Abstract
Topologically complex proteins fold by multiple routes as a result of hard-to-fold regions of the proteins. Oftentimes these regions are introduced into the protein scaffold for function and increase frustration in the otherwise smooth-funneled landscape. Interestingly, while functional regions add complexity to folding landscapes, they may also contribute to a unique behavior referred to as hysteresis. While hysteresis is predicted to be rare, it is observed in various proteins, including proteins containing a unique peptide cyclization to form a fluorescent chromophore as well as proteins containing a knotted topology in their native fold. Here, hysteresis is demonstrated to be a consequence of the decoupling of unfolding events from the isomerization or hula-twist of a chromophore in one protein and the untying of the knot in a second protein system. The question now is- can hysteresis be a marker for the interplay of landscapes where complex folding and functional regions overlap?
Keywords: Energy Landscape, Protein Folding, Knotted Proteins, Interplay
The crux of the protein folding problem lies in the elucidation of a dominant folding pathway from the astronomical number of protein conformations that the Levinthal Paradox1 predicts from a completely random search. The current view is that a typical protein, even a “simple folder” like RNAseA2, has numerous folding pathways. One consequence of this is that the native state, and thus, the folding pathway, is minimally affected by most mutations in the primary sequence of a protein3. The idea of a protein having an energy bias towards the native state, multiple pathways to reach that native state, and resilience to mutation has led to the funneled energy landscape theory. Funneled energy landscape theory, or, “folding funnels”, predicts that an unfolded protein will have many folding pathways available, although only one or few dominate the folding process4,5. To function properly, robust folding is a necessity, and funneled folding has evolutionary pressure to be devoid of deep traps, leading to a smooth, minimally frustrated landscape. However, in larger and multi-domain proteins, folding may become more complicated where intermediates, both kinetic and thermodynamic, may be detected. These intermediates are a result of “roughness” within the folding funnel. If a minimally frustrated funnel is the sole “goal” in protein folding, where does observed frustration come from?
Larger, more topologically complex proteins, may fold by multiple routes as a result of hard-to-fold functional regions of the proteins6. These functional regions introduce frustration or roughness to the otherwise smooth, funneled landscape. While in some cases, functional regions are defined by single amino acids or group of interacting amino acids located in loops or active sites that relay a signal, in other instances function may not be as clearly defined, as structural features that impart function are not optimized for folding. However, evidence is accumulating that functional regions of natural proteins do not significantly aid folding and might, in fact, interfere with it; in other words, folding and function may conflict within the context of a smooth energy landscape as conserved and/or functional regions may not be optimized for folding,7,5,8. Thus, it is possible that the most difficult or stressful regions to fold in a protein are functional sites9,7. Details within the protein sequence help to determine the folding route to preserve the functional fold. Thus, functional loops that cause complexity and trapping can modulate folding routes and rates10,11. While functional regions show little effect on folding of smaller, more two state-like proteins12, such interactions make the overall fold topology significantly more complicated for larger ones10.
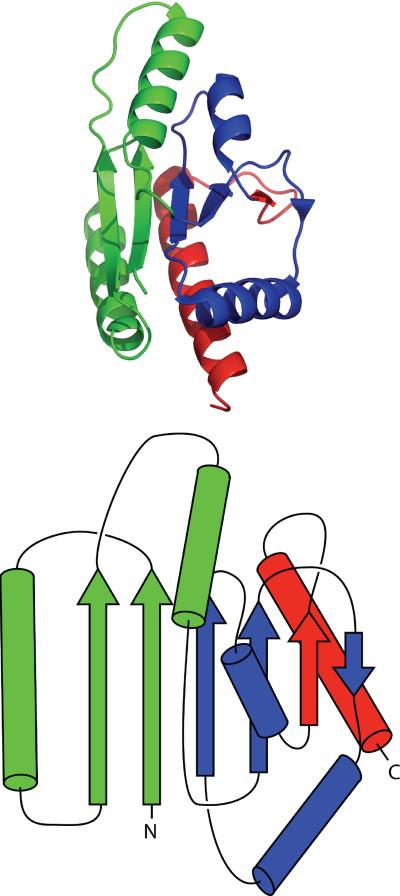
Green fluorescent protein (GFP) is a 228-residue protein originally discovered in the jellyfish Aequorea Victoria13 that now is popularly used as a fluorescent label14. GFP has been employed in several novel techniques including acting as a reporter for gene expression15, organelle tracking in vivo, and folding16,17. Reengineering of GFP has also led to split-GFP protein-protein interaction reporters18,19,20, and pH reporters21. Mutations of residues near the chromophore have created a spectrum of colors for use in labeling22, as well as utilization of fluorescent proteins from other organisms23,24. GFP consists of an 11-stranded β-barrel surrounding an α-helix containing the fluorescent chromophore (Fig. 1)25. The lid of the barrel contains 3 distorted α-helices on one side, and an exceedingly distorted helix on the other. The barrel is thought to sequester the chromophore from bulk solvent to avoid fluorescent quenching. The unique structure of GFP creates a large average loop length, or contact order26; high contact order is linked to folding complexity and slow folding27.
Figure 1.
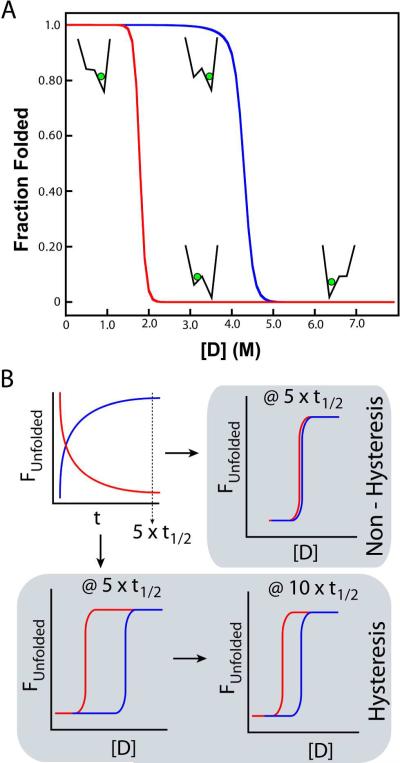
(A) A rough energy landscape can lead to observed hysteresis. Non-coincidence of equilibrium unfolding (blue) and refolding (red) curves are evidence of a complex energy landscape. Hysteresis is a property in which a system does not immediately respond to the stresses applied to it, which may arise from a bifurcation in the energy landscape, which leads to a bi-stable system. Here, the observed equilibrium is dependent not only on the final conditions, but also the initial conditions (memory of the system). A simplified landscape is shown at selected parts of the hysteresis curves to show how a hysteresis cycle can arise. (B) Hysteresis is a kinetic effect manifesting itself within “equilibrium” data. At 5 half-lives, folding (red) and unfolding (blue) are considered complete. In a non-hysteretic (simple) folding scheme (upper right schematic), equilibrium curves overlay and are stable. In proteins that exhibit hysteresis (lower left panel), “equilibrium” folding and unfolding curves are non-equivalent after 5 half-lives, and may continue to drift (lower right) as a second, non-folding kinetic step limits denaturation.
The fluorescent moiety in GFP is a p-hydroxybenzylidene-imidazolinone chromophore formed from cyclization, dehydration, and oxidation reactions of the polypeptide backbone28,29. After folding, chromophore formation occurs to release strain on a “kink” within the internal α-helix30; presumably, the α-helix kink is formed from interactions between the barrel staves and the central helix. While the highly conjugated rings of the chromophore lead to fluorescence, the ring is thought to exist in a non-planar state within the barrel31. Furthermore, the chromophore is thought to isomerize through a “hula-twist” cis-trans isomerization32, which maintains the volume of the chromophore, allowing isomerization within the barrel. This process is linked to kindling33,34 and photo-switchable proteins, in which fluorescence can be turned on and off by the absorbance of certain wavelengths35,36.
Initial work on GFP folding highlighted slow observed unfolding/refolding kinetics and non-coincident equilibrium transition curves37,38 as well as slow (over months) changes in transition curves39. These results are indicative of a rough folding energy landscape in GFP with potentially large folding barriers. Even in a robustly folding GFP variant, denaturation must occur at 95°C in 6M urea; once unfolded, GFP will remain unfolded in 6M urea. Using this denaturation protocol, GFP may exist in both a folded or unfolded state in 6M urea, depending on whether it has been heated first40. The implication is significant as it suggests that GFP folding exhibits hysteresis, a unique and important phenomenon indicative of coupled events in folding. A rigorous check of both unfolding and refolding transitions of GFP verifies this occurrence (Fig.2)41.
Figure 2.
GFP exists as a highly regular β-barrel surrounding the fluorescent chromophore (green). A splay diagram presents and numbers the β-strands as discussed in the text.
While the folding of GFP has been comprehensively reviewed previously42, recent studies of GFP folding may have further, functional implications43,19,44,45,46,47,39. A rigorous examination of GFP in which each β-strand is “left out” shows that β-strand 7 is the most amenable to removal and later annealing47. These results show that GFP is able to fold down a path in which β-strand 7 is the final step in barrel closure. Consistent with this finding, it has been recently shown that GFP is resistant to mechanical degradation where pulling and degradation48,49,50 from either β-strands 7 or 11 indicate a near-fully folded molecule where either β-strand participates in the final folding step51. A similar experiment in pulling from β-strand 6 indicated a loss of the tertiary fold51.
The overall topology of GFP and its link to folding and function was studied by “rewiring” GFP via exchanging the loops of the lid of the protein to radically change how the barrel staves were linked to each other, and thus, the overall topology52. Here, rewired variants of β-strands 1–6 were non-fluorescent, and appeared to misfold according to CD spectra, consistent with previous results45. Furthermore, disruption of β-strands 1–6 likely perturbed the lid of GFP, which is linked to chromophore formation41,53, and locking of the barrel into a final, correct native state54. Rewiring the remaining β-strands of GFP, while maintaining the native connections of β-strands 1–6, showed native-like fluorescence, given long enough linkers between the β-sheets. Interestingly, the circularly permutated rewired GFP also withstood breaks on either end of the central α-helix, implying that helix strain required for chromophore formation came from the outer barrel, and is not transmitted through the helix by the lids. More recent work on denaturation of GFP55 indicate that chromophore formation is coupled to early folding events; here, lack of a mature chromophore led to more two-state like behavior in folding. Thus, chromophore formation/stabilization, the functional aspect for this molecule, appears to be important first steps in the folding of GFP. Similar results have been observed in other large, complex proteins where functional regions add roughness to the folding landscape and are early determinants in route selection10,56. Interestingly, while functional regions add complexity to folding landscapes, they may also contribute to a unique kinetic behavior related to the stability of the system.
Hysteresis is a property in which a system does not immediately respond to the stresses applied to it, which may arise from a bifurcation in the energy landscape, as observed in pulling experiments of GFP48. This, in turn, leads to a bi-stable system, where the observed equilibrium is dependent not only on the final conditions, but also the initial conditions (memory of the system). Simply stated, hysteresis is a situation in which the state of the system depends not only on its current conditions, but also its history; thus, a hysteretic system is pathway dependent. Hysteresis is a kinetic effect manifesting itself within “equilibrium” data. Typically, one measures the folding and unfolding kinetics over a range of denaturant concentrations by multiple techniques and waits greater than 5 half-lives times the slowest reaction (>97%) before measuring the thermodynamic stability of a protein57. It is problematic in folding studies, as full reversibility is a requirement for many analyses. However, observable hysteresis results from a separation of time-scales between two required processes in folding or unfolding, revealing a further level of complexity within the free energy landscape. Hysteresis occurs when experimental observation lies between these time scales, and is abrogated when observation lies outside of the two time-scales (e.g. hysteresis disappears at infinite time). Thus, experiments are conducted at a `pseudo-equilibrium' state for hysteresis-dependent proteins.
Initially, it was suggested that the non-coincidence of equilibrium transition curves in GFP was linked to very slow folding and unfolding kinetics39. However, upon further analysis, folding kinetics simulations predicted equilibration after two months in which both the unfolding and refolding transition curves would shift towards the center of the hysteresis “zone”58, approaching superimposablity. When testing chromophore fluorescence, native tryptophan fluorescence, or circular dichroism of GFP, hysteresis was observed, suggesting that hysteresis is linked to the native structure, not just one probe. Upon closer inspection at apparent equilibrium, the unfolding curve continued to shift towards the refolding curve over three months, while the refolding curve remained constant; evidence that another, non-folding process limits denaturant-induced destabilization. Remarkably, all hysteresis was abolished, and the unfolding transition coincides with the refolding transition, when the chromophore was not present either through catalytic mutation (R96A mutant) or structural requirements (M88Y/Y74M mutant) clearly indicating that hysteresis is linked to the chromophore41. These results further suggest a functional relationship with the chromophore. Further examination of the complex folding landscape of GFP revealed that hysteresis arises from attempting to form the β-barrel around a chromophore, where the barrel requires a precise isomerization and torsion of the chromophore; in the absence of the chromophore (de novo folding) folding is greatly simplified, and hysteresis is abolished, as the strain of isomerization is eliminated. Interestingly, fluorescent proteins tend to contain chromophores in a cis isomerization while structurally analogous but non-fluorescent GFP-like proteins contain a chromophore in a trans isomerization53, a key structural difference in the `functional' relationship of these molecules. To this end, chromophore packing and the imposed strain in the native state, and the resulting hysteresis, are linked in both the folding and functional aspects of GFP. Consistent with this hypothesis, mutations that disrupt the formation of the chromophore lead to a well-folded protein, but abolish the observed hysteretic effect30,41.
While hysteresis is rare, it has been observed in the folding of other systems, although typically in multi-domain or multimeric proteins59,60,61,62,63. In these complex systems, differences in energies of the folding transition ensemble lead to hysteresis as unfolding are controlled by domain transitions, while refolding occurs much more cooperatively. Titin, a modular repeat protein, exhibits hysteresis in single-molecule stretching experiments, where hysteresis is observed in stretch-release cycles attributed to domain unfolding/refolding60,64. Collagen, another single-domain protein, exhibits hysteresis in both thermally and chemically induced transitions, linked to proline-rich regions62; refolding requires slow annealing from loop rearrangement while denaturation is very cooperative once collagen stability is broken. Both proteins exhibit similar hysteresis characteristics: unfolding and refolding occur through different pathways; one direction in a single, global transition while the other direction transitions in a domain-wise or stepwise process. Other instances of hysteresis have also been observed in aggregation and association events in proteins65,66,67 where stable conformations exist on the free energy landscape, consistent with a rough or possibly dual-basin landscape. In agreement with the principle of hysteresis, proteins that are highly stable with complex topologies, like GFP, do not appear to easily regain their native states once disruption of their tertiary structure has taken place54. While the current views of hysteresis are predicated on native states that are well represented in biology (i.e., β-barrels, triple helix), do other protein topologies share this property?
The existence of proteins containing a unique, intricate, knotted topology in their native fold was once considered unlikely or even impossible, and knotted topologies were discarded from structure prediction and electron density68. Since then, proteins with simple and complicated knots, some with up to 6 crossing points, have been discovered and characterized69. Today several distinct protein families with conserved knotted topology are recognized70. While the functional role of knotting is not yet resolved, some studies indicated its role in stabilization of functional regions71,72,73,70. YibK and YbeA, two knot-containing methyltransferase proteins, have been extensively studied both in silico74, and in vitro75,76,77. Molecular dynamics simulations with minimal energetic frustration show multiple basins and off-pathway intermediates74,78. Interestingly, these non-native basins correspond to near-native structures without threaded knots75, analogous to the dual-basin energy landscape observed in GFP58, which has high contact order but does not contain a knot. In silico, both backtracking and large topological barriers are evidence of a complex energy landscape and are implicated in the folding of both YibK and YbeA74.
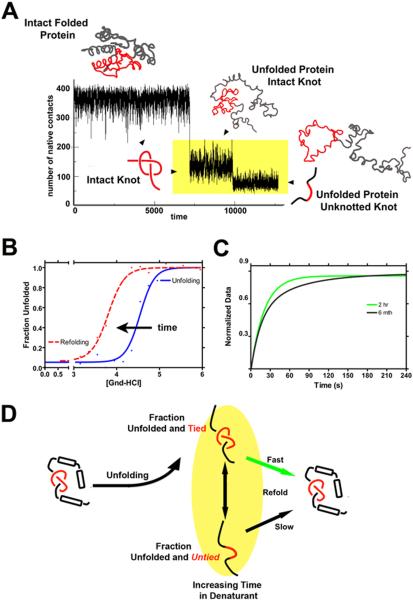
More recently, characterization and identification of the folding mechanism of knotted proteins highlight complex interactions76,79,80,81,82,71,72,83. Interestingly, a protein can self-tie from a newly translated polypeptide chain despite the complexity of the knotted structures, where knot formation is a post-translational, rate-limiting step in folding84. While it has been suggested that threading the knot may be impossible to observe during in vitro denaturation studies, as the knot persists in the denatured state75, the inability to untie the knot results in a significant gap in the full description of the free energy landscape for knotted proteins. Initial results using a structurally homologous YibK-like methyltransferase (PDB 1O6D) (Fig.3) in kinetic unfolding simulations with structure-based models85,74 are unambiguous and demonstrate that unfolding of 1O6D to a fully unfolded, untied conformation is achieved in a stepwise process (Fig.4A). Unfolding of the secondary structure elements precedes and is decoupled from untying of the knotted protein backbone. The untangling process is at least an order of magnitude slower than protein unfolding. These simulations clearly show, in agreement with translational experiments, that untying of the knot is the rate-limiting step in knot formation/folding. This indicates that given enough time, the protein can be unfolded and the knot untied (Fig.4A). Moreover, recent work has directly highlighted how complex the unfolding landscape is for knotted proteins86. A similar mechanism of unfolding is observed in all-atom simulations77. Interestingly, the single molecule approach with SBMs to investigate the energy landscape of knotted protein consistently reveals a step-wise unfolding process for several proteins with the homologous α/β fold86. All these simulations suggest that experiments performed under previous conditions only probe the reversibility of unfolding/refolding of the knotted chain87,75. Furthermore, they demonstrate that extended times in denaturing conditions are necessary to probe knot untying and true equilibration between native-knotted and unfolded-untied is extremely difficult to achieve. Consistent with this prediction and the apparent hysteric behavior, 1O6D exhibits an unconventional landscape where unfolding and refolding equilibrium studies are nonsuperimposable (Fig.4B), despite exhaustive sample incubation (Fig.4C). Here, hysteresis is speculated to be a consequence of the decoupling of unfolding events (loss of secondary/tertiary structure) from the untying of the knotted protein backbone (Fig. 4D). This is analogous to observations of GFP, in which folding is decoupled from chromophore isomerization, leading to two kinetic steps with distinctly different timescales.
Figure 3.
The thermophilic methyltransferase 1O6D is a knotted protein. A splay diagram simplifies the structure to focus on the knotted topology.
Figure 4.
Theoretical and Experimental Data both Suggest Time-Dependent Unfolding of the Knotted Polypeptide Chain (A) The mechanism of unfolding shows two distinct steps, where unfolding of the secondary structure occurs first, followed by untying of the knot. The unfolding and untying events appear on distinctly different timescales and are highlighted in yellow. (B) Denaturant-induced unfolding (blue) and refolding (red dashes) measured by circular dichroism (CD) spectroscopy. The unfolding (blue) and refolding (red dashes) transitions for 1O6D show apparent hysteresis (the nonsuperimposability of the curves), consistent with the uncoupling of unfolding and untying of the knotted protein and the shift in the folded ensemble. The fit of the data was to a two-state model. Given enough time, these curves would coalesce. (C) Observed experimental refolding kinetics as a function of time in the denatured state, monitored by CD spectroscopy. Protein was unfolded at 6.0M denaturant (Gnd-HCl) for the given amount of time, and refolding was initiated by dilution to a final Gnd-HCl concentration of 3.2M. As predicted, changes are observed in the folding kinetics, consistent with untying the knot in the unfolded ensemble, and occur over a period of 6 months. The fit of the data was to a single-state (green trace) and two-state (black trace) model, respectively. (D) A schematic drawing of the “double-jump” experiment used in (C) to test the effect of the persistence of the knot in the denatured state on the refolding kinetics. In this scenario, extended times in the denatured state are necessary for untying of the unfolded protein.
While the symptoms of hysteresis may be diverse, the results are driven by the free energy landscape. As evolution drives robust folding, forming a minimally frustrated landscape where both kinetic and thermodynamic traps are small compared to the overall depth of the native basin, hysteresis presents itself as a second basin, or trap, clashing with the minimal-frustration concept. However, as landscape roughness has been recently linked to protein function10, the `smoothness' of the folding funnel may have been redefined. In fact, recently, examples of functional regions adding frustration have been demonstrated with both experimentally10 and theoretically11,88 lending credence to the idea of overlapping landscapes (folding and function). For GFP, differences in the folding/unfolding pathway is linked to protein stability, proline and chromophore isomerization. For titin, different routes to unfold and refold multiple domain chain is related to viscoelastic properties which allow muscle fiber to increase the range of protein extension and increase flexibility60. In the case of aggregation or assembly, a second basin of the free energy landscape exists as an off-pathway trap89. While not linked directly to function, in vivo folding may utilize molecular chaperones to avoid off-pathway aggregation. The knotted region within knot proteins also appears to play a functional role in the topology of the protein71,72,73,70. As the study of protein structure and folding shifts into more complicated topologies, the potential for complex energy landscapes increases. While evolution favors minimally frustrated and simpler landscapes, protein topology and/or function add complexity or roughness, leading to phenomena such as hysteresis. Here, the presence of hysteresis in folding is on a complex energy landscape with folding linked to a slow search step toward the active conformation, acting as a marker for processes like isomerization or backbone knotting. During this second step, folding is no longer limited by a conventional folding mechanism, but the required slow search to native. Interestingly, isomerization and post-translational modifications are not the only factors that alter the energy landscapes of proteins and result in hysteresis. Synonymous or “silent” single nucleotide polymorphisms in coding DNA (which does not change the amino acid sequence of the protein product) can result in a protein with altered substrate specificity90. We suggest this protein, too, will exhibit a hysteretic or dual-basin energy landscape as rare codons influence translation rate, impact protein folding91,92,93,94,95,96, and ultimately function. It will be interesting to explore the question of whether barrier controlled, fast-folding followed by a prefactor-controlled slow search step to native58 is more common than previously believed.
Acknowledgements
We thank the Jennings and Onuchic Labs for discussions of the work. This work was supported by NIH grant GM 2RO1 GM054038 (PAJ, DTC), NSF PHY-1212312 (PAJ, DTC), the Center for Theoretical Biological Physics sponsored by the NSF (Grant PHY-0822283) and NSF-MCB-1051438 (JNO, DTC), the Molecular Biophysics Training Grant GM08326 (BTA) and the NSRA F32GM-905652 (BTA), and by the Foundation for Polish Science (JIS). JNO is a CPRIT Scholar in Cancer Research sponsored by the Cancer Prevention and Research Institute of Texas.
REFERENCES
- 1.Levinthal C. Are There Pathways for Protein Folding? Extrait du Journal De Chimie Physique. 1968;65(1):44–7. [Google Scholar]
- 2.Weissman JS. All roads lead to Rome? The multiple pathways of protein folding. Chem Biol. 1995;2(5):255–60. doi: 10.1016/1074-5521(95)90044-6. [DOI] [PubMed] [Google Scholar]
- 3.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nature structural biology. 1997;4(1):10–9. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 4.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: the energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 5.Onuchic JN, Wolynes PG. Theory of protein folding. Current opinion in structural biology. 2004;14(1):70–5. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Gosavi S, Chavez LL, Jennings PA, Onuchic JN. Topological frustration and the folding of interleukin-1 beta. Journal of molecular biology. 2006;357(3):986–96. doi: 10.1016/j.jmb.2005.11.074. [DOI] [PubMed] [Google Scholar]
- 7.Jager C, Welzel T, Meyer-Zaika W, Epple M. A solid-state NMR investigation of the structure of nanocrystalline hydroxyapatite. Magnetic resonance in chemistry: MRC. 2006;44(6):573–80. doi: 10.1002/mrc.1774. [DOI] [PubMed] [Google Scholar]
- 8.Onuchic JN, Nymeyer H, Garcia AE, Chahine J, Socci ND. The energy landscape theory of protein folding: insights into folding mechanisms and scenarios. Advances in protein chemistry. 2000;53:87–152. doi: 10.1016/s0065-3233(00)53003-4. [DOI] [PubMed] [Google Scholar]
- 9.Gruebele M. Downhill protein folding: evolution meets physics. Comptes rendus biologies. 2005;328(8):701–12. doi: 10.1016/j.crvi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Capraro DT, Roy M, Onuchic JN, Gosavi S, Jennings PA. beta-Bulge triggers route-switching on the functional landscape of interleukin-1beta. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(5):1490–3. doi: 10.1073/pnas.1114430109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gosavi S, Whitford PC, Jennings PA, Onuchic JN. Extracting function from a beta-trefoil folding motif. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(30):10384–10389. doi: 10.1073/pnas.0801343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haglund E, Lindberg MO, Oliveberg M. Changes of protein folding pathways by circular permutation. Overlapping nuclei promote global cooperativity. The Journal of biological chemistry. 2008;283(41):27904–15. doi: 10.1074/jbc.M801776200. [DOI] [PubMed] [Google Scholar]
- 13.Shimomura O, Johnson FH, Saiga Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol. 1962;59:223–39. doi: 10.1002/jcp.1030590302. [DOI] [PubMed] [Google Scholar]
- 14.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–44. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 15.Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332(6028):475–8. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waldo GS. Improving protein folding efficiency by directed evolution using the GFP folding reporter. Methods Mol Biol. 2003;230:343–59. doi: 10.1385/1-59259-396-8:343. [DOI] [PubMed] [Google Scholar]
- 17.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat Biotechnol. 1999;17(7):691–5. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 18.Cabantous S, Pedelacq JD, Mark BL, Naranjo C, Terwilliger TC, Waldo GS. Recent advances in GFP folding reporter and split-GFP solubility reporter technologies. Application to improving the folding and solubility of recalcitrant proteins from Mycobacterium tuberculosis. J Struct Funct Genomics. 2005;6(2–3):113–9. doi: 10.1007/s10969-005-5247-5. [DOI] [PubMed] [Google Scholar]
- 19.Wilson CG, Magliery TJ, Regan L. Detecting protein-protein interactions with GFP-fragment reassembly. Nat Methods. 2004;1(3):255–62. doi: 10.1038/nmeth1204-255. [DOI] [PubMed] [Google Scholar]
- 20.Lindman S, Hernandez-Garcia A, Szczepankiewicz O, Frohm B, Linse S. In vivo protein stabilization based on fragment complementation and a split GFP system. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(46):19826–31. doi: 10.1073/pnas.1005689107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394(6689):192–5. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 22.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(26):12501–4. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiedenmann J, Schenk A, Rocker C, Girod A, Spindler KD, Nienhaus GU. A far-red fluorescent protein with fast maturation and reduced oligomerization tendency from Entacmaea quadricolor (Anthozoa, Actinaria) Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11646–51. doi: 10.1073/pnas.182157199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17(10):969–73. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 25.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273(5280):1392–5. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 26.Kamagata K, Arai M, Kuwajima K. Unification of the folding mechanisms of non-two-state and two-state proteins. Journal of molecular biology. 2004;339(4):951–65. doi: 10.1016/j.jmb.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. Journal of molecular biology. 1998;277(4):985–94. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 28.Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry. 1993;32(5):1212–8. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 29.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111(2):229–33. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 30.Barondeau DP, Putnam CD, Kassmann CJ, Tainer JA, Getzoff ED. Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(21):12111–6. doi: 10.1073/pnas.2133463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tozzini V, Nifosi R. Ab initio molecular dynamics of the green fluorescent protein (GFP) chromophore: An insight into the photoinduced dynamics of green fluorescent proteins. Journal of Physical Chemistry B. 2001;105(24):5797–5803. [Google Scholar]
- 32.Maddalo SL, Zimmer M. The role of the protein matrix in green fluorescent protein fluorescence. Photochem Photobiol. 2006;82(2):367–72. doi: 10.1562/2005-04-11-RA-485. [DOI] [PubMed] [Google Scholar]
- 33.Chudakov DM, Feofanov AV, Mudrik NN, Lukyanov S, Lukyanov KA. Chromophore environment provides clue to “kindling fluorescent protein” riddle. The Journal of biological chemistry. 2003;278(9):7215–9. doi: 10.1074/jbc.M211988200. [DOI] [PubMed] [Google Scholar]
- 34.Quillin ML, Anstrom DM, Shu X, O'Leary S, Kallio K, Chudakov DM, Remington SJ. Kindling fluorescent protein from Anemonia sulcata: dark-state structure at 1.38 A resolution. Biochemistry. 2005;44(15):5774–87. doi: 10.1021/bi047644u. [DOI] [PubMed] [Google Scholar]
- 35.Habuchi S, Ando R, Dedecker P, Verheijen W, Mizuno H, Miyawaki A, Hofkens J. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(27):9511–6. doi: 10.1073/pnas.0500489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andresen M, Wahl MC, Stiel AC, Grater F, Schafer LV, Trowitzsch S, Weber G, Eggeling C, Grubmuller H, Hell SW, Jakobs S. Structure and mechanism of the reversible photoswitch of a fluorescent protein. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(37):13070–4. doi: 10.1073/pnas.0502772102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enoki S, Saeki K, Maki K, Kuwajima K. Acid denaturation and refolding of green fluorescent protein. Biochemistry. 2004;43(44):14238–48. doi: 10.1021/bi048733+. [DOI] [PubMed] [Google Scholar]
- 38.Fukuda H, Arai M, Kuwajima K. Folding of green fluorescent protein and the cycle3 mutant. Biochemistry. 2000;39(39):12025–32. doi: 10.1021/bi000543l. [DOI] [PubMed] [Google Scholar]
- 39.Huang JR, Craggs TD, Christodoulou J, Jackson SE. Stable intermediate states and high energy barriers in the unfolding of GFP. Journal of molecular biology. 2007 doi: 10.1016/j.jmb.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 40.Campanini B, Bologna S, Cannone F, Chirico G, Mozzarelli A, Bettati S. Unfolding of Green Fluorescent Protein mut2 in wet nanoporous silica gels. Protein science: a publication of the Protein Society. 2005;14(5):1125–33. doi: 10.1110/ps.041190805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andrews BT, Schoenfish AR, Roy M, Waldo G, Jennings PA. The rough energy landscape of superfolder GFP is linked to the chromophore. Journal of molecular biology. 2007;373(2):476–90. doi: 10.1016/j.jmb.2007.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu ST, Blaser G, Jackson SE. The folding, stability and conformational dynamics of beta-barrel fluorescent proteins. Chem Soc Rev. 2009;38(10):2951–65. doi: 10.1039/b908170b. [DOI] [PubMed] [Google Scholar]
- 43.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol. 2005;23(1):102–7. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 44.Demidov VV, Dokholyan NV, Witte-Hoffmann C, Chalasani P, Yiu HW, Ding F, Yu Y, Cantor CR, Broude NE. Fast complementation of split fluorescent protein triggered by DNA hybridization. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(7):2052–6. doi: 10.1073/pnas.0511078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(20):11241–6. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang YM, Bystroff C. Complementation and reconstitution of fluorescence from circularly permuted and truncated green fluorescent protein. Biochemistry. 2009;48(5):929–40. doi: 10.1021/bi802027g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y, Huang X, Cai J, Ye F, Guan L, Liu H, Qin Q. Construction of green fluorescent protein-tagged recombinant iridovirus to assess viral replication. Virus Res. 2011;160(1–2):221–9. doi: 10.1016/j.virusres.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 48.Mickler M, Dima RI, Dietz H, Hyeon C, Thirumalai D, Rief M. Revealing the bifurcation in the unfolding pathways of GFP by using single-molecule experiments and simulations. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(51):20268–73. doi: 10.1073/pnas.0705458104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dietz H, Rief M. Exploring the energy landscape of GFP by single-molecule mechanical experiments. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(46):16192–7. doi: 10.1073/pnas.0404549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertz M, Kunfermann A, Rief M. Navigating the folding energy landscape of green fluorescent protein. Angew Chem Int Ed Engl. 2008;47(43):8192–5. doi: 10.1002/anie.200802987. [DOI] [PubMed] [Google Scholar]
- 51.Nager AR, Baker TA, Sauer RT. Stepwise unfolding of a beta barrel protein by the AAA+ ClpXP protease. Journal of molecular biology. 2011;413(1):4–16. doi: 10.1016/j.jmb.2011.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reeder PJ, Huang YM, Dordick JS, Bystroff C. A rewired green fluorescent protein: folding and function in a nonsequential, noncircular GFP permutant. Biochemistry. 2010;49(51):10773–9. doi: 10.1021/bi100975z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ong WJ, Alvarez S, Leroux IE, Shahid RS, Samma AA, Peshkepija P, Morgan AL, Mulcahy S, Zimmer M. Function and structure of GFP-like proteins in the protein data bank. Mol Biosyst. 2011;7(4):984–92. doi: 10.1039/c1mb05012e. [DOI] [PubMed] [Google Scholar]
- 54.Andrews BT, Roy M, Jennings PA. Chromophore packing leads to hysteresis in GFP. Journal of molecular biology. 2009;392(1):218–27. doi: 10.1016/j.jmb.2009.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reddy G, Liu Z, Thirumalai D. Denaturant-dependent folding of GFP. Proceedings of the National Academy of Sciences of the United States of America. 2012 doi: 10.1073/pnas.1201808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Capraro DT, Gosavi S, Roy M, Onuchic JN, Jennings PA. Folding circular permutants of IL-1beta: route selection driven by functional frustration. PloS one. 2012;7(6):e38512. doi: 10.1371/journal.pone.0038512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jennings PA, Saalau-Bethell SM, Finn BE, Chen XW, Matthews CR. Mutational analysis of protein folding mechanisms. Methods in enzymology. 1991;202:113–26. doi: 10.1016/0076-6879(91)02009-x. [DOI] [PubMed] [Google Scholar]
- 58.Andrews BT, Gosavi S, Finke JM, Onuchic JN, Jennings PA. The dual-basin landscape in GFP folding. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(34):12283–12288. doi: 10.1073/pnas.0804039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh S, Zlotnick A. Observed hysteresis of virus capsid disassembly is implicit in kinetic models of assembly. The Journal of biological chemistry. 2003;278(20):18249–55. doi: 10.1074/jbc.M211408200. [DOI] [PubMed] [Google Scholar]
- 60.Minajeva A, Kulke M, Fernandez JM, Linke WA. Unfolding of titin domains explains the viscoelastic behavior of skeletal myofibrils. Biophysical journal. 2001;80(3):1442–51. doi: 10.1016/S0006-3495(01)76116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Markevich NI, Hoek JB, Kholodenko BN. Signaling switches and bistability arising from multisite phosphorylation in protein kinase cascades. Journal of Cell Biology. 2004;164(3):353–359. doi: 10.1083/jcb.200308060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis JM, Bachinger HP. Hysteresis in the triple helix-coil transition of type III collagen. The Journal of biological chemistry. 1993;268(34):25965–72. [PubMed] [Google Scholar]
- 63.Benitez-Cardoza CG, Rojo-Dominguez A, Hernandez-Arana A. Temperature-induced denaturation and renaturation of triosephosphate isomerase from Saccharomyces cerevisiae: evidence of dimerization coupled to refolding of the thermally unfolded protein. Biochemistry. 2001;40(30):9049–58. doi: 10.1021/bi010528w. [DOI] [PubMed] [Google Scholar]
- 64.Kellermayer MS, Smith SB, Bustamante C, Granzier HL. Complete unfolding of the titin molecule under external force. J Struct Biol. 1998;122(1–2):197–205. doi: 10.1006/jsbi.1998.3988. [DOI] [PubMed] [Google Scholar]
- 65.Preissner R, Egner U, Saenger W. Occurrence of bifurcated three-center hydrogen bonds in proteins. FEBS letters. 1991;288(1–2):192–6. doi: 10.1016/0014-5793(91)81032-4. [DOI] [PubMed] [Google Scholar]
- 66.Zhuang P, Blackburn MN, Peterson CB. Characterization of the denaturation and renaturation of human plasma vitronectin. I. Biophysical characterization of protein unfolding and multimerization. The Journal of biological chemistry. 1996;271(24):14323–32. doi: 10.1074/jbc.271.24.14323. [DOI] [PubMed] [Google Scholar]
- 67.Kosinski-Collins MS, King J. In vitro unfolding, refolding, and polymerization of human gammaD crystallin, a protein involved in cataract formation. Protein science: a publication of the Protein Society. 2003;12(3):480–90. doi: 10.1110/ps.0225503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mansfield ML. Are there knots in proteins? Nature structural biology. 1994;1(4):213–4. doi: 10.1038/nsb0494-213. [DOI] [PubMed] [Google Scholar]
- 69.Bolinger D, Sulkowska JI, Hsu HP, Mirny LA, Kardar M, Onuchic JN, Virnau P. A Stevedore's protein knot. PLoS computational biology. 2010;6(4):e1000731. doi: 10.1371/journal.pcbi.1000731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sulkowska JI, Rawdon EJ, Millett KC, Onuchic JN, Stasiak A. Conservation of complex knotting and slipknotting patterns in proteins. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(26):E1715–23. doi: 10.1073/pnas.1205918109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeates TO, Norcross TS, King NP. Knotted and topologically complex proteins as models for studying folding and stability. Current opinion in chemical biology. 2007;11(6):595–603. doi: 10.1016/j.cbpa.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Virnau P, Mirny LA, Kardar M. Intricate knots in proteins: Function and evolution. PLoS computational biology. 2006;2(9):e122. doi: 10.1371/journal.pcbi.0020122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sulkowska JI, Sulkowski P, Szymczak P, Cieplak M. Stabilizing effect of knots on proteins. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(50):19714–9. doi: 10.1073/pnas.0805468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sulkowska JI, Sulkowski P, Onuchic J. Dodging the crisis of folding proteins with knots. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(9):3119–24. doi: 10.1073/pnas.0811147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mallam AL, Jackson SE. Probing nature's knots: the folding pathway of a knotted homodimeric protein. Journal of molecular biology. 2006;359(5):1420–36. doi: 10.1016/j.jmb.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 76.Wallin S, Zeldovich KB, Shakhnovich EI. The folding mechanics of a knotted protein. Journal of molecular biology. 2007;368(3):884–93. doi: 10.1016/j.jmb.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tuszynska I, Bujnicki JM. Predicting atomic details of the unfolding pathway for YibK, a knotted protein from the SPOUT superfamily. Journal of biomolecular structure & dynamics. 27(4):511–20. doi: 10.1080/07391102.2010.10507335. [DOI] [PubMed] [Google Scholar]
- 78.Prentiss MC, Wales DJ, Wolynes PG. The energy landscape, folding pathways and the kinetics of a knotted protein. PLoS computational biology. 6:e1000835. doi: 10.1371/journal.pcbi.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Taylor WR, Lin K. Protein knots: A tangled problem. Nature. 2003;421(6918):25. doi: 10.1038/421025a. [DOI] [PubMed] [Google Scholar]
- 80.Sulkowska JI, Noel JK, Onuchic JN. Energy landscape of knotted protein folding. Proceedings of the National Academy of Sciences of the United States of America. 2012 doi: 10.1073/pnas.1201804109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Skrbic T, Micheletti C, Faccioli P. The role of non-native interactions in the folding of knotted proteins. PLoS computational biology. 2012;8(6):e1002504. doi: 10.1371/journal.pcbi.1002504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Faisca PF, Travasso RD, Charters T, Nunes A, Cieplak M. The folding of knotted proteins: insights from lattice simulations. Physical biology. 2010;7(1):16009. doi: 10.1088/1478-3975/7/1/016009. [DOI] [PubMed] [Google Scholar]
- 83.Sayre TC, Lee TM, King NP, Yeates TO. Protein stabilization in a highly knotted protein polymer. Protein engineering, design & selection: PEDS. 2011;24(8):627–30. doi: 10.1093/protein/gzr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mallam AL, Jackson SE. Knot formation in newly translated proteins is spontaneous and accelerated by chaperonins. Nature chemical biology. 2011;8(2):147–53. doi: 10.1038/nchembio.742. [DOI] [PubMed] [Google Scholar]
- 85.Noel JK, Whitford PC, Sanbonmatsu KY, Onuchic JN. SMOG@ctbp: simplified deployment of structure-based models in GROMACS. Nucleic acids research. 38(Web Server issue):W657–61. doi: 10.1093/nar/gkq498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sulkowska JI, Sulkowski P, Szymczak P, Cieplak M. Tightening of knots in proteins. Physical review letters. 2008;100(5):058106. doi: 10.1103/PhysRevLett.100.058106. [DOI] [PubMed] [Google Scholar]
- 87.Mallam AL, Jackson SE. Folding studies on a knotted protein. Journal of molecular biology. 2005;346(5):1409–21. doi: 10.1016/j.jmb.2004.12.055. [DOI] [PubMed] [Google Scholar]
- 88.Ferreiro DU, Hegler JA, Komives EA, Wolynes PG. Localizing frustration in native proteins and protein assemblies. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(50):19819–24. doi: 10.1073/pnas.0709915104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Preissner KT, Jenne D. Structure of vitronectin and its biological role in haemostasis. Thromb Haemost. 1991;66(1):123–32. [PubMed] [Google Scholar]
- 90.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–8. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 91.Brunak S, Engelbrecht J. Protein structure and the sequential structure of mRNA: alpha-helix and beta-sheet signals at the nucleotide level. Proteins. 1996;25(2):237–52. doi: 10.1002/(SICI)1097-0134(199606)25:2<237::AID-PROT9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 92.Siemion IZ. Compositional frequencies of amino acids in the proteins and the genetic code. Bio Systems. 1994;32(3):163–70. doi: 10.1016/0303-2647(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 93.Adzhubei AA, Adzhubei IA, Krasheninnikov IA, Neidle S. Non-random usage of `degenerate' codons is related to protein three-dimensional structure. FEBS letters. 1996;399(1–2):78–82. doi: 10.1016/s0014-5793(96)01287-2. [DOI] [PubMed] [Google Scholar]
- 94.Gupta SK, Majumdar S, Bhattacharya TK, Ghosh TC. Studies on the relationships between the synonymous codon usage and protein secondary structural units. Biochemical and biophysical research communications. 2000;269(3):692–6. doi: 10.1006/bbrc.2000.2351. [DOI] [PubMed] [Google Scholar]
- 95.Cortazzo P, Cervenansky C, Marin M, Reiss C, Ehrlich R, Deana A. Silent mutations affect in vivo protein folding in Escherichia coli. Biochemical and biophysical research communications. 2002;293(1):537–41. doi: 10.1016/S0006-291X(02)00226-7. [DOI] [PubMed] [Google Scholar]
- 96.Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS letters. 1999;462(3):387–91. doi: 10.1016/s0014-5793(99)01566-5. [DOI] [PubMed] [Google Scholar]