Abstract
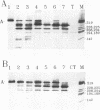
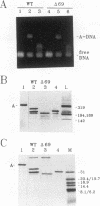
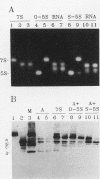
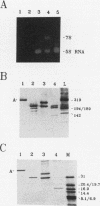
Transcription factor IIIA (TFIIIA) employs an array of nine N-terminal zinc fingers to bind specifically to both 5S RNA and 5S DNA. The binding of TFIIIA to 5S RNA and 5S DNA was studied by using a protease footprinting technique. Brief treatment of free or bound TFIIA with trypsin or chymotrypsin generated fragments which were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Fragments retaining the N terminus of TFIIA were identified by immunoblotting with an antibody directed against the N terminus of TFIIIA. Proteolytic footprinting of TFIIIA complexed with 5S DNA derivatives reinforced other evidence that the three N-terminal zinc fingers of TFIIIA bind most tightly to 5S DNA. Proteolytic footprinting of TFIIIA in reconstituted 7S ribonucleoprotein particles revealed different patterns of trypsin sensitivity for TFIIIA bound to oocyte versus somatic 5S RNA. Trypsin cleaved TFIIIA between zinc fingers 3 and 4 more readily when the protein was bound to somatic 5S RNA than when it was bound to oocyte 5S RNA. A tryptic fragment of TFIIIA containing zinc fingers 4 through 7 remained tightly associated with somatic 5S RNA. Zinc fingers 4 through 7 may represent a tightly binding site for 5S RNA in the same sense that fingers 1 through 3 represent a tightly binding site for 5S DNA.
Full text
PDF

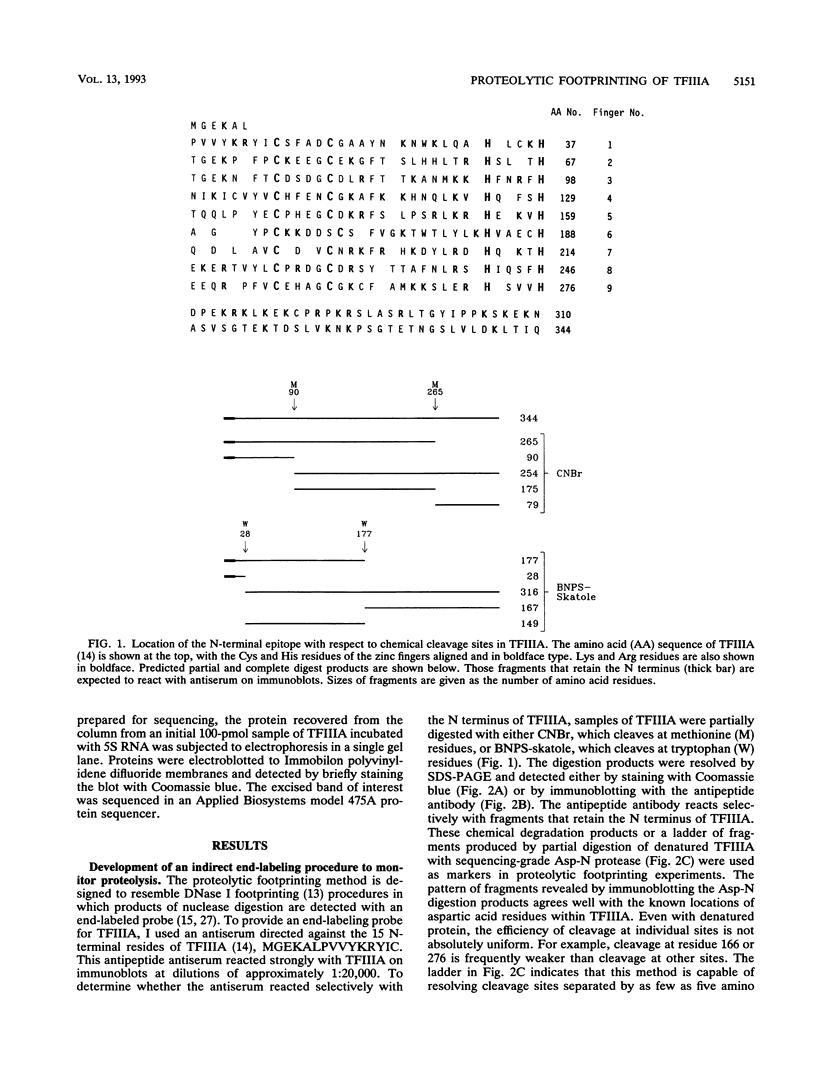
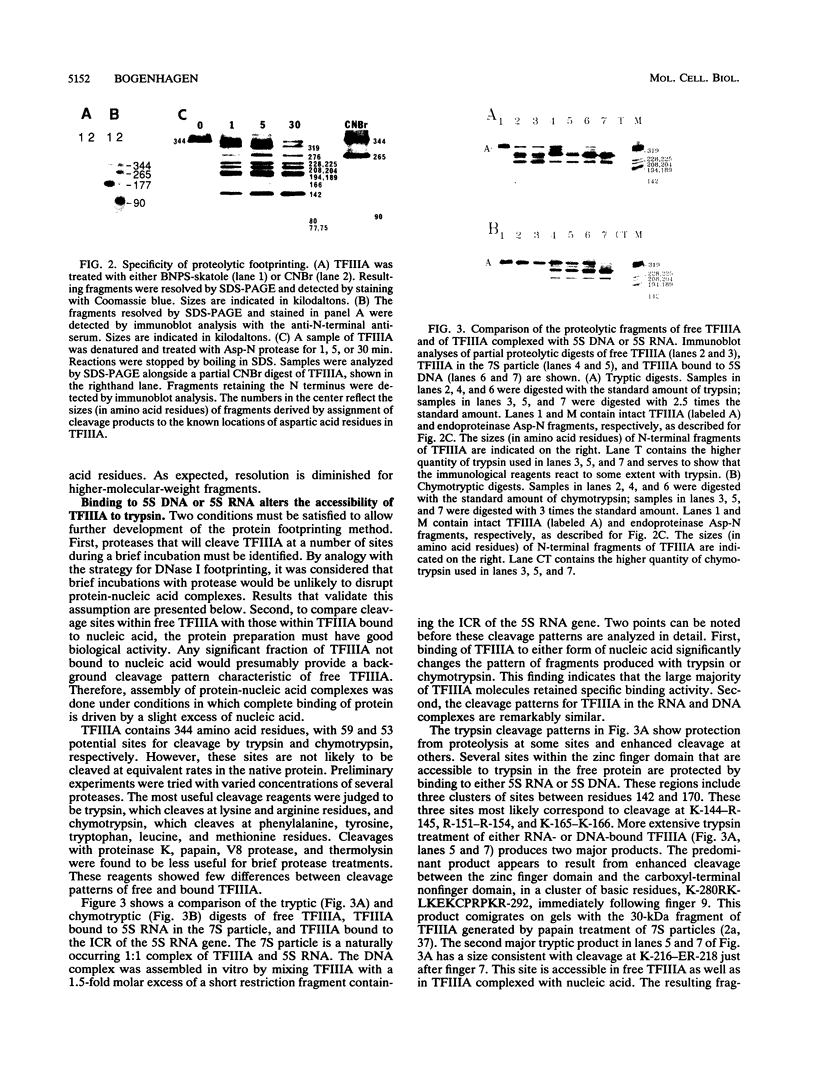






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bieker J. J., Roeder R. G. Physical properties and DNA-binding stoichiometry of a 5 S gene-specific transcription factor. J Biol Chem. 1984 May 25;259(10):6158–6164. [PubMed] [Google Scholar]
- Bogenhagen D. F., Sands M. S. Binding of TFIIIA to derivatives of 5S RNA containing sequence substitutions or deletions defines a minimal TFIIIA binding site. Nucleic Acids Res. 1992 Jun 11;20(11):2639–2645. doi: 10.1093/nar/20.11.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F. The intragenic control region of the Xenopus 5 S RNA gene contains two factor A binding domains that must be aligned properly for efficient transcription initiation. J Biol Chem. 1985 May 25;260(10):6466–6471. [PubMed] [Google Scholar]
- Christensen J. H., Hansen P. K., Lillelund O., Thøgersen H. C. Sequence-specific binding of the N-terminal three-finger fragment of Xenopus transcription factor IIIA to the internal control region of a 5S RNA gene. FEBS Lett. 1991 Apr 9;281(1-2):181–184. doi: 10.1016/0014-5793(91)80388-j. [DOI] [PubMed] [Google Scholar]
- Clemens K. R., Liao X., Wolf V., Wright P. E., Gottesfeld J. M. Definition of the binding sites of individual zinc fingers in the transcription factor IIIA-5S RNA gene complex. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10822–10826. doi: 10.1073/pnas.89.22.10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens K. R., Wolf V., McBryant S. J., Zhang P., Liao X., Wright P. E., Gottesfeld J. M. Molecular basis for specific recognition of both RNA and DNA by a zinc finger protein. Science. 1993 Apr 23;260(5107):530–533. doi: 10.1126/science.8475383. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W. Peptide mapping in one dimension by limited proteolysis of sodium dodecyl sulfate-solubilized proteins. Methods Enzymol. 1983;96:222–229. doi: 10.1016/s0076-6879(83)96020-2. [DOI] [PubMed] [Google Scholar]
- Darby M. K., Joho K. E. Differential binding of zinc fingers from Xenopus TFIIIA and p43 to 5S RNA and the 5S RNA gene. Mol Cell Biol. 1992 Jul;12(7):3155–3164. doi: 10.1128/mcb.12.7.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Río S., Setzer D. R. High yield purification of active transcription factor IIIA expressed in E. coli. Nucleic Acids Res. 1991 Nov 25;19(22):6197–6203. doi: 10.1093/nar/19.22.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairall L., Rhodes D. A new approach to the analysis of DNase I footprinting data and its application to the TFIIIA/5S DNA complex. Nucleic Acids Res. 1992 Sep 25;20(18):4727–4731. doi: 10.1093/nar/20.18.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg A. M., King B. O., Roeder R. G. Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell. 1984 Dec;39(3 Pt 2):479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- Gralla J. D. Rapid "footprinting" on supercoiled DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanas J. S., Bogenhagen D. F., Wu C. W. Cooperative model for the binding of Xenopus transcription factor A to the 5S RNA gene. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2142–2145. doi: 10.1073/pnas.80.8.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanas J. S., Duke A. L., Gaskins C. J. Conformation states of Xenopus transcription factor IIIA. Biochemistry. 1989 May 2;28(9):4083–4088. doi: 10.1021/bi00435a068. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Tullius T. D. Structure of the TFIIIA-5 S DNA complex. J Mol Biol. 1992 Sep 20;227(2):407–417. doi: 10.1016/0022-2836(92)90897-s. [DOI] [PubMed] [Google Scholar]
- Hayes J., Tullius T. D., Wolffe A. P. A protein-protein interaction is essential for stable complex formation on a 5 S RNA gene. J Biol Chem. 1989 Apr 15;264(11):6009–6012. [PubMed] [Google Scholar]
- Honda B. M., Roeder R. G. Association of a 5S gene transcription factor with 5S RNA and altered levels of the factor during cell differentiation. Cell. 1980 Nov;22(1 Pt 1):119–126. doi: 10.1016/0092-8674(80)90160-9. [DOI] [PubMed] [Google Scholar]
- Jue R. A., Doolittle R. F. Determination of the relative positions of amino acids by partial specific cleavages of end-labeled proteins. Biochemistry. 1985 Jan 1;24(1):162–170. doi: 10.1021/bi00322a023. [DOI] [PubMed] [Google Scholar]
- Klenow H., Henningsen I. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by limited proteolysis. Proc Natl Acad Sci U S A. 1970 Jan;65(1):168–175. doi: 10.1073/pnas.65.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lerner R. A., Green N., Alexander H., Liu F. T., Sutcliffe J. G., Shinnick T. M. Chemically synthesized peptides predicted from the nucleotide sequence of the hepatitis B virus genome elicit antibodies reactive with the native envelope protein of Dane particles. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3403–3407. doi: 10.1073/pnas.78.6.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. B., Clemens K. R., Tennant L., Wright P. E., Gottesfeld J. M. Specific interaction of the first three zinc fingers of TFIIIA with the internal control region of the Xenopus 5 S RNA gene. J Mol Biol. 1992 Feb 20;223(4):857–871. doi: 10.1016/0022-2836(92)90248-i. [DOI] [PubMed] [Google Scholar]
- Lindsley J. E., Wang J. C. Proteolysis patterns of epitopically labeled yeast DNA topoisomerase II suggest an allosteric transition in the enzyme induced by ATP binding. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10485–10489. doi: 10.1073/pnas.88.23.10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey G. A., Bogenhagen D. F. TFIIIA binds with equal affinity to somatic and major oocyte 5S RNA genes. Genes Dev. 1988 Feb;2(2):205–214. doi: 10.1101/gad.2.2.205. [DOI] [PubMed] [Google Scholar]
- Miller J. R., Cartwright E. M., Brownlee G. G., Fedoroff N. V., Brown D. D. The nucleotide sequence of oocyte 5S DNA in Xenopus laevis. II. The GC-rich region. Cell. 1978 Apr;13(4):717–725. doi: 10.1016/0092-8674(78)90221-0. [DOI] [PubMed] [Google Scholar]
- Munro S., Pelham H. R. Use of peptide tagging to detect proteins expressed from cloned genes: deletion mapping functional domains of Drosophila hsp 70. EMBO J. 1984 Dec 20;3(13):3087–3093. doi: 10.1002/j.1460-2075.1984.tb02263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H. R., Brown D. D. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R. C., Doering J. L., Brown D. D. Characterization of two xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980 May;20(1):131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- Porter R. R. Structural studies of immunoglobulins. Science. 1973 May 18;180(4087):713–716. doi: 10.1126/science.180.4087.713. [DOI] [PubMed] [Google Scholar]
- Romaniuk P. J., de Stevenson I. L., Ehresmann C., Romby P., Ehresmann B. A comparison of the solution structures and conformational properties of the somatic and oocyte 5S rRNAs of Xenopus laevis. Nucleic Acids Res. 1988 Mar 25;16(5):2295–2312. doi: 10.1093/nar/16.5.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands M. S., Bogenhagen D. F. The carboxyterminal zinc fingers of TFIIIA interact with the tip of helix V of 5S RNA in the 7S ribonucleoprotein particle. Nucleic Acids Res. 1991 Apr 25;19(8):1791–1796. doi: 10.1093/nar/19.8.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Z. G., Windsor W. T., Liao Y. D., Wu C. W. Purification of Xenopus transcription factor IIIA and 5 S RNA from 7 S ribonucleoprotein particle by ammonium sulfate precipitation. Anal Biochem. 1988 Jan;168(1):156–163. doi: 10.1016/0003-2697(88)90023-1. [DOI] [PubMed] [Google Scholar]
- Sheshberadaran H., Payne L. G. Protein footprinting method for studying antigen-antibody interactions and epitope mapping. Methods Enzymol. 1989;178:746–764. doi: 10.1016/0076-6879(89)78049-6. [DOI] [PubMed] [Google Scholar]
- Smith D. R., Jackson I. J., Brown D. D. Domains of the positive transcription factor specific for the Xenopus 5S RNA gene. Cell. 1984 Jun;37(2):645–652. doi: 10.1016/0092-8674(84)90396-9. [DOI] [PubMed] [Google Scholar]
- Theunissen O., Rudt F., Guddat U., Mentzel H., Pieler T. RNA and DNA binding zinc fingers in Xenopus TFIIIA. Cell. 1992 Nov 13;71(4):679–690. doi: 10.1016/0092-8674(92)90601-8. [DOI] [PubMed] [Google Scholar]
- Wakefield L., Gurdon J. B. Cytoplasmic regulation of 5S RNA genes in nuclear-transplant embryos. EMBO J. 1983;2(9):1613–1619. doi: 10.1002/j.1460-2075.1983.tb01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Romby P., Romaniuk P. J., Ebel J. P., Ehresmann C., Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol. 1989 May 20;207(2):417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]
- You Q. M., Romaniuk P. J. The effects of disrupting 5S RNA helical structures on the binding of Xenopus transcription factor IIIA. Nucleic Acids Res. 1990 Sep 11;18(17):5055–5062. doi: 10.1093/nar/18.17.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]