies also demonstrated that miRNA blockade in vivo can have an enduring effect [35]. Additional studies revealed yet another surprising role for miRNA-122 in Hepatitis C (HCV) infection. In vitro studies demonstrated that miRNA-122 expression is a requirement for HCV replication through direct binding to sites within the 5′UTR of HCV [68]. Following these results, Santaris Pharma introduced a locked nucleic acid (LNA) miRNA-122 antisense sequence and preliminary experiments in non-human primates demonstrated a significant reduction of HCV titer in the higher dose group [69]. Also of note, the effect was maintained over several weeks while therapy was administered. In addition, there was no evidence of liver toxicity or HCV mutations. These preliminary results formed the basis of a recently concluded phase 2a study led by Santaris Pharma, published in abstract form at the 2011 Annual meeting of the American Association for the Study of Liver Diseases (AASLD). The authors reported that 4 of the 9 patients on the high dose of anti-miR-122 therapy had no detectable HCV titer 4 weeks after initiation of therapy.
These studies also shed some light onto other aspects of miRNA functioning. Interestingly, miR-122 binds to the 5′UTR of HCV, not to the typical 3′UTR location. In addition, miR-122 binding appears to stimulate, rather than inhibit the expression of HCV [68]. Another interesting observation stemmed from measuring and correlating miRNA-122 levels in serum of HCV patients with serum HCV titers [70]. Interestingly, there was no correlation between serum levels of HCV and miRNA-122. In other words, if the first step in identifying a miRNA that may inhibit HCV were a profiling study, miRNA-122 would not have been found. This study raises an important question: is the current protocol involving profiling human tissues to identify dysregulated miRNA species with the purpose of finding candidate miRNAs for therapeutics the best approach? Would a typical drug screening approach, where knowledge regarding miRNA levels in diseased tissue compared to normal tissue is irrelevant, work better?
A significant area of concern with further developing miRNA-122 blockade for human use stems from the observation that miRNA-122 has low levels of expression in HCC [71]. Down-regulation of miRNA-122 is observed in patients with HCC and over expression of miRNA-122 inhibits tumor proliferation in HCC cell lines [72]. Moreover, two recent studies demonstrated that miR-122 deletion in mice results in hepatosteatosis, fibrosis and HCC development [73, 74]. In our view, these seemingly contradictory results do not necessarily suggest the presence of off target effects, but they rather underline the paramount importance of tissue and temporal specific miRNA manipulation. In our opinion, these studies argue that miRNA-122 levels may play roles of vastly different magnitude on a variety of processes (growth, stemness, fibrosis, cancer development, Hepatitis C infection) dependent on cell type as well as on developmental stage.
Fibrosis
Therapeutic miRNA-21 blockade in heart fibrosis and miRNA-21 upregulation in abdominal aortic aneurysms (AAA)
In response to injury, fibroblasts produce extracellular matrix as a normal, physiologic response. However, uncontrolled matrix production leads to fibrosis and to a variety of fibrosis-related complications, such as idiopathic pulmonary fibrosis, liver cirrhosis, scleroderma, bowel strictures or bone marrow failure. Transforming growth factor beta (TGF-b) is thought to be a central mediator of fibrosis, and miRNA-21 is a known modulator of TGF-b pathway. Several groups, therefore, studied the utility of miRNA-21 blockade in various fibrosis models. In a mouse model of lung fibrosis, reminiscent of human idiopathic pulmonary fibrosis, antisense blockade of miRNA-21 was shown to diminish the magnitude of the fibrotic process [75]. In a different study, miRNA-21 was found to be upregulated in activated fibroblasts of the failing heart, with a direct impact onto the extent of heart fibrosis [76]. Based on these findings, an in vivo study of a mouse model of pressure-overload-induced heart disease, found that administration of a miRNA-21 antisense construct reduces the extent of heart fibrosis and overall heart function [76]. In sharp contrast, a different study in a mouse model of abdominal aortic aneurysm (AAA) found that miRNA-21 blockade increased the size of the AAA and conversely, miRNA-21 overexpression limited the expansion of AAA [21]. Although these findings are encouraging for the potential utility of miRNA-21 overexpression for patients with AAA, clear concerns exist regarding potential side effects from upregulating miRNA-21 in liver (where high levels are associated with HCC) or heart (where high levels are associated with increased fibrosis).
Lipid metabolism
Therapeutic miRNA-33 blockade
MiRNA-33a and miRNA-33b are located within the intron of sterol regulatory element-binding proteins (SREBP) 2 and SREBP1, respectively. SREBPs are key genes for cholesterol synthesis and uptake. Interestingly MiRNA-33a and miRNA-33b were also found to play a pivotal role in lipid metabolism, through interaction with adenosine triphosphate-binding cassette transporter A1 (ABCA1) and ABCG1, regulators of high density lipoprotein synthesis [77]. Therefore, systemic antagonism of miRNA-33 is expected to improve lipid metabolism and reduce plaque formation in vessels. Mice on a western diet injected with LNA-antisense to miRNA-33 demonstrated an increased in plasma levels of high density lipoprotein (HDL) [77]. Moreover, in vivo inhibition of miRNA-33 was found to have a reduction in atherosclerotic plaques size and lipid content, through raising ABCA1 levels in the liver, as well as in the plaque macrophages [32].
Spinal and bulbar muscular atrophy (SBMA)
Therapeutic miRNA-196a upregulation in SBMA
SBMA, also known as Kennedy-Alter-Sung disease, is an inherited neurodegenerative disorder that results in proximal muscle weakness, atrophy, contractions and bulbar involvement. So far, there are no curative treatments for SBMA. A recent study evaluated AAV-mediated delivery of miRNA-196a in a mouse model of SBMA [20]. CUGBP, Elav-like family member2 (CELF2) is known to bind to the mutated androgen receptor and stabilize it. MiRNA-196a was found to be overexpressed in spinal cords of SBMA mice compared to controls, perhaps as a protective, albeit insufficient, mechanism. In addition, miRNA-196 binds to CELF2 and represses it, resulting in a lower amount of mutated androgen receptor, with amelioration of SBMA. An AAV vector expressing MiRNA-196a delivered in vivo to SBMA mice induced clinical improvement and represents an exciting new therapeutic venue to be explored in SBMA clinical trials.
Conclusion
MiRNA based therapeutics is an emerging field showing significant promise. Studies in mice, non-human primates and early trials in humans clearly demonstrate that there is potential for developing miRNAs into valuable therapeutics. There are, however, multiple areas of concern that deserve further investigation. For example, it is not entirely clear if miRNAs should be viewed similarly to siRNAs or chemical compounds in terms of off-target effects. miRNAs appear to be built-in regulatory molecules, designed to exert global modulation through moderate effects on a multitude of targets. Therefore, development of miRNAs into therapeutics may be less prone to true off-target effects. Nonetheless, while true off target effects could be rare, the same cannot be stated regarding side effects. There are several examples (miRNA-21, miRNA-122, and others) where downregulation in a diseased tissue appears beneficial, but data from a different tissue or disease indicate severe potential side effects of decreasing same miRNA levels, probably in particular if chronic treatment is necessary. Other areas of further research include efficient in vivo delivery. In vivo miRNA upregulation has been attempted by employing viral vectors, however, care must be exercised, similar to other viral-based in vivo gene therapy efforts. In addition, while some specificity can be attained based on the AAV serotype, further efforts are necessary to increase delivery specificity to the organ of interest alone, or, even to the cell of interest. However, given the relatively short period of time since discovery of miRNAs, the progress appears sufficient to justify optimism regarding developing novel therapeutics based on miRNAs.
Footnotes
Disclosure No potential conflicts of interest relevant to this article were reported.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-y. 0092-8674(93)90529-Y [pii] [DOI] [PubMed] [Google Scholar]
- 2*.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. S0092-8674(09)00008-7 [pii] This paper is an excellent review regarding current understanding of miRNA target recognition and mechanism of action. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faller M, Guo F. MicroRNA biogenesis: there’s more than one way to skin a cat. Biochim Biophys Acta. 2008;1779(11):663–7. doi: 10.1016/j.bbagrm.2008.08.005. S1874-9399(08)00177-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–5. doi: 10.1038/nature03049. nature03049 [pii] [DOI] [PubMed] [Google Scholar]
- 5.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6. doi: 10.1101/gad.1158803. 1158803 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117(1):69–81. doi: 10.1016/s0092-8674(04)00261-2. S0092867404002612 [pii] [DOI] [PubMed] [Google Scholar]
- 7.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110(5):563–74. doi: 10.1016/s0092-8674(02)00908-x. S009286740200908X [pii] [DOI] [PubMed] [Google Scholar]
- 8.van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res. 110(3):496–507. doi: 10.1161/CIRCRESAHA.111.247916. 110/3/496 [pii] [DOI] [PubMed] [Google Scholar]
- 9.Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ, Hou JH, et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 13(1):R2. doi: 10.1186/bcr2803. bcr2803 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guessous F, Zhang Y, Kofman A, Catania A, Li Y, Schiff D, et al. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 9(6):1031–6. doi: 10.4161/cc.9.6.10987. 10987 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinger ME, Mercer TR, Mattick JS. RNAs as extracellular signaling molecules. J Mol Endocrinol. 2008;40(4):151–9. doi: 10.1677/JME-07-0160. 40/4/151 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Bader AG, Brown D, Stoudemire J, Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 18(12):1121–6. doi: 10.1038/gt.2011.79. gt201179 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–38. doi: 10.1038/nrd2742. nrd2742 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takeshita F, Patrawala L, Osaki M, Takahashi RU, Yamamoto Y, Kosaka N, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol Ther. 18(1):181–7. doi: 10.1038/mt.2009.207. mt2009207 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeshita F, Minakuchi Y, Nagahara S, Honma K, Sasaki H, Hirai K, et al. Efficient delivery of small interfering RNA to bone-metastatic tumors by using atelocollagen in vivo. Proc Natl Acad Sci U S A. 2005;102(34):12177–82. doi: 10.1073/pnas.0501753102. 0501753102 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 19(6):1116–22. doi: 10.1038/mt.2011.48. mt201148 [pii] This paper employs lipid nanoparticles to deliver miRNAs in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 70(14):5923–30. doi: 10.1158/0008-5472.CAN-10-0655. 0008-5472.CAN-10-0655 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y, Crawford M, Yu B, Mao Y, Nana-Sinkam SP, Lee LJ. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol Pharm. 8(4):1381–9. doi: 10.1021/mp2002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, et al. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14(11):1271–7. doi: 10.1038/nm.1880. nm.1880 [pii] [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki Y, Adachi H, Katsuno M, Minamiyama M, Jiang YM, Huang Z, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 18(7):1136–41. doi: 10.1038/nm.2791. nm.2791 [pii] [DOI] [PubMed] [Google Scholar]
- 21.Maegdefessel L, Azuma J, Toh R, Deng A, Merk DR, Raiesdana A, et al. MicroRNA-21 blocks abdominal aortic aneurysm development and nicotine-augmented expansion. Sci Transl Med. 4(122):122ra22. doi: 10.1126/scitranslmed.3003441. 4/122/122ra22 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137(6):1005–17. doi: 10.1016/j.cell.2009.04.021. S0092-8674(09)00446-2 [pii] This study describes the successful use of adenoassociated viruses to transduce miRNA in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 29(11):1580–7. doi: 10.1038/onc.2009.445. onc2009445 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27(6):2240–52. doi: 10.1128/MCB.02005-06. MCB.02005-06 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czech MP. MicroRNAs as therapeutic targets. N Engl J Med. 2006;354(11):1194–5. doi: 10.1056/NEJMcibr060065. 354/11/1194 [pii] [DOI] [PubMed] [Google Scholar]
- 26.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10(3):544–50. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–9. doi: 10.1038/nature04303. nature04303 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, et al. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31(11):2705–16. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3(2):87–98. doi: 10.1016/j.cmet.2006.01.005. S1550-4131(06)00029-5 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9(9):1034–48. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, et al. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009;37(1):70–7. doi: 10.1093/nar/gkn904. gkn904 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 121(7):2921–31. doi: 10.1172/JCI57275. 57275 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS ONE. 2008;3(5):e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432(7014):173–8. doi: 10.1038/nature03121. nature03121 [pii] [DOI] [PubMed] [Google Scholar]
- 35.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–9. doi: 10.1038/nature06783. nature06783 [pii] [DOI] [PubMed] [Google Scholar]
- 36.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 43(4):371–8. doi: 10.1038/ng.786. ng.786 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wahlestedt C, Salmi P, Good L, Kela J, Johnsson T, Hokfelt T, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proc Natl Acad Sci U S A. 2000;97(10):5633–8. doi: 10.1073/pnas.97.10.5633. 97/10/5633 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Good L, Nielsen PE. Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc Natl Acad Sci U S A. 1998;95(5):2073–6. doi: 10.1073/pnas.95.5.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fabani MM, Abreu-Goodger C, Williams D, Lyons PA, Torres AG, Smith KG, et al. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 38(13):4466–75. doi: 10.1093/nar/gkq160. gkq160 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 16(11):2043–50. doi: 10.1261/rna.2414110. rna.2414110 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JB, Hong J, Bonner DK, Poon Z, Hammond PT. Self-assembled RNA interference microsponges for efficient siRNA delivery. Nat Mater. 11(4):316–22. doi: 10.1038/nmat3253. nmat3253 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z. The principles of MiRNA-masking antisense oligonucleotides technology. Methods Mol Biol. 676:43–9. doi: 10.1007/978-1-60761-863-8_3. [DOI] [PubMed] [Google Scholar]
- 43.Marquez RT, McCaffrey AP. Advances in microRNAs: implications for gene therapists. Hum Gene Ther. 2008;19(1):27–38. doi: 10.1089/hum.2007.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh S, Narang AS, Mahato RI. Subcellular fate and off-target effects of siRNA, shRNA, and miRNA. Pharm Res. 28(12):2996–3015. doi: 10.1007/s11095-011-0608-1. [DOI] [PubMed] [Google Scholar]
- 45.Wilson PA, Plucinski M. A simple Bayesian estimate of direct RNAi gene regulation events from differential gene expression profiles. BMC Genomics. 12:250. doi: 10.1186/1471-2164-12-250. 1471-2164-12-250 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seinen E, Burgerhof JG, Jansen RC, Sibon OC. RNAi experiments in D. melanogaster: solutions to the overlooked problem of off-targets shared by independent dsRNAs. PLoS ONE. 5(10) doi: 10.1371/journal.pone.0013119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamanaka S, Campbell NR, An F, Kuo SC, Potter JJ, Mezey E, et al. Coordinated effects of microRNA-494 induce G(2)/M arrest in human cholangiocarcinoma. Cell Cycle. 11(14):2729–38. doi: 10.4161/cc.21105. 21105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 1812(5):592–601. doi: 10.1016/j.bbadis.2011.02.002. S0925-4439(11)00031-7 [pii] [DOI] [PubMed] [Google Scholar]
- 49.Cho WC. MicroRNAs as therapeutic targets and their potential applications in cancer therapy. Expert Opin Ther Targets. 16(8):747–59. doi: 10.1517/14728222.2012.696102. [DOI] [PubMed] [Google Scholar]
- 50.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Zhu X, Gu J, Hu H, Dong D, Yao J, et al. Anti-miR-21 oligonucleotide enhances chemosensitivity of leukemic HL60 cells to arabinosylcytosine by inducing apoptosis. Hematology. 15(4):215–21. doi: 10.1179/102453310X12647083620840. [DOI] [PubMed] [Google Scholar]
- 52.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 18(3):282–93. doi: 10.1016/j.ccr.2010.08.013. S1535-6108(10)00310-7 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamanaka S, Olaru AV, An F, Luvsanjav D, Jin Z, Agarwal R, et al. MicroRNA-21 inhibits Serpini1, a gene with novel tumour suppressive effects in gastric cancer. Dig Liver Dis. 44(7):589–96. doi: 10.1016/j.dld.2012.02.016. S1590-8658(12)00071-0 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steele CW, Oien KA, McKay CJ, Jamieson NB. Clinical potential of microRNAs in pancreatic ductal adenocarcinoma. Pancreas. 40(8):1165–71. doi: 10.1097/MPA.0b013e3182218ffb. 00006676-201111000-00002 [pii] [DOI] [PubMed] [Google Scholar]
- 55.Wong ST, Zhang XQ, Zhuang JT, Chan HL, Li CH, Leung GK. MicroRNA-21 inhibition enhances in vitro chemosensitivity of temozolomide-resistant glioblastoma cells. Anticancer Res. 32(7):2835–41. 32/7/2835 [pii] [PubMed] [Google Scholar]
- 56.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67(19):8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. 67/19/8994 [pii] [DOI] [PubMed] [Google Scholar]
- 57.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361(15):1437–47. doi: 10.1056/NEJMoa0901282. 361/15/1437 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim H, Huang W, Jiang X, Pennicooke B, Park PJ, Johnson MD. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc Natl Acad Sci U S A. 107(5):2183–8. doi: 10.1073/pnas.0909896107. 0909896107 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, Han C, Wu T. MicroRNA-26a promotes cholangiocarcinoma growth by activating beta-catenin. Gastroenterology. 143(1):246–56. e8. doi: 10.1053/j.gastro.2012.03.045. S0016-5085(12)00493-3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee CT, Risom T, Strauss WM. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 2007;26(4):209–18. doi: 10.1089/dna.2006.0545. [DOI] [PubMed] [Google Scholar]
- 61.Shi XB, Tepper CG, de Vere White RW. Cancerous miRNAs and their regulation. Cell Cycle. 2008;7(11):1529–38. doi: 10.4161/cc.7.11.5977. 5977 [pii] [DOI] [PubMed] [Google Scholar]
- 62.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. nrc1840 [pii] [DOI] [PubMed] [Google Scholar]
- 63.Nadiminty N, Tummala R, Lou W, Zhu Y, Zhang J, Chen X, et al. MicroRNA let-7c suppresses androgen receptor expression and activity via regulation of Myc expression in prostate cancer cells. J Biol Chem. 287(2):1527–37. doi: 10.1074/jbc.M111.278705. M111.278705 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barh D, Malhotra R, Ravi B, Sindhurani P. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr Oncol. 17(1):70–80. doi: 10.3747/co.v17i1.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong Q, Meng P, Wang T, Qin W, Wang F, Yuan J, et al. MicroRNA let-7a inhibits proliferation of human prostate cancer cells in vitro and in vivo by targeting E2F2 and CCND2. PLoS ONE. 5(4):e10147. doi: 10.1371/journal.pone.0010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–64. doi: 10.4161/cc.7.6.5834. 5834 [pii] [DOI] [PubMed] [Google Scholar]
- 67.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105(10):3903–8. doi: 10.1073/pnas.0712321105. 0712321105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–81. doi: 10.1126/science.1113329. 309/5740/1577 [pii] [DOI] [PubMed] [Google Scholar]
- 69*.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 327(5962):198–201. doi: 10.1126/science.1178178. science.1178178 [pii] This study, along with Reference 68, formed the basis for anti-Hepatitis C miRNA-122-based therapeutics, including the first Phase II clinical studies in humans to employ miRNA based therapeutics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15(1):31–3. doi: 10.1038/nm.1902. nm.1902 [pii] [DOI] [PubMed] [Google Scholar]
- 71.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99(3):671–8. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 72.Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang Q, et al. MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wnt/beta-catenin pathway. Liver Int. 32(5):752–60. doi: 10.1111/j.1478-3231.2011.02750.x. [DOI] [PubMed] [Google Scholar]
- 73.Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 122(8):2884–97. doi: 10.1172/JCI63455. 63455 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 122(8):2871–83. doi: 10.1172/JCI63539. 63539 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 207(8):1589–97. doi: 10.1084/jem.20100035. jem.20100035 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76*.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456(7224):980–4. doi: 10.1038/nature07511. nature07511 [pii] This study clarifies the involvement of miRNA-21 in fibrosis, with particular emphasis on heart fibrosis and failure. [DOI] [PubMed] [Google Scholar]
- 77*.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 328(5985):1566–9. doi: 10.1126/science.1189123. science.1189123 [pii] This paper reveals the cooperation between miRNA-33 and its mother gene in regulating lipid metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee YM, Lee JY, Ho CC, Hong QS, Yu SL, Tzeng CR, et al. miRNA-34b as a tumor suppressor in estrogen-dependent growth of breast cancer cells. Breast Cancer Res. 13(6):R116. doi: 10.1186/bcr3059. bcr3059 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cai C, Ashktorab H, Pang X, Zhao Y, Sha W, Liu Y, et al. MicroRNA-211 expression promotes colorectal cancer cell growth in vitro and in vivo by targeting tumor suppressor CHD5. PLoS ONE. 7(1):e29750. doi: 10.1371/journal.pone.0029750. PONE-D-11-19620 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thomas M, Lange-Grunweller K, Weirauch U, Gutsch D, Aigner A, Grunweller A, et al. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene. 31(7):918–28. doi: 10.1038/onc.2011.278. onc2011278 [pii] [DOI] [PubMed] [Google Scholar]
- 81.Jin Z, Selaru FM, Cheng Y, Kan T, Agarwal R, Mori Y, et al. MicroRNA-192 and -215 are upregulated in human gastric cancer in vivo and suppress ALCAM expression in vitro. Oncogene. 30(13):1577–85. doi: 10.1038/onc.2010.534. onc2010534 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nishida N, Mimori K, Fabbri M, Yokobori T, Sudo T, Tanaka F, et al. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 17(9):2725–33. doi: 10.1158/1078-0432.CCR-10-2132. 1078-0432.CCR-10-2132 [pii] [DOI] [PubMed] [Google Scholar]
- 83.Kwak HJ, Kim YJ, Chun KR, Woo YM, Park SJ, Jeong JA, et al. Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene. 30(21):2433–42. doi: 10.1038/onc.2010.620. onc2010620 [pii] [DOI] [PubMed] [Google Scholar]
- 84.Zheng YS, Zhang H, Zhang XJ, Feng DD, Luo XQ, Zeng CW, et al. MiR-100 regulates cell differentiation and survival by targeting RBSP3, a phosphatase-like tumor suppressor in acute myeloid leukemia. Oncogene. 31(1):80–92. doi: 10.1038/onc.2011.208. onc2011208 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu X, Sempere LF, Ouyang H, Memoli VA, Andrew AS, Luo Y, et al. MicroRNA-31 functions as an oncogenic microRNA in mouse and human lung cancer cells by repressing specific tumor suppressors. J Clin Invest. 120(4):1298–309. doi: 10.1172/JCI39566. 39566 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of zeste homolog 2. J Thorac Oncol. 6(4):671–8. doi: 10.1097/JTO.0b013e318208eb35. [DOI] [PubMed] [Google Scholar]
- 87.Lou Y, Yang X, Wang F, Cui Z, Huang Y. MicroRNA-21 promotes the cell proliferation, invasion and migration abilities in ovarian epithelial carcinomas through inhibiting the expression of PTEN protein. Int J Mol Med. 26(6):819–27. doi: 10.3892/ijmm_00000530. [DOI] [PubMed] [Google Scholar]
- 88.Wendler A, Keller D, Albrecht C, Peluso JJ, Wehling M. Involvement of let-7/miR-98 microRNAs in the regulation of progesterone receptor membrane component 1 expression in ovarian cancer cells. Oncol Rep. 25(1):273–9. [PubMed] [Google Scholar]
- 89.Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA, Vasile E, et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 70(11):4528–38. doi: 10.1158/0008-5472.CAN-09-4467. 0008-5472.CAN-09-4467 [pii] [DOI] [PubMed] [Google Scholar]
- 90.Srivastava SK, Bhardwaj A, Singh S, Arora S, Wang B, Grizzle WE, et al. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. 32(12):1832–9. doi: 10.1093/carcin/bgr223. bgr223 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jalava SE, Urbanucci A, Latonen L, Waltering KK, Sahu B, Janne OA, et al. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene. 31(41):4460–71. doi: 10.1038/onc.2011.624. onc2011624 [pii] [DOI] [PubMed] [Google Scholar]
- 92.Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 17(2):211–5. doi: 10.1038/nm.2284. nm.2284 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]


