Abstract
Calcium signals, pivotal in controlling cell function, can be generated by calcium entry channels activated by plasma membrane depolarization or depletion of internal calcium stores. We reveal a regulatory link between these two channel subtypes mediated by the ubiquitous calcium-sensing STIM proteins. STIM1 activation by store depletion or mutational modification strongly suppresses voltage-operated calcium (CaV1.2) channels while activating store-operated Orai channels. Both actions are mediated by the short STIM-Orai activating region (SOAR) of STIM1. STIM1 interacts with CaV1.2 channels and localizes within discrete endoplasmic reticulum/plasma membrane junctions containing both CaV1.2 and Orai1 channels. Hence, STIM1 interacts with and reciprocally controls two major calcium channels hitherto thought to operate independently. Such coordinated control of the widely expressed CaV1.2 and Orai channels has major implications for Ca2+ signal generation in excitable and nonexcitable cells.
Ca2+ entry channels, crucial in providing cellular Ca2+ signals, are controlled by sensing mechanisms, including membrane voltage, surface receptors, and Ca2+-sensing STIM proteins in the endoplasmic reticulum (ER) (1). The coordinated operation of these transduction processes is key to controlling cell function. STIM proteins are dynamic ER Ca2+ sensors that aggregate when ER is depleted of Ca, then rapidly translocate into ER-plasma membrane (PM) junctions, where they interact with and activate the highly Ca2+-selective Orai family of PM channels (2, 3). We determined that STIM proteins also mediate inhibitory control of voltage-activated CaV1.2 channels. This action was independent of Orai channel function or changes in cytosolic Ca2+ and was mediated by a direct action of STIM1 on the CaV1.2 α1C subunit. Thus, STIM1 reciprocally controls Orai and CaV1.2 channels, indicating a hitherto unknown and potentially crucial regulatory link between receptor-induced Ca2+ store depletion and control of voltage-activated Ca2+ signals.
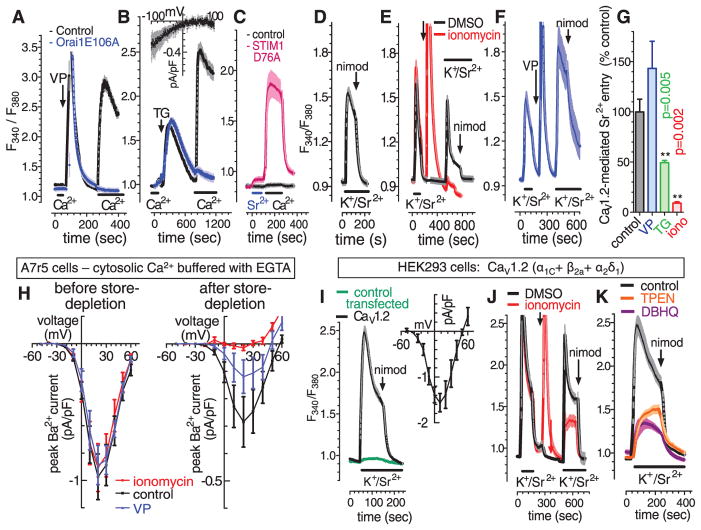
We studied STIM1-mediated Ca2+ entry signals in A7r5 clonal vascular smooth muscle cells (VSMC). Ca2+ store depletion by vasopressin (VP) or ER Ca2+ pump blockade by thapsigargin (TG) induced a large Ca2+ influx across the PM when Ca2+ was added externally (Fig. 1, A and B). However, the inwardly rectifying Ca2+-release activated Ca2+ (CRAC) current mediating this Ca2+ entry was barely measurable (Fig. 1B, inset) because only a small number of Ca2+ ions flow through these highly selective channels. Expression in A7r5 cells of the STIM1-D76A mutant defective in sensing ER Ca2+ (1) selectively activated Ca2+ entry (almost no entry of Sr2+) (Fig. 1C), which required no store emptying. Expression of the E106A Orai1 mutant lacking a functional pore and acting as a dominant negative on Orai channels (4, 5) completely eliminated VP- or TG-induced Ca2+ entry (Fig. 1A and B), revealing that entry is exclusively Orai-mediated.
Fig. 1.
Store-dependent control of Orai and CaV1.2 channels. Fura-2 F340/F380 ratiometric responses to cytosolic Ca2+ in A7r5 VSMCs in response to 100 nM VP (A) or 2 μM TG (B), in control or Orai1-E106A-CFP–transfected cells in nominally Ca2+ free or 3 mM external Ca2+ (bars). (Inset) Current-voltage curve for CRAC channels in control A7r5 cells. (C) Fura-2 responses in control and STIM1-D76A–transfected A7r5 cells (bars, 3 mM external Sr2+ or Ca2+). (D) Fura-2 responses to application of external 134 mM K+ with 3 mM Sr2+ (bars, K+/Sr2+); 2 μM nimodipine (arrow). (E) Fura-2 responses to two sequential K+/Sr2+ pulses (bars) with addition of dimethyl sulfoxide (DMSO) or 2 μM ionomycin (arrow), and subsequent 2 μM nimodipine addition. (F) As in (E), except with the addition of 100 nM VP. (G) Statistics for results in (E), (F), and TG (trace not shown) (n = 6, paired t test). (H) I/V curve for whole-cell Ba2+ current through CaV1.2 channels in A7r5 cells with cytosolic Ca2+ clamped to 0.1 μM (10 mM EGTA; 3.4 mM CaCl2), either before (left) or 5 min after store depletion with 2 μM ionomycin, 100 nM VP, or control (right). (I to K) Fura-2 responses to K+/Sr2+ (bars) in HEK293 cells expressing three CaV1.2 subunits (α1C+ β2a+ α2δ1) or control-transfected cells. (I) K+/Sr2+ responses in CaV1.2-transfected and control-transfected cells; 2 μM nimodipine (arrow); inset, I/V curve for CaV1.2-transfected cells (n = 4). (J) Fura-2 responses to sequential K+/Sr2+ pulses (bars) with additions of DMSO or 2 μM ionomycin (arrow), and 2 μM nimodipine (arrows). (K) K+/Sr2 responses after DBHQ (10 μM, 10 min, present throughout) or TPEN (250 μM, 5 min, removed at K+/Sr2+ addition).
Canonical transient receptor potential (TRPC) channels, widely reported to contribute to store-induced Ca2+ entry (1, 6), are highly expressed in VSMCs (7). In A7r5 cells, a large nonselective current activated by VP and mediated by TRPCs (7) was not influenced by store depletion or by expression of the dominant Orai1-E106A mutant or the constitutively active STIM1-D76A mutant, reinforcing recent evidence (8) against a role of STIM proteins or Ca2+ stores in TRPC channel function. In contrast, endogenous CaV1.2 channels did respond to emptying of intracellular Ca2+ stores. In A7r5 cells, Sr2+ entry through CaV1.2 channels was activated by a depolarizing pulse of high extracellular [K+] and blocked by the CaV1.2-specific blocker nimodipine (Fig. 1D). If 2 μM ionomycin or 2 μM TG was added to deplete Ca2+ stores, a subsequent depolarizing pulse resulted in greatly decreased CaV1.2-mediated entry of Sr2+ (Fig. 1, E and G). However, although VP treatment also induced Ca2+ store depletion, CaV1.2-mediated Sr2+ entry was instead enhanced (Fig. 1, F and G), likely due to activation of CaV1.2 channels by Ca2+- and diacylglcycerol-induced protein kinase C stimulation (9, 10). To more directly examine the actions of store emptying on CaV1.2 channels, we measured CaV1.2 currents by patch-clamping cells with cytosolic Ca2+ buffered at physiological resting levels with the Ca2+ chelator EGTA to prevent any Ca2+-dependent changes in CaV1.2 function. Compared with control CaV1.2 peak current (0.28 ± 0.08 pA/pF), store depletion by ionomycin inhibited CaV1.2 current by 96% (0.01 ± 0.02 pA/pF; n = 6, P = 0.001) (Fig. 1H). Store depletion with VP also reduced current by 58% (0.11 ± 0.06 pA/pF; n = 6, P = 0.004). Thus, Ca2+ buffering revealed a consistent store depletion–mediated inhibition of CaV1.2 channels. We further examined the influence of Ca2+ store depletion on CaV1.2 channels expressed in human embryonic kidney (HEK293) cells. Nimodipine-sensitive, depolarization-induced Sr2+ entry (Fig. 1I) with typical CaV1.2 current-voltage relationship (inset) was observed in HEK293 cells expressing the three CaV1.2 component subunits α1C, β2a, and α2δ1. Ca2+ store depletion with ionomycin or ER Ca2+ pump blocker di-tertbutylhydroquinone (DBHQ) substantially reduced CaV1.2-mediated Sr2+ entry compared with control cells (Fig. 1, J and K). Application of the cell-permeant intraluminal ER-Ca2+ chelator, N, N,N′,N′-tetrakis(2-pyridylmethyl)-ethylenediamine (TPEN), caused a similar decrease (Fig. 1K). TPEN reduces ER-Ca2+ with no increase in cytosolic Ca2+ (11), confirming that store depletion inhibits CaV1.2 channels independent of cytosolic Ca2+ changes.
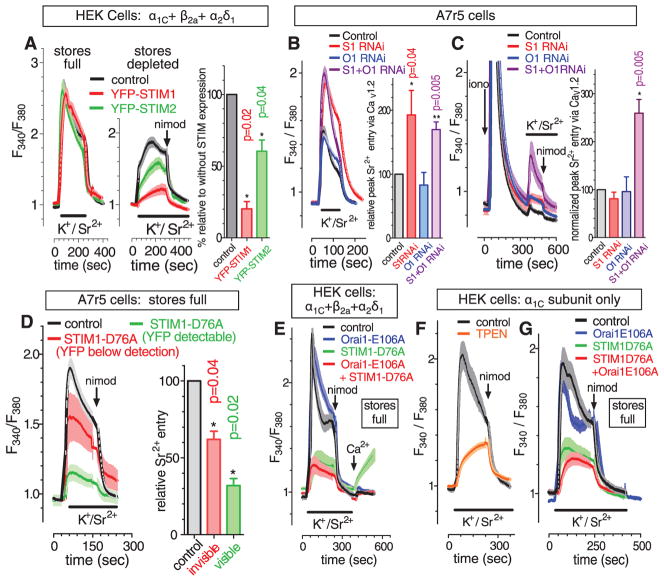
The ER Ca2+ sensors, STIM1 and STIM2, are triggered by ER Ca2+ depletion to translocate and activate PM Orai channels (1). We investigated the role of STIM proteins in store depletion–induced inhibition of CaV1.2 channels. Using CaV1.2 (α1C + β2a + α2δ1)–transfected HEK293 cells, overexpressed yellow fluorescent protein–fusion constructs (YFP-STIM1 or YFP-STIM2) (fig. S1, A to D) had little effect on CaV1.2-mediated Sr2+ entry in store-replete cells (Fig. 2A). However, YFP-STIM1 expression greatly increased the inhibitory effect of ionomycin-induced store depletion on CaV1.2-mediated Sr2+ entry (Fig. 2A). Similar expression of YFP-STIM2 had a smaller effect. STIM2 is similarly poorer in coupling to activate Orai channels (1, 12, 13). Although native STIM1 is expressed in both ER and PM, YFP-STIM1 is exclusively expressed in ER (1, 14); thus, the action of STIM1 on CaV1.2 function is mediated by ER-STIM1.
Fig. 2.
STIM proteins mediate store depletion–induced control of CaV1.2 channels. (A) HEK293 cells expressing α1C + β2a+ α2δ1 CaV1.2 channel subunits; fura-2 responses to K+/Sr2+ pulses (bars) before (left) and after (middle) store emptying with 2 μM ionomycin and subsequent 2 μM nimodipine addition (arrow). Statistics (right), paired t test, n = 6. (B) Store-replete A7r5 cells treated either with STIM1 RNAi, Orai1 RNAi, STIM1+Orai1 RNAi, or control RNAi; left, fura-2 responses to K+/Sr2+ pulse (bar); right, statistics (paired t test, n = 8). (C) The same RNAi-treated cells used in (B) were then store-depleted with 2 μM ionomycin (arrow); left, fura-2 responses to K+/Sr2+ pulse (bar); right, statistics (paired t test, n = 8). (D) Store-replete A7r5 cells either control-transfected or transfected with YFP-STIM1-D76A (cells with detectable or undetectable YPP on a single coverslip); left, fura-2 responses to K+/Sr2+ pulse (bar) and 2 μM nimodipine (arrow); right, statistics (paired t test, n = 8). (E) HEK293 cells expressing α1C + β2a+ α2δ1 CaV1.2 channel subunits cotransfected with either Orai1-E106A-CFP, YFP-STIM1-D76A, Orai1-E106A-CFP + YFP-STIM1-D76A, or control plasmid; fura-2 responses to K+/Sr2+ pulse (bar), 2 μM nimodipine (arrow) and 3 mM Ca2+ (arrow). (F) HEK293 cells expressing α1C alone; fura-2 response to K+/Sr2 pulse (bar) and 2 μM nimodipine (arrow) in cells either TPEN-treated (250 μM, 5 min, removed at K+/Sr2+ addition) or untreated. (G) HEK293 cells expressing α1C cotransfected with either Orai1-E106A-CFP, YFP-STIM1-D76A, Orai1-E106A-CFP + YFP-STIM1-D76A, or control plasmid; fura-2 response to K+/Sr2+ pulse (bar), and 2 μM nimodipine (arrow).
We examined how STIM1 and Orai1 RNA interference (RNAi) influenced endogenous CaV1.2 channels in A7r5 cells. STIM1-siRNA (small interfering RNA) increased CaV1.2-mediated Sr2+ entry in store-replete cells (Fig. 2B), suggesting that some endogenous STIM1 exists in junctions and constitutively inhibits CaV1.2. Orai1-siRNA had little effect. STIM1-siRNA had little effect on store depletion–induced inhibition of CaV1.2 channels (Fig. 2C). However, although RNAi reduced STIM1 expression by ~80% (fig. S2A), store emptying may efficiently cause the remaining 20% to enter ER-PM junctions, perhaps explaining the differential effectiveness of STIM1-RNAi on CaV1.2 activity in store-replete versus store-depleted cells. A combination of STIM1- and Orai1-siRNA resulted in a substantial reduction in the inhibition of CaV1.2 channels by store depletion (Fig. 2C). After store depletion, Orai1 traps STIM1 within ER-PM junctions (15); thus, with limited STIM1 in RNAi-treated cells, endogenous Orai1 may be necessary to bring STIM1 to the PM where it can alter CaV1.2 channels.
STIM1 EF-hand mutants fail to bind Ca2+ (16–18), aggregate and translocate into ER-PM junctions, and constitutively activate Orai1 channels without requiring changes in ER Ca2+ (18). Expression of YFP-STIM1-D76A EF-hand mutant in A7r5 cells led to a constitutive reduction in the function of endogenous CaV1.2 channels (Fig. 2D). Cells with visible YFP expression had >70% reduced CaV1.2-mediated Sr2+ entry, and almost all mutant STIM1 was within clearly discernible junctions (fig. S2B). There was little difference in resting cytosolic [Ca2+] in D76A-expressing cells (fig. S2C), consistent with studies on cells stably expressing D76A (19). Cells with no detectable fluorescence had 40% reduced CaV1.2 function (Fig. 2D) and no change in resting Ca2+ (fig. S2C). Hence, the action of STIM1 on CaV1.2 channel function is independent of both luminal and cytosolic Ca2+ changes.
The same inhibitory effect of STIM1-D76A was observed on Sr2+ entry through CaV1.2 channels expressed in HEK cells (Fig. 2E). Coexpression of the dominant negative Orai1-E106A mutant did not alter the STIM1-D76A–induced inhibition of CaV1.2 channels, although it completely blocked the constitutive Ca2+ entry observed after addition of Ca2+ in STIM1-D76A–expressing cells after washout of the K+/Sr2+ solution (Fig. 2E, red versus green traces). Hence, we conclude that the inhibitory effect of STIM1 on CaV1.2 channels does not require Orai channel function and does not reflect any passage of ions through store-operated channels.
CaV1.2 channels comprise three subunits: the α1C pore-forming moiety and the β and α2δ1 auxiliary subunits (10). STIM1-mediated CaV1.2 inhibition did not require the auxiliary proteins. As for HEK293 cells expressing all three CaV1.2 subunits, cells expressing only the α1C subunit had depolarization-induced Sr2+ entry inhibited by TPEN (Fig. 2F), enhanced with STIM1 or STIM2 overexpression (fig. S3, A to C) and mimicked by STIM1-D76A expression independently of Orai function (Fig. 2G). STIM1-D76A did not alter α1C expression or vice versa, and α1C did not alter STIM1-D76A localization (fig. S3, D and E).
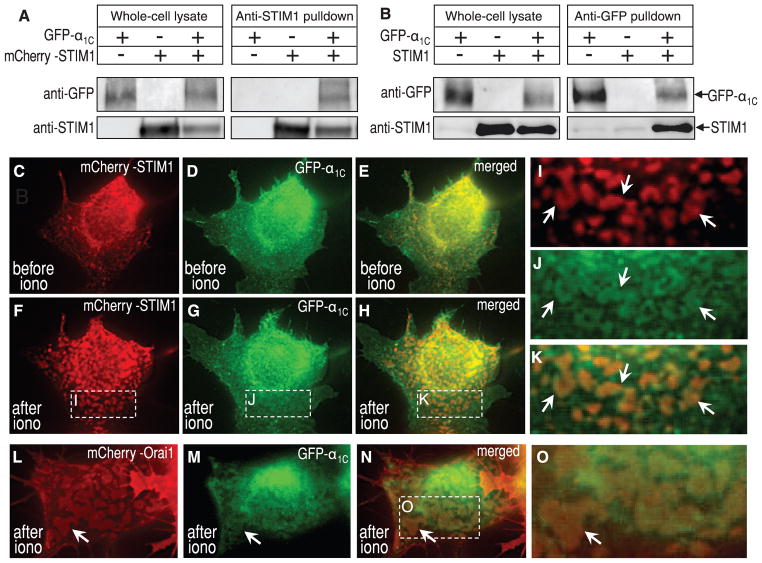
To address whether CaV1.2 α1C interacts with STIM1, we undertook coimmunoprecipitation studies using green fluorescent protein (GFP)–tagged α1C and either native or mCherry-tagged STIM1. Coexpressed in HEK cells, antibody to STIM1 precipitates contained GFP-α1C (Fig. 3A). Moreover, precipitates of GFP-α1C with antibody to GFP revealed bound STIM1 (Fig. 3B). Although interactions between the two proteins exist, store depletion did not detectably alter this association. We undertook high-resolution imaging studies to examine distribution of STIM1, CaV1.2 channels, and Orai channels at the PM (19). We expressed N-tagged GFP-α1C (20), β2a and α2δ1 subunits, and N-tagged mCherry-STIM1 (19). High-resolution imaging of the ER-PM junctions adjacent to the coverslip revealed precise colocalization between mCherry-STIM1 and GFP-α1C induced by emptying Ca2+ stores with ionomycin (Fig. 3, C to K). Ionomycin-mediated store depletion for 5 min induced mCherry-STIM1 to move into clearly defined ER-PM junctional areas (Fig. 3, C and F). GFP-CaV1.2 moved into the same areas (Fig. 3, D, G, and H). High magnification of the boxed areas (Fig. 3, I to K) reveals the precise nature of the colocalization of the two proteins after store depletion. There were no areas of STIM1 not colocalized with α1C, although there were areas of α1C extending beyond the STIM1-defined junctional areas. Small areas with colocalized STIM-CaV1.2 were evident in store-replete cells (Fig. 3, C to E). This indicates a possible constitutive role of STIM1 in attenuating CaV1.2 activity, consistent with the effect of STIM1 RNAi on CaV1.2 function (Fig. 2B). We also coexpressed mCherry-Orai1 [exclusively PM-localized (18)] with GFP-α1C in stable STIM1-expressing HEK cells, revealing areas of Orai1-CaV1.2 colocalization after store depletion (Fig. 3, L to O). Such Orai1-localization is exclusively STIM-associated (1, 18); hence, ER-PM junctions appear to contain a complex of Orai1, CaV1.2, and STIM proteins.
Fig. 3.
Interactions between STIM1 and CaV1.2 α1C, and store-dependent co-localization of STIM1, CaV1.2, and Orai1 proteins. (A) HEK293 cells coexpressing mCherry-STIM1 and GFP-α1C; whole-cell lysates (left) and antibody to STIM1 immunoprecipitates were probed with antibody to GFP and antibody to STIM1. (B) HEK293 cells coexpressing GFP-α1C and STIM1; whole-cell lysates (left) and antibody to GFP immunoprecipitates were probed with antibody to GFP and antibody to STIM1. (C to O) High-resolution imaging focused on the ER-PM interface in HEK293 cells. (C to H) Distribution of expressed mCherry-STIM1 and GFP-α1C (coexpressed with β2a + α2δ1 subunits), examined in cells either just before (C and E) or 5 min after (F to H) Ca2+ store depletion with 2 μM ionomycin. (I to K) Magnification of the STIM, α1C, and merged images, respectively. (L and M) Distribution of coexpressed mCherry-Orai1 and GFP-α1C (with β2a + α2δ1 subunits) in cells 5 min after ionomycin-induced emptying. (O) Magnified area of the merged image from (N).
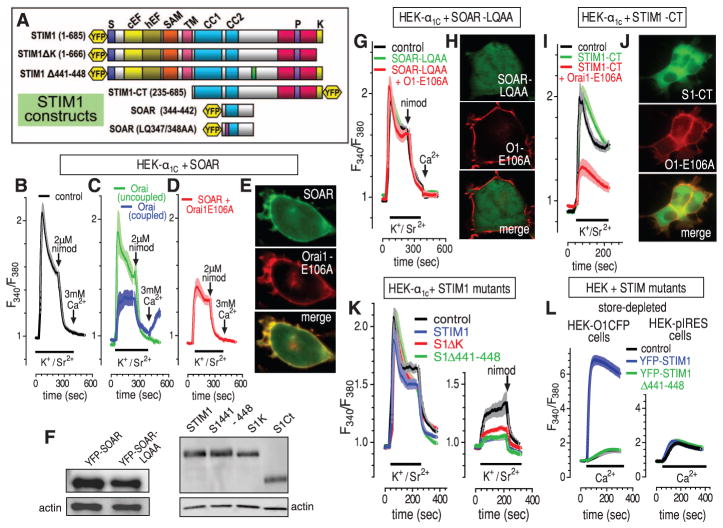
We investigated whether STIM1 domains coupling to Orai were also coupling to α1C and whether Orai might participate in STIM-α1C coupling. The smallest units of STIM1 to bind and activate Orai1 are the 344-442 STIM-Orai activating region (SOAR) (21) and 342-448 Ca2+-activating domain (CAD) (15) (Fig. 4A). We transfected YFP-SOAR into HEK293 cells expressing α1C (Fig. 4, B to F). CaV1.2 channels were activated with K+/Sr2+, then blocked with nimodipine, followed by Ca2+ addition to observe Orai function. In non-SOAR–expressing cells, there was full α1C channel activity and no Orai function (Fig. 4B). In cells cotransfected with SOAR (Fig. 4C), a clear correlation existed between Orai activation, α1C inhibition, and SOAR expression (Fig. 4, C and D, and fig. S4A). Cells with full α1C-mediated Sr2+ entry had no Orai-induced Ca2+ entry (green), whereas cells with a large reduction in α1C function showed Orai-induced Ca2+ entry (blue). The Orai-coupled cells had ~50% greater SOAR levels (fig. S4B); thus, SOAR coupling to both α1C and Orai exactly correlated. Coexpressing Orai1-E106A with α1C and SOAR (Fig. 4D) caused SOAR to be more consistently PM-associated (Fig. 4E), and α1C channel activity was inhibited in all cells (Fig. 4D and fig. S4A). Thus, Orai may assist SOAR associating with the PM and inhibit α1C; however, the Orai channel need not be functional to pull SOAR toward α1C and inhibit α1C function. Expression of the LQ347/348AA SOAR mutant (Fig. 4F) devoid of coupling to Orai1 (21) did not inhibit α1C function, with or without Orai1-E106A (Fig. 4G), and did not associate with Orai1 at the PM (Fig. 4H). We expressed the STIM1 C terminus (YFP-STIM1-CT; 235-685) (Fig. 4, F, I, and J), which is largely cytosolic (15, 18). Coexpressed with Orai channels, some STIM1-CTassociates with Orai1 (Fig. 4J). STIM1-CT expressed with α1C caused no change in CaV1.2 activity consistent with its cytosolic location (18). However, coexpressed with Orai1-E106A, the STIM1-CT gave substantial reduction in α1C activity (Fig. 4I), indicating that the association between STIM1-CT and Orai1 was sufficient for STIM1-CT to inhibit α1C.
Fig. 4.
Defining the molecular domains of STIM1 mediating CaV1.2 channel inhibition. (A) STIM1 constructs used. (B to D) Fura-2 responses to K+/Sr2+ pulses (bars), 2 μM nimodipine, and 3 mM Ca2+ (arrows) in α1C-expressing HEK293 cells without cotransfection (B) or with cotransfection of YFP-SOAR (C) or YFP-SOAR + Orai1-E106A-CFP (D). Statistics shown in fig. S4A. In (C), Orai-coupled (n = 97; cells with Orai-mediated Ca2+ entry) and Orai-uncoupled (n = 135; cells with no Orai-mediated Ca2+ entry) are defined in the text. Their relative YFP-SOAR expression is shown in fig. S4B. (E) Localization of YFP-SOAR + Orai1-E106A-CFP cotransfected cells. (F) Western analysis of STIM construct expression. (G) Fura-2 responses in α1C-expressing HEK293 cells cotransfected with YFP-SOAR-LQ347/348AA alone or with Orai-E106A-CFP. (H) Imaging of the mutant SOAR + Orai–expressing cells in (G). (I) Fura-2 responses in α1C-expressing HEK293 cells cotransfected with YFP-STIM1-CT alone or with Orai-E106A-CFP. (J) Imaging of the STIM-CT + Orai–expressing cells in (I). (K) Fura-2 responses in α1C-expressing HEK293 cells before (left) and after (right) store depletion with 2 μM ionomycin; cells were cotransfected with either normal STIM1, the STIM1-ΔK truncation (Δ667-685), the STIM1-Δ441-448 deletion, or empty plasmid (statistics shown in fig. S4C). (L) Fura-2 responses to 1 mM Ca2+ addition for Orai1-CFP–expressing HEK293 cells (left) or control internal ribosomal entry site plasmid (pIRES)–HEK cells (right) transfected with YFP-STIM1-wild-type or YFP-STIM1-Δ441-448.
The far C-terminal K-rich region of STIM1, although not essential for Orai1 activation, assists STIM1-induced association with PM junctions (15). Compared with wild-type STIM1, STIM1ΔK (Δ667-685) (Fig. 4, A and F) was less effective in inhibiting α1C function after store depletion (Fig. 4K and fig. S4C), indicating that the K-rich region enhances PM docking of STIM1 and association with α1C. The STIM1Δ441-448 deletion (Fig. 4A) does not activate Orai1 channels and does not block endogenous STIM1-mediated Ca2+ influx (Fig. 4L) and is likely defective in Orai1-binding. However, STIM1Δ441-448 does still inhibit α1C-mediated Sr2+ entry almost identical to wild-type STIM1 (Fig. 4K and fig. S4C). Thus, the site on STIM1 coupling to inhibit CaV1.2 channels is not identical to that mediating Orai channel activation. Moreover, the action of STIM1 on CaV1.2 channels does not require Orai channel activation, despite Orai channels assisting STIM1’s approach to CaV1.2 channels.
The work herein provides evidence that the CaV1.2 channel is an authentic target of STIM proteins, in addition to their well-described Orai targets. The reciprocal control mediated by STIM on the Orai and CaV1.2 channel targets may have functional implications in many cells expressing both channels. Although we reveal that the action of STIM1 on CaV1.2 channels does not require the function of Orai channels, we do reveal that the actions of STIM1 on Orai1 and CaV1.2 channels are closely connected, both spatially and functionally. Orai channels are very effective at trapping STIM1 in puncta (15) and appear to enhance STIM1 recruitment to the vicinity of CaV1.2 channels in the PM. CaV1.2 proteins are widely expressed not only in excitable cells but also nonexcitable cells—for example, immune cells including T cells, B cells, dendritic cells, and mast cells (22, 23). STIM proteins may have important roles in suppressing CaV1.2 function in immune cells, and STIM-mediated reciprocal control of CaV1.2 and Orai channels could be a decisive mechanism for controlling Ca2+ signals. STIM proteins are highly expressed in many excitable tissues, including smooth muscle cells (24, 25), neurons (26), and skeletal muscle (27), where they have been implicated in mediating not only Ca2+ signals but also fundamental control over growth, differentiation, and apoptosis (25–27). The finding of reciprocal control of the two major and widely expressed Ca2+ channels by a single Ca2+-sensing regulatory protein should enhance understanding of Ca2+ signal transduction.
Supplementary Material
Acknowledgments
Rabbit smooth muscle α1C, β2a, and α2-δ1 DNA were gifts from M. Davis, University of Missouri School of Medicine. This work was supported by NIH grants HL55426 and AI058173 (D.L.G.), a Novartis Institutes for Biomedical Research fellowship (Y.W.), the American Heart Association (J.S.), NSFC 30871011, Tianjin S&T Support Project 08ZCKFSH04500, and “973” program 2010CB945001 (X.P.T.).
Footnotes
References and Notes
- 1.Deng X, Wang Y, Zhou Y, Soboloff J, Gill DL. J Biol Chem. 2009;284:22501. doi: 10.1074/jbc.R109.018655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis RS. Nature. 2007;446:284. doi: 10.1038/nature05637. [DOI] [PubMed] [Google Scholar]
- 3.Cahalan MD. Nat Cell Biol. 2009;11:669. doi: 10.1038/ncb0609-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soboloff J, et al. J Biol Chem. 2006;281:20661. doi: 10.1074/jbc.C600126200. [DOI] [PubMed] [Google Scholar]
- 5.Lis A, et al. Curr Biol. 2007;17:794. doi: 10.1016/j.cub.2007.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan JP, et al. Channels (Austin) 2009;3:221. doi: 10.4161/chan.3.4.9198. [DOI] [PubMed] [Google Scholar]
- 7.Soboloff J, et al. J Biol Chem. 2005;280:39786. doi: 10.1074/jbc.M506064200. [DOI] [PubMed] [Google Scholar]
- 8.DeHaven WI, et al. J Physiol. 2009;587:2275. doi: 10.1113/jphysiol.2009.170431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shistik E, Ivanina T, Blumenstein Y, Dascal N. J Biol Chem. 1998;273:17901. doi: 10.1074/jbc.273.28.17901. [DOI] [PubMed] [Google Scholar]
- 10.Dai S, Hall DD, Hell JW. Physiol Rev. 2009;89:411. doi: 10.1152/physrev.00029.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofer AM, Fasolato C, Pozzan T. J Cell Biol. 1998;140:325. doi: 10.1083/jcb.140.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soboloff J, et al. Curr Biol. 2006;16:1465. doi: 10.1016/j.cub.2006.05.051. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y, et al. J Biol Chem. 2009;284:19164. doi: 10.1074/jbc.C109.010900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baba Y, et al. Proc Natl Acad Sci USA. 2006;103:16704. doi: 10.1073/pnas.0608358103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park CY, et al. Cell. 2009;136:876. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liou J, et al. Curr Biol. 2005;15:1235. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spassova MA, et al. Proc Natl Acad Sci USA. 2006;103:4040. doi: 10.1073/pnas.0510050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Proc Natl Acad Sci USA. 2009;106:7391. [Google Scholar]
- 19.Hewavitharana T, et al. J Biol Chem. 2008;283:26252. doi: 10.1074/jbc.M802239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabner M, Dirksen RT, Beam KG. Proc Natl Acad Sci USA. 1998;95:1903. doi: 10.1073/pnas.95.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan JP, et al. Nat Cell Biol. 2009;11:337. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matza D, Flavell RA. Immunol Rev. 2009;231:257. doi: 10.1111/j.1600-065X.2009.00805.x. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki Y, Inoue T, Ra C. Mol Immunol. 2010;47:640. doi: 10.1016/j.molimm.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Deng X, Hewavitharana T, Soboloff J, Gill DL. Clin Exp Pharmacol Physiol. 2008;35:1127. doi: 10.1111/j.1440-1681.2008.05018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potier M, et al. FASEB J. 2009;23:2425. doi: 10.1096/fj.09-131128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berna-Erro A, et al. Sci Signal. 2009;2:ra67. doi: 10.1126/scisignal.2000522. [DOI] [PubMed] [Google Scholar]
- 27.Stiber J, et al. Nat Cell Biol. 2008;10:688. doi: 10.1038/ncb1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.