Abstract
Porcine reproductive and respiratory syndrome (PRRS) has devastated pig industries worldwide for many years. It is caused by a small RNA virus (PRRSV), which targets almost exclusively pig monocytes or macrophages. In the present study, five SAGE (serial analysis of gene expression) libraries derived from 0 hour mock-infected and 6, 12, 16 and 24 hours PRRSV-infected porcine alveolar macrophages (PAMs) produced a total 643,255 sequenced tags with 91,807 unique tags. Differentially expressed (DE) tags were then detected using the Bayesian framework followed by gene/mRNA assignment, arbitrary selection and manual annotation, which determined 699 DE genes for reactome analysis. The DAVID, KEGG and REACTOME databases assigned 573 of the DE genes into six biological systems, 60 functional categories and 504 pathways. The six systems are: cellular processes, genetic information processing, environmental information processing, metabolism, organismal systems and human diseases as defined by KEGG with modification. Self-organizing map (SOM) analysis further grouped these 699 DE genes into ten clusters, reflecting their expression trends along these five time points. Based on the number one functional category in each system, cell growth and death, transcription processes, signal transductions, energy metabolism, immune system and infectious diseases formed the major reactomes of PAMs responding to PRRSV infection. Our investigation also focused on dominant pathways that had at least 20 DE genes identified, multi-pathway genes that were involved in 10 or more pathways and exclusively-expressed genes that were included in one system. Overall, our present study reported a large set of DE genes, compiled a comprehensive coverage of pathways, and revealed system-based reactomes of PAMs infected with PRRSV. We believe that our reactome data provides new insight into molecular mechanisms involved in host genetic complexity of antiviral activities against PRRSV and lays a strong foundation for vaccine development to control PRRS incidence in pigs.
Introduction
Porcine reproductive and respiratory syndrome (PRRS), also known as Mystery Swine Disease, Blue Ear Disease, Porcine Endemic Abortion and Respiratory Syndrome (PEARS) and Swine Infertility Respiratory Syndrome (SIRS), was first reported in USA in 1987 and in Europe in 1990 [1], [2]. Since then, PRRS has devastated the pig industries of many countries and has become the most economically important disease in pigs worldwide. A recent study estimated that PRRS costs the pork industry $664 million per year in the United States of America (http://www.pork.org/News).
The disease is caused by a small RNA virus (PRRSV) classified in the order Nidovirales, family Arteriviridae, and genus Arterivirus. PRRSV causes severe reproductive failure of the sow, including third-trimester abortions, early farrowing with stillborns, mummies, neonatal death and weak piglets, agalactia and mastitis, and prolonged anoestrus and delayed return to estrus post-weaning. Respiratory disease is the major clinical sign in neonatal pigs and is characterized by fever, interstitial pneumonia, eyelid edema, periocular edema, blue discoloration of the ears and shaking [3], [4]. The mortality in neonatal pigs infected with PRRSV can reach 100%. In growing/finishing pigs, subclinical infection is much more common. Some PRRSV-infected boars demonstrate a loss of libido, lethargy, lowered sperm volume and decreased fertility.
PRRSV has remarkable genetic variation with two distinct genetic and antigenic groups: Type 1 (European) and Type 2 (North American), which only share 60% nucleotide identity [5]. In 2006, previously unparalleled large-scale outbreaks of highly-pathogenic PRRS, also named “Blue Ear” or “high fever” disease, occurred in China. It spread to more than 10 provinces (autonomous cities or regions) and affected over 2 million pigs with about 400,000 fatal cases [6]. Best estimates suggest that at least 50 million pigs were affected [7]. Since then, highly-pathogenic PRRS outbreaks were also reported in 2007 and 2008 in other Asian countries, such as Vietnam and the Philippines [8]. These data clearly indicate that PRRSV is able to mutate, thus causing challenges in effective vaccine development. For example, while modified live-attenuated vaccines and inactivated vaccines against PRRSV have been available for many years, none of them can prevent respiratory infection, transmission, or pig-to-pig transmission of virus. In particular, modified-live vaccines are generally effective against homologous strains but variable in success against heterologous strains, while efficacy of inactivated vaccines in the field is more limited and restricted to homologous strains [9]. In addition, PRRSV has developed diverse mechanisms to evade porcine antiviral immune responses [10]. Once the virus infects pig tissues, it has several mechanisms to evade the pig’s immune system, causing a several week delay in protective antibody production [11]–[13]. In the absence of control efforts, the virus will persist indefinitely in swine herds.
PRRSV targets almost exclusively pig monocytes or macrophages [14], [15]. The entry of PRRSV into porcine alveolar macrophages (PAMs) is proposed to include four steps [16]. First, the PRRSV virion attaches to heparan sulphate glycosaminoglycans on the macrophage surface. Second, the virus then forms a more stable binding with the sialoadhesin receptor via sialic acid residues associated with M/GP5 glycoprotein complexes present in the viral envelope. Third, following attachment to sialoadhesin, the virus–receptor complex is endocytosed via clathrin-coated vesicles. Once endocytosed, viral genome release is dependent on endosomal acidification. There appears to be involvement of CD163 with viral genome release that is possible through interactions with the viral glycoproteins, GP2 and GP4 and that is dependent upon a function CD-163 scavenger receptor cysteine rich domain 5 being present. In addition, several proteases have been implicated in this final step of PRRSV entry into macrophages. Once the genome is released into the cytoplasm of the host cell, virus transcriptional and translational events required for the formation of new virions are initiated. Here we report the reactome dynamics of PAMs in response to PRRSV infection in vitro, following serial analysis of gene expression (SAGE) [17], in order to reveal the host transcriptional events in response to virus replication and cellular resistance, thus providing new insights into molecular mechanisms involved in the cellular complexity of antiviral activities against PRRSV.
Results
Reactome of PAMs Infected with PRRSV: Snapshots
In SAGE analysis, a set of “tag” fragments (13–15 bp in size) derived from restriction positions of cDNA molecules are pooled, collected, sequenced and assigned to genes/transcripts. Five SAGE libraries constructed from the 0 hour mock-infected and 6, 12, 16 and 24 hour PRRSV-infected cells produced a total of 643,255 sequenced tags, which allowed identification of 91,807 unique tags among these five time points (Figure 1). As PAMs were infected with PRRSV, we anticipated the existence of viral mRNA tags in the cells. In fact, the virus complete genome sequence contains a total of 74 cut sites for restriction enzyme NlaIII. Using the complete genome sequence of PRRSV strain SD1-100 (GQ914997.1) as a reference, we discovered a total of 78 tentative virus tags, including 46 derived from the sense strand and 32 from the antisense strand (Table S1). The total count for all of these virus-specific tags was 0 in the 0 hour mock-infected cells, but reached 267, 11,270, 7,854 and 3,770 copies in the 6, 12, 16 and 24 hour PRRSV-infected cell libraries, respectively. The most abundantly expressed tag was the 3′-most cut site (CGGCCGAAAT) (Table S1), having 225 (84.27% of 267), 9,500 (84.29% of 11270), 6,902 (87.88% of 7,854) and 3,622 (96.07% of 3,770) copies sequenced in PAMs infected with PRRSV for 6, 12, 16 and 24 hours. Virus tags accounted for 9.16% of total tags (9,500/103,662 tags) at 12 hours post infection; therefore, we deleted all virus tags from each library and re-calculated the number of tags per million (TPM) for each host gene tag.
Figure 1. Identification and characterization of tags/genes differentially expressed between the 0 hour mock-infected and the 6, 12, 16 and 24 hours PRRSV-infected PAM cells.
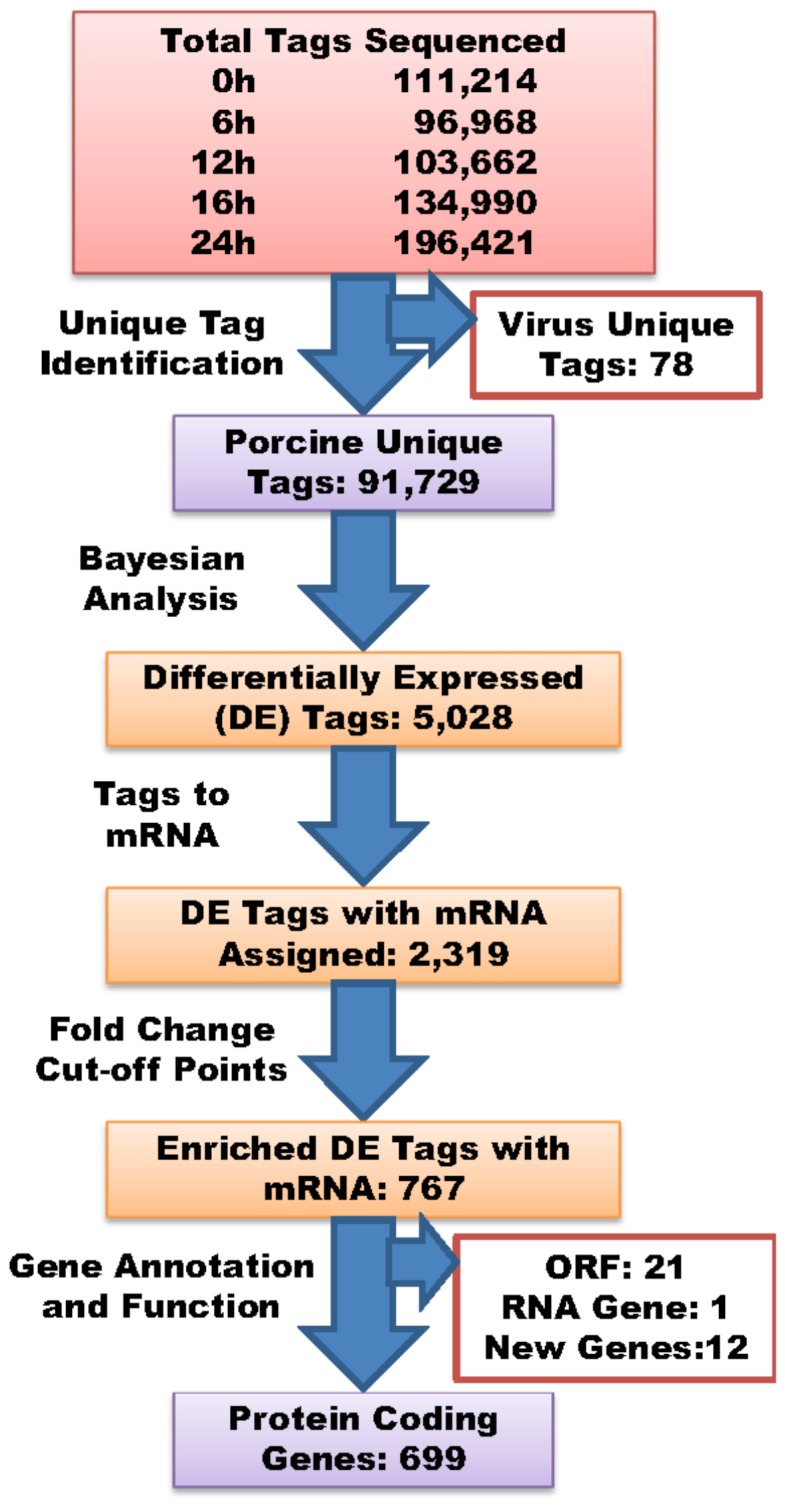
Compared to the 0 hour mock-infected cells, Bayesian analysis revealed that PRRSV-infected cells had 891, 972, 1,230 and 1,323 down- and 1,201, 1,199, 1,276 and 1,042 up-regulated DE tags at 6, 12, 16 and 24 hours post infection, respectively. These up- and down-regulated DE tags at all four time points post infection in fact represented only 5,028 tags, and included 2,716 DE tags at one, 1,066 at two, 697 at three and 549 at four of four time points, respectively (Table S2). Among them, only 2,319 tags had unique mRNAs and/or genes assigned (Figure 1). After the aforementioned cut-off points for each DE gene were employed 767 tags with mRNA and/or genes assigned remained for further analysis (Figure 1).
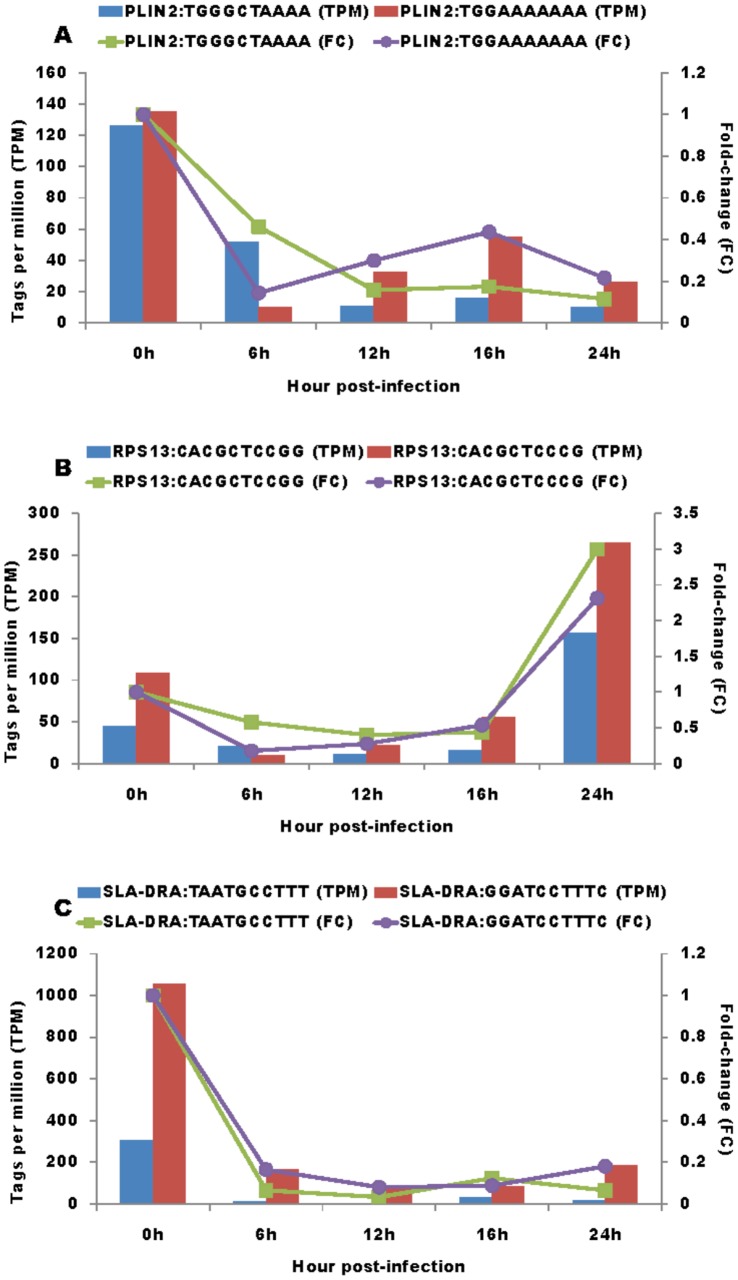
Manual annotation of these 767 tags with mRNA sequences revealed that they represented a total of 733 genes, and included 700 genes with one tag collected from one unique mRNA sequence, 32 genes with two tags collected from two different mRNA sequences and one gene with three tags collected from three different mRNA sequences, respectively. For those genes that had two or three tags, we further determined whether they represented the true 3′-most tags or not. Interestingly, true cases were confirmed for two tags in ARG1 (arginase, liver), SLA-DQA1 (MHC class II, DQ alpha 1), TIMP2 (TIMP metallopeptidase inhibitor 2) and TOB1 (transducer of ERBB2, 1) genes (Figure 2A–D) due to different mRNA isoforms and in PLIN2 (perilipin 2), RPS13 (ribosomal protein S13) and SLA-DRA (MHC class II, DR-alpha) genes (Figure 3A–C) due to nucleotide polymorphisms. Although TPM were variable, trends in fold changes were similar between the two isoforms or two alleles of each gene.
Figure 2. Fold change in TPM for genes with multiple tags due to mRNA isoforms.
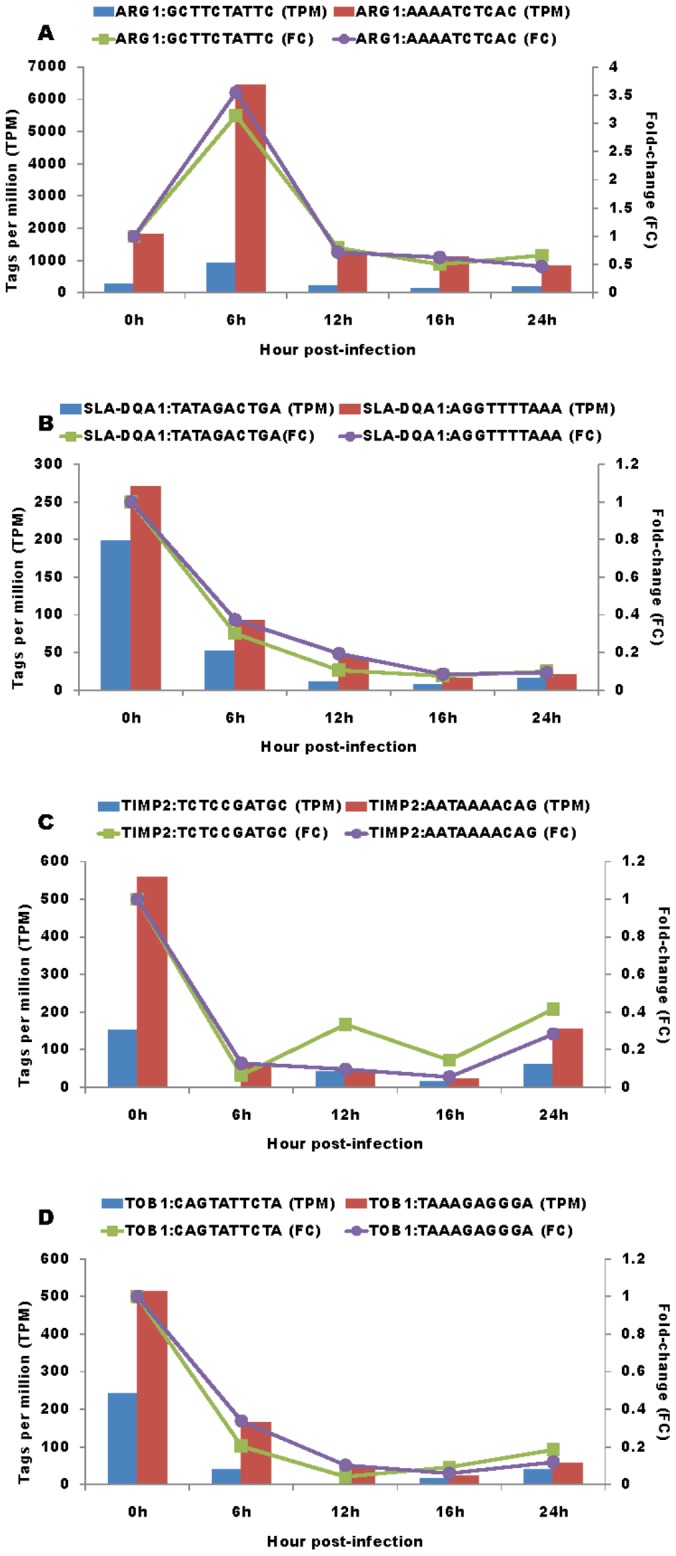
TPM and fold changes for two tags in ARG1 (A), SLA-DQA1 (B), TIMP2 (C) and TOB1 (D) representing different mRNA isoforms at 0, 6, 12, 16 and 24 hours post-infection.
Figure 3. Fold change in TPM for genes with multiple tags due to nucleotide polymorphisms.
TPM and fold changes for two tags in PLIN2 (A), RPS13 (B) and SLA-DRA (C) genes representing different alleles at 0, 6, 12, 16 and 24 hours post-infection.
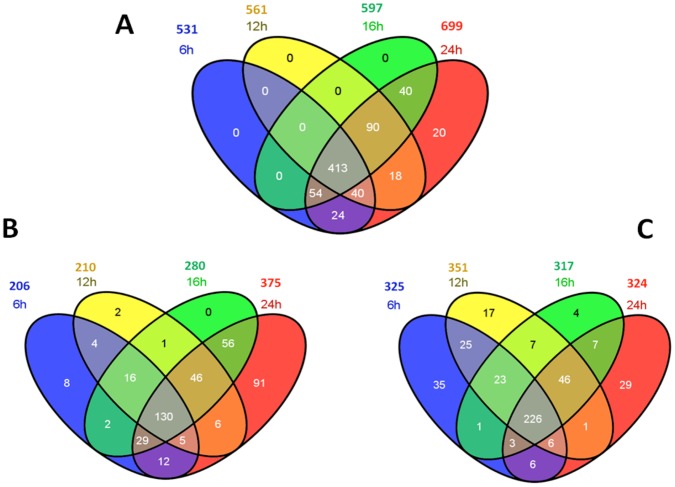
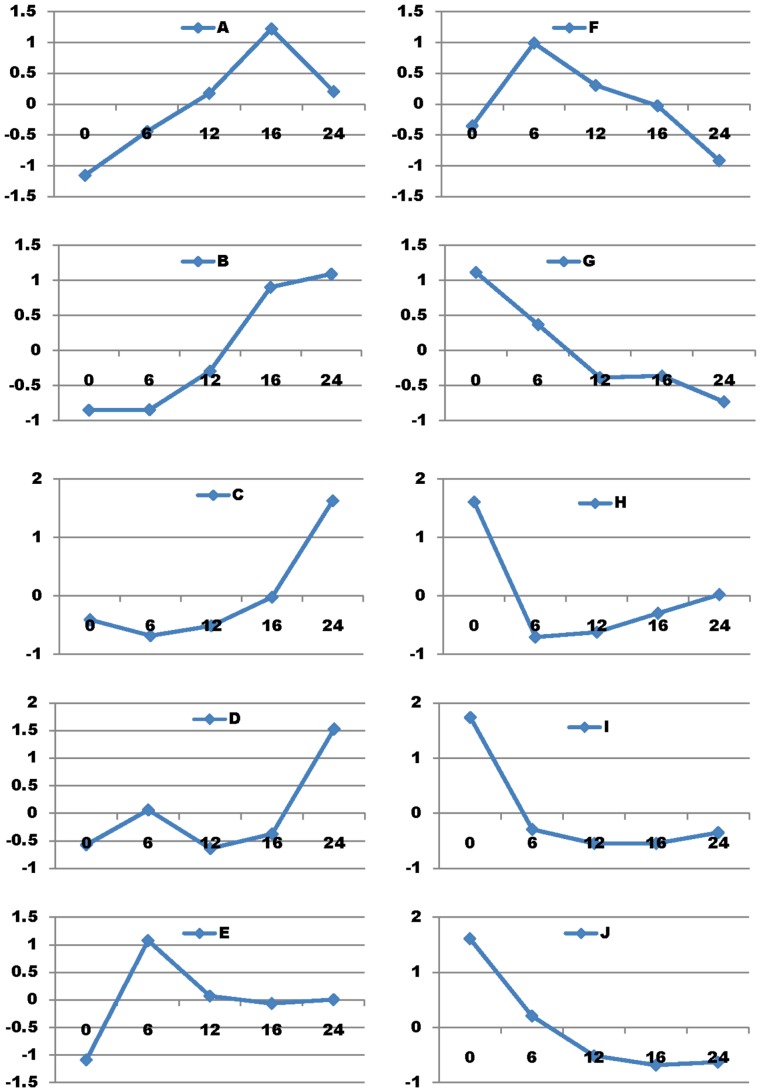
Of these 733 pig genes (Figure 1), 699 also had orthologs identified as protein coding genes, while 21 were open reading frame genes (functionally unknown), and one was a non-coding RNA mitochondrial gene in humans. The remaining 12 genes were pig species-specific, including 11 novel genes and a porcine endogenous retrovirus PERV-MSL gene. Except for one novel pig gene (AK351197.1) that was missing both the genomic DNA sequence and location, the rest of the 10 novel genes all had complete genomic DNA sequences with clones mapped to Sus scrofa chromosomes (SSCs) 2, 3, 5, 7, 9, 10, 12 and 13, respectively. Compared to the 0 hour mock-infected cells, PRRSV infection induced differential expression of 531, 561, 597, 699 genes (Figure 4A) at 6, 12, 16 and 24 hours post infection, including 206, 210, 280 and 375 genes that were up-regulated (Figure 4B) and 325, 351, 317 and 324 genes that were down-regulated (Figure 4C), respectively at these four time points. Overall, among these 699 DE genes, 226 (63.5%) and 130 (36.5%) were consistently down- or up-regulated, respectively at all four infected time points. Self-organizing map (SOM) method of analysis assigned these 699 DE genes to 10 clusters (Figure 5) based on their expression trends regardless of fold-change magnitudes along these five time points (0 h, 6 h, 12 h, 16 h and 24 h) (Table S3). However, only 573 genes were assigned to pathways, specifically 72 (12.56%) in cluster A, 37 (6.46%) in B, 121 (21.12%) in C, 39 (6.81%) in D, 30 (5.24%) in E, 29 (5.06%) in F, 27 (4.71%) in G, 71 (12.39%) in H, 93 (16.23%) in I and 54 (9.42%) in J.
Figure 4. Summary of differentially expressed genes in PAMs infected with PRRSV.
All genes (A), up-regulated genes (B) and down-regulated genes at four time-points post-infection (C).
Figure 5. Ten expression trend clusters of 699 DE genes derived from PAMs during PRRSV infection.
Reactome of PAMs Infected with PRRSV: Cellular Processes
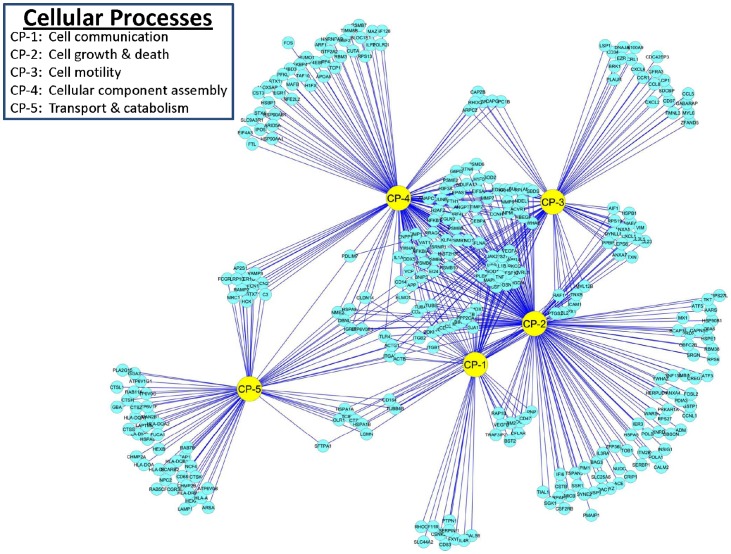
The GO, KEGG and REACTOME databases identified 329 DE genes that were involved in cellular processes of PAMs infected with PRRSV (Figure 6). Specific functions included: 1) cell communication, 2) cell growth and death, 3) cell motility, 4) cell organization and biogenesis, and 5) transport and catabolism. Many genes in the system functioned in two or more sub-category pathways; however, there were smaller clusters of genes that contributed to only one cellular process. The largest number of DE genes (191) were broadly involved in cell growth and death and were specifically linked to pathways associated with cell cycle, division, proliferation, growth, cell size regulation, apoptosis, anti-apoptosis, induction and regulation of apoptosis, and regulation of endothelial, fibroblast and smooth muscle cell proliferation. PAMs infected with PRRSV had 153 DE genes that were involved in pathways related to cell organization and biogenesis, which were most notably associated with extracellular matrix organization, macromolecular complex assembly, membrane organization and protein complex assembly, macromolecular/protein complex assembly or disassembly, cellular component biogenesis, organization and size, and macromolecule metabolic/biosynthetic processes. There were 88 DE genes in PRRSV-infected PAMs that are important for cell motility and contributed to pathways related to cell migration, motility, motion and shape; actin cytoskeleton and filament organization; and chemotaxis. Seventy-seven genes important for cellular transport and catabolism were DE in PRRSV-infected PAMs. Most of these DE genes were associated with pathways involved in autophagocytosis, including endocytosis, and lysosomal and phagosomal processes. Cell communication in PAMs infected with PRRSV appears to be quite important because there were 65 DE genes involved in pathways related to cell adhesion, cell junction, cell activation, and cell-cell communication pathways. The 329 DE genes related to cellular process networks of PAMs infected with PRRSV are shown in Figure 6 and are summarized in Table 1.
Figure 6. DE gene distributions and interactions among functional categories associated with Cellular Processes in PAMs infected with PRRSV.
Table 1. Pathway summary of DE genes that are related to six biological systems of PAMs infected with PRRSV.
| Total | Down | Down% | Up | Up% | ||
| Cellular Process | ||||||
| Cell Communication | Adhesion - Focal adhesion | 14 | 9 | 64 | 5 | 36 |
| Cell Communication | Adhesion - heterophilic cell adhesion | 4 | 3 | 75 | 1 | 25 |
| Cell Communication | Adhesion - negative regulation of cell adhesion | 5 | 1 | 20 | 4 | 80 |
| Cell Communication | Adhesion - positive regulation of cell adhesion | 3 | 2 | 67 | 1 | 33 |
| Cell Communication | Adhesion - regulation of cell adhesion | 7 | 5 | 71 | 2 | 29 |
| Cell Communication | cell activation - positive regulation of cell activation | 13 | 11 | 85 | 2 | 15 |
| Cell Communication | cell activation - regulation of cell activation | 4 | 1 | 25 | 3 | 75 |
| Cell Communication | Communication - Cell-Cell communication | 8 | 5 | 63 | 3 | 38 |
| Cell Communication | Communication - positive regulation of cell communication | 24 | 16 | 67 | 8 | 33 |
| Cell Communication | Junction - Adherens junction | 5 | 4 | 80 | 1 | 20 |
| Cell Communication | Junction - Cell junction organization | 6 | 5 | 83 | 1 | 17 |
| Cell Communication | Junction - Gap junction | 6 | 4 | 67 | 2 | 33 |
| Cell Communication | Junction - Gap junction trafficking and regulation | 5 | 5 | 100 | 0 | 0 |
| Cell Communication | Junction - Tight junction | 9 | 6 | 67 | 3 | 33 |
| Cell Growth and Death | Apoptosis - anti-apoptosis | 34 | 23 | 68 | 11 | 32 |
| Cell Growth and Death | apoptosis - anti-apoptosis: positive regulation | 5 | 4 | 80 | 1 | 20 |
| Cell Growth and Death | apoptosis - anti-apoptosis: regulation of anti-apoptosis | 2 | 2 | 100 | 0 | 0 |
| Cell Growth and Death | Apoptosis - Apoptotic execution phase | 7 | 2 | 29 | 5 | 71 |
| Cell Growth and Death | Apoptosis - apoptotic mitochondrial changes | 7 | 3 | 43 | 4 | 57 |
| Cell Growth and Death | Apoptosis - apoptotic nuclear changes | 4 | 0 | 0 | 4 | 100 |
| Cell Growth and Death | Apoptosis - negative regulation of apoptosis | 36 | 20 | 56 | 16 | 44 |
| Cell Growth and Death | apoptosis - positive regulation of apoptosis | 28 | 14 | 50 | 14 | 50 |
| Cell Growth and Death | apoptosis - regulation of apoptosis | 23 | 10 | 43 | 13 | 57 |
| Cell Growth and Death | apoptosis - regulation of neuron apoptosis | 8 | 4 | 50 | 4 | 50 |
| Cell Growth and Death | apoptosis and induction of apoptosis | 81 | 39 | 48 | 42 | 52 |
| Cell Growth and Death | Cell cycle - Cell cycle | 28 | 17 | 61 | 11 | 39 |
| Cell Growth and Death | Cell cycle regulation - positive regulation of cell cycle | 7 | 5 | 71 | 2 | 29 |
| Cell Growth and Death | Cell cycle regulation - Regulation of cell cycle | 25 | 12 | 48 | 13 | 52 |
| Cell Growth and Death | Cell cycle, division and proliferation - Meiosis | 12 | 9 | 75 | 3 | 25 |
| Cell Growth and Death | Cell division - positive regulation of cell division | 6 | 2 | 33 | 4 | 67 |
| Cell Growth and Death | Cell division - regulation of cell division | 1 | 1 | 100 | 0 | 0 |
| Cell Growth and Death | cell growth - negative regulation of cell growth | 9 | 4 | 44 | 5 | 56 |
| Cell Growth and Death | cell growth - regulation of cell growth | 13 | 5 | 38 | 8 | 62 |
| Cell Growth and Death | Cell proliferation - cell proliferation | 31 | 17 | 55 | 14 | 45 |
| Cell Growth and Death | Cell proliferation - homeostasis of number of cells | 10 | 3 | 30 | 7 | 70 |
| Cell Growth and Death | Cell proliferation - negative regulation of cell proliferation | 50 | 32 | 64 | 18 | 36 |
| Cell Growth and Death | Cell proliferation - regulation of cell proliferation | 5 | 3 | 60 | 2 | 40 |
| Cell Growth and Death | cell size - regulation of cell size | 18 | 9 | 50 | 9 | 50 |
| Cell Growth and Death | endothelial cell - positive regulation of proliferation | 4 | 2 | 50 | 2 | 50 |
| Cell Growth and Death | fibroblast proliferation - positive regulation | 4 | 2 | 50 | 2 | 50 |
| Cell Growth and Death | fibroblast proliferation - regulation of fibroblast proliferation | 1 | 0 | 0 | 1 | 100 |
| Cell growth and Death | smooth muscle cell - positive regulation of proliferation | 6 | 5 | 83 | 1 | 17 |
| Cell Growth and Death | smooth muscle cell - regulation of proliferation | 2 | 2 | 100 | 0 | 0 |
| Cell Motility | cell migration - positive regulation of cell migration | 8 | 5 | 63 | 3 | 38 |
| Cell Motility | cell migration and motility | 24 | 15 | 63 | 9 | 38 |
| Cell Motility | cell motion | 47 | 28 | 60 | 19 | 40 |
| Cell Motility | cell motion - positive regulation of cell motion | 9 | 5 | 56 | 4 | 44 |
| Cell Motility | cell shape - regulation of cell shape | 5 | 3 | 60 | 2 | 40 |
| Cell Motility | chemotaxis | 16 | 8 | 50 | 8 | 50 |
| Cell Motility | cytoskeleton - actin cytoskeleton organization | 30 | 19 | 63 | 11 | 37 |
| Cell Motility | cytoskeleton - Regulation of actin cytoskeleton | 22 | 13 | 59 | 9 | 41 |
| Cell Motility | filamen - regulation of actin filament depolymerization | 5 | 3 | 60 | 2 | 40 |
| Cell Motility | filament - actin filament organization | 7 | 6 | 86 | 1 | 14 |
| Cell Motility | filament - actin filament-based process | 23 | 16 | 70 | 7 | 30 |
| Cell Motility | filament - regulation of actin filament length | 7 | 3 | 43 | 4 | 57 |
| cell organization and biogenesis | component size - regulation of cellular component size | 24 | 11 | 46 | 13 | 54 |
| cell organization and biogenesis | macromolecular complex assembly | 37 | 19 | 51 | 18 | 49 |
| cell organization and biogenesis | macromolecule - negative regulation of macromolecule biosynthetic/metabolic process | 24 | 15 | 63 | 9 | 38 |
| cell organization and biogenesis | macromolecule - positive regulation of macromolecule biosynthetic/metabolic process | 36 | 19 | 53 | 17 | 47 |
| cell organization and biogenesis | membrane organization | 31 | 10 | 32 | 21 | 68 |
| cell organization and biogenesis | protein complex assembly | 31 | 16 | 52 | 15 | 48 |
| cell organization and biogenesis | macromolecule - regulation of macromolecule biosynthetic/metabolic process | 19 | 9 | 47 | 10 | 53 |
| cell organization and biogenesis | component organization - positive regulation of cellular component organization | 18 | 12 | 67 | 6 | 33 |
| cell organization and biogenesis | component biogenesis - regulation of cellular component biogenesis | 16 | 6 | 38 | 10 | 63 |
| cell organization and biogenesis | component organization - negative regulation of cellular component organization | 13 | 6 | 46 | 7 | 54 |
| cell organization and biogenesis | organelle organization - positive regulation of organelle organization | 10 | 9 | 90 | 1 | 10 |
| cell organization and biogenesis | protein complex - regulation of protein complex assembly | 9 | 2 | 22 | 7 | 78 |
| cell organization and biogenesis | organelle organization - regulation of organelle organization | 8 | 4 | 50 | 4 | 50 |
| cell organization and biogenesis | protein complex - regulation of protein complex disassembly | 7 | 4 | 57 | 3 | 43 |
| cell organization and biogenesis | Extracellular matrix organization | 5 | 3 | 60 | 2 | 40 |
| Transport and Catabolism | endocytosis | 26 | 12 | 46 | 14 | 54 |
| Transport and Catabolism | Lysosome | 25 | 8 | 32 | 17 | 68 |
| Transport and Catabolism | Phagosome | 38 | 21 | 55 | 17 | 45 |
| 1160 | 638 | 522 | ||||
| Genetic Information Processing | ||||||
| Folding, Sorting and Degradation | Degradation of the extracellular matrix | 5 | 3 | 60 | 2 | 40 |
| Folding, Sorting and Degradation | endopeptidase - regulation of endopeptidase activity | 13 | 9 | 69 | 4 | 31 |
| Folding, Sorting and Degradation | glycosylation - Asparagine N-linked glycosylation | 5 | 3 | 60 | 2 | 40 |
| Folding, Sorting and Degradation | nucleocytoplasmic transport | 14 | 9 | 64 | 5 | 36 |
| Folding, Sorting and Degradation | nucleocytoplasmic transport - positive regulation of nucleocytoplasmic transport | 4 | 4 | 100 | 0 | 0 |
| Folding, Sorting and Degradation | nucleocytoplasmic transport - regulation of nucleocytoplasmic transport | 3 | 2 | 67 | 1 | 33 |
| Folding, Sorting and Degradation | post-Golgi vesicle-mediated transport | 7 | 1 | 14 | 6 | 86 |
| Folding, Sorting and Degradation | proteasomal ubiquitin-dependent protein catabolic process | 18 | 6 | 33 | 12 | 67 |
| Folding, Sorting and Degradation | Protein folding | 23 | 18 | 78 | 5 | 22 |
| Folding, Sorting and Degradation | protein import - regulation of protein import into nucleus | 6 | 5 | 83 | 1 | 17 |
| Folding, Sorting and Degradation | protein import into nucleus | 13 | 9 | 69 | 4 | 31 |
| Folding, Sorting and Degradation | protein localization | 52 | 25 | 48 | 27 | 52 |
| Folding, Sorting and Degradation | protein localization - regulation of protein localization | 16 | 10 | 63 | 6 | 38 |
| Folding, Sorting and Degradation | protein localization in organelle | 13 | 8 | 62 | 5 | 38 |
| Folding, Sorting and Degradation | Protein processing in endoplasmic reticulum | 24 | 18 | 75 | 6 | 25 |
| Folding, Sorting and Degradation | protein targeting | 18 | 9 | 50 | 9 | 50 |
| Folding, Sorting and Degradation | protein transport - intracellular protein transport | 68 | 33 | 49 | 35 | 51 |
| Folding, Sorting and Degradation | protein transport - negative regulation of intracellular Protein transport | 13 | 9 | 69 | 4 | 31 |
| Folding, Sorting and Degradation | Protein transport - regulation of intracellular protein transport | 12 | 9 | 75 | 3 | 25 |
| Folding, Sorting and Degradation | protein ubiquitination - positive regulation of protein ubiquitination | 11 | 2 | 18 | 9 | 82 |
| Folding, Sorting and Degradation | SNARE interactions in vesicular transport | 6 | 1 | 17 | 5 | 83 |
| Replication and Repair | DNA repair | 6 | 3 | 50 | 3 | 50 |
| Replication and Repair | DNA replication | 17 | 10 | 59 | 7 | 41 |
| Replication and Repair | DNA replication - Regulation of DNA replication | 11 | 4 | 36 | 7 | 64 |
| Transcription | DNA binding - negative regulation of DNA binding | 6 | 3 | 50 | 3 | 50 |
| Transcription | DNA binding - positive regulation of DNA binding | 9 | 7 | 78 | 2 | 22 |
| Transcription | DNA binding - regulation of DNA binding | 2 | 2 | 100 | 0 | 0 |
| Transcription | Gene Expression | 74 | 25 | 34 | 49 | 66 |
| Transcription | gene expression - positive regulation of gene expression | 33 | 19 | 58 | 14 | 42 |
| Transcription | gene expression - posttranscriptional regulation of gene expression | 24 | 15 | 63 | 9 | 38 |
| Transcription | mRNA stability | 10 | 6 | 60 | 4 | 40 |
| Transcription | mRNA stability - regulation of mRNA stability | 7 | 5 | 71 | 2 | 29 |
| Transcription | mRNA Stability - Regulation of mRNA Stability by Proteins that Bind AU-rich Elements | 17 | 10 | 59 | 7 | 41 |
| Transcription | NF-kappaB - positive regulation of I-kappaB kinase/NF-kappaB cascade | 14 | 11 | 79 | 3 | 21 |
| Transcription | NF-kappaB - positive regulation of NF-kappaB transcription factor activity | 6 | 6 | 100 | 0 | 0 |
| Transcription | NF-kappaB - regulation of NF-kappaB import into nucleus | 4 | 3 | 75 | 1 | 25 |
| Transcription | Nonsense-Mediated Decay | 33 | 5 | 15 | 28 | 85 |
| Transcription | Processing of Capped Intron-Containing Pre-mRNA | 10 | 7 | 70 | 3 | 30 |
| Transcription | RNA biosynthetic process | 18 | 8 | 44 | 10 | 56 |
| Transcription | RNA Polymerases I, II and III Transcription | 7 | 2 | 29 | 5 | 71 |
| Transcription | Spliceosome | 11 | 11 | 100 | 0 | 0 |
| Transcription | transcription factor - positive regulation of transcription factor activity | 8 | 7 | 88 | 1 | 13 |
| Transcription | transcription factor - regulation of transcription factor import into nucleus | 5 | 4 | 80 | 1 | 20 |
| Transcription | transcription factor - regulation of transcription factor activity | 5 | 3 | 60 | 2 | 40 |
| Translation | Post-translational protein modification | 8 | 5 | 63 | 3 | 38 |
| Translation | ribosomal small subunit biogenesis | 5 | 1 | 20 | 4 | 80 |
| Translation | Ribosome | 35 | 4 | 11 | 31 | 89 |
| Translation | ribosome biogenesis | 10 | 3 | 30 | 7 | 70 |
| Translation | RNA transport | 9 | 4 | 44 | 5 | 56 |
| Translation | SRP-dependent cotranslational protein targeting to membrane | 33 | 3 | 9 | 30 | 91 |
| Translation | translation | 53 | 12 | 23 | 41 | 77 |
| Translation | translation - positive regulation of translation | 4 | 1 | 25 | 3 | 75 |
| Translation | translation - regulation of translation | 11 | 7 | 64 | 4 | 36 |
| Translation | translation elongation | 37 | 6 | 16 | 31 | 84 |
| Translation | Translation Initiation | 35 | 5 | 14 | 30 | 86 |
| Translation | translation initiation - regulation of translational initiation | 5 | 4 | 80 | 1 | 20 |
| Translation | Translation Termination | 31 | 3 | 10 | 28 | 90 |
| 957 | 427 | 45 | 530 | 55 | ||
| Environmental Information Processing | ||||||
| Membrane Transport | Golgi vesicle transport | 14 | 3 | 21 | 11 | 79 |
| Membrane Transport | membrane docking | 6 | 3 | 50 | 3 | 50 |
| Membrane Transport | Membrane Trafficking | 16 | 9 | 56 | 7 | 44 |
| Membrane Transport | secretion - negative regulation of secretion | 6 | 4 | 67 | 2 | 33 |
| Membrane Transport | transport - Aquaporin-mediated transport | 3 | 0 | 0 | 3 | 100 |
| Membrane Transport | transport - SLC-mediated transmembrane transport | 7 | 3 | 43 | 4 | 57 |
| Membrane Transport | transport - Transmembrane transport of small molecules | 22 | 7 | 32 | 15 | 68 |
| Signal Transduction | signal transduction - positive regulation of signal transduction | 20 | 13 | 65 | 7 | 35 |
| Signal Transduction | signal transduction - Ras protein signal transduction | 10 | 3 | 30 | 7 | 70 |
| Signal Transduction | signal transduction - small GTPase mediated signal transduction | 21 | 10 | 48 | 11 | 52 |
| Signal Transduction | signaling - Calcium signaling pathway | 7 | 3 | 43 | 4 | 57 |
| Signal Transduction | signaling - cytokine-mediated signaling pathway | 7 | 5 | 71 | 2 | 29 |
| Signal Transduction | Signaling - ER-nuclear signaling pathway | 7 | 4 | 57 | 3 | 43 |
| Signal Transduction | signaling - platelet-derived growth factor receptor signaling pathway | 4 | 3 | 75 | 1 | 25 |
| Signal Transduction | Signaling by EGFR | 8 | 5 | 63 | 3 | 38 |
| Signal Transduction | Signaling by ErbB | 12 | 9 | 75 | 3 | 25 |
| Signal Transduction | Signaling by FGFR | 8 | 6 | 75 | 2 | 25 |
| Signal Transduction | Signaling by GPCR | 30 | 20 | 67 | 10 | 33 |
| Signal Transduction | Signaling by Jak-STAT | 6 | 3 | 50 | 3 | 50 |
| Signal Transduction | Signaling by MAPK | 26 | 22 | 85 | 4 | 15 |
| Signal Transduction | Signaling by mTOR | 5 | 0 | 0 | 5 | 100 |
| Signal Transduction | Signaling by NGF | 20 | 14 | 70 | 6 | 30 |
| Signal Transduction | Signaling by PDGF | 4 | 2 | 50 | 2 | 50 |
| Signal Transduction | Signaling by SCF-KIT | 5 | 3 | 60 | 2 | 40 |
| Signal Transduction | Signaling by VEGF | 9 | 4 | 44 | 5 | 56 |
| Signal Transduction | Signaling by Wnt | 14 | 7 | 50 | 7 | 50 |
| Signaling Molecules and Interaction | Cell adhesion molecules (CAMs) | 15 | 11 | 73 | 4 | 27 |
| Signaling Molecules and Interaction | cytokine biosynthetic - positive regulation of cytokine biosynthetic process | 7 | 7 | 100 | 0 | 0 |
| Signaling Molecules and Interaction | cytokine biosynthetic - regulation of cytokine biosynthetic process | 1 | 1 | 100 | 0 | 0 |
| Signaling Molecules and Interaction | cytokine production - negative regulation of cytokine production | 3 | 2 | 67 | 1 | 33 |
| Signaling Molecules and Interaction | cytokine production - positive regulation of cytokine production | 7 | 4 | 57 | 3 | 43 |
| Signaling Molecules and Interaction | cytokine production - regulation of cytokine production | 7 | 5 | 71 | 2 | 29 |
| Signaling Molecules and Interaction | Cytokine-cytokine receptor interaction | 23 | 12 | 52 | 11 | 48 |
| Signaling Molecules and Interaction | ECM-receptor interaction | 5 | 2 | 40 | 3 | 60 |
| Signaling Molecules and Interaction | GPCR ligand binding | 21 | 14 | 67 | 7 | 33 |
| 386 | 223 | 58 | 163 | 42 | ||
| Metabolism | ||||||
| amide metabolism | cellular amide metabolic process | 12 | 6 | 50 | 6 | 50 |
| Amino Acid Metabolism | Arginine and proline metabolism | 5 | 4 | 80 | 1 | 20 |
| Amino Acid Metabolism | Glutathione metabolism | 5 | 2 | 40 | 3 | 60 |
| Amino Acid Metabolism | Metabolism of amino acids and derivatives | 12 | 3 | 25 | 9 | 75 |
| Biosynthesis of Other Secondary Metabolites | secondary metabolic process | 12 | 5 | 42 | 7 | 58 |
| Carbohydrate Metabolism | alcohol biosynthetic process | 10 | 5 | 50 | 5 | 50 |
| Carbohydrate Metabolism | Amino sugar and nucleotide sugar metabolism | 7 | 4 | 57 | 3 | 43 |
| Carbohydrate Metabolism | carbohydrate biosynthetic process | 11 | 5 | 45 | 6 | 55 |
| Carbohydrate Metabolism | carbohydrate catabolic process | 17 | 4 | 24 | 13 | 76 |
| Carbohydrate Metabolism | catabolic process - negative regulation of catabolic process | 4 | 4 | 100 | 0 | 0 |
| Carbohydrate Metabolism | catabolic process - positive regulation of catabolic process | 4 | 3 | 75 | 1 | 25 |
| Carbohydrate Metabolism | catabolic process - regulation of catabolic process | 3 | 2 | 67 | 1 | 33 |
| Carbohydrate Metabolism | gluconeogenesis | 7 | 4 | 57 | 3 | 43 |
| Carbohydrate Metabolism | glucose import - regulation of glucose import | 5 | 3 | 60 | 2 | 40 |
| Carbohydrate Metabolism | glucose metabolic process | 21 | 6 | 29 | 15 | 71 |
| Carbohydrate Metabolism | glucose transport - negative regulation of glucose transport | 4 | 4 | 100 | 0 | 0 |
| Carbohydrate Metabolism | glucose transport - regulation of glucose transport | 2 | 0 | 0 | 2 | 100 |
| Carbohydrate Metabolism | glutathione metabolic process | 5 | 1 | 20 | 4 | 80 |
| Carbohydrate Metabolism | Glycolysis/Gluconeogenesis | 12 | 2 | 17 | 10 | 83 |
| Carbohydrate Metabolism | Pentose phosphate pathway | 7 | 1 | 14 | 6 | 86 |
| Carbohydrate Metabolism | pentose-phosphate shunt | 4 | 0 | 0 | 4 | 100 |
| Carbohydrate Metabolism | pyruvate metabolic process | 10 | 5 | 50 | 5 | 50 |
| Energy Metabolism | ATP biosynthetic process | 13 | 4 | 31 | 9 | 69 |
| Energy Metabolism | Biological oxidations | 9 | 5 | 56 | 4 | 44 |
| Energy Metabolism | cell redox homeostasis | 14 | 6 | 43 | 8 | 57 |
| Energy Metabolism | cellular respiration | 14 | 2 | 14 | 12 | 86 |
| Energy Metabolism | electron transport chain | 18 | 4 | 22 | 14 | 78 |
| Energy Metabolism | energy coupled proton transport, down electrochemical gradient | 9 | 1 | 11 | 8 | 89 |
| Energy Metabolism | energy derivation by oxidation of organic compounds | 15 | 2 | 13 | 13 | 87 |
| Energy Metabolism | generation of precursor metabolites and energy | 45 | 8 | 18 | 37 | 82 |
| Energy Metabolism | Integration of energy metabolism | 8 | 3 | 38 | 5 | 63 |
| Energy metabolism | mitochondrial ATP synthesis coupled electron transport | 10 | 0 | 0 | 10 | 100 |
| Energy metabolism | mitochondrial electron transport, NADH to ubiquinone | 6 | 0 | 0 | 6 | 100 |
| Energy Metabolism | Mitochondrial Protein Import | 5 | 0 | 0 | 5 | 100 |
| Energy metabolism | mitochondrial transport | 7 | 4 | 57 | 3 | 43 |
| Energy metabolism | mitochondrion organization | 13 | 6 | 46 | 7 | 54 |
| Energy metabolism | monooxygenase - regulation of monooxygenase activity | 4 | 4 | 100 | 0 | 0 |
| Energy metabolism | NAD metabolic process | 5 | 3 | 60 | 2 | 40 |
| Energy Metabolism | nitrogen compound - positive regulation of nitrogen compound metabolic process | 40 | 25 | 63 | 15 | 38 |
| Energy Metabolism | nitrogen compound biosynthetic process | 21 | 10 | 48 | 11 | 52 |
| Energy Metabolism | oxidation reduction | 48 | 13 | 27 | 35 | 73 |
| Energy Metabolism | Oxidative phosphorylation | 59 | 19 | 32 | 40 | 68 |
| Energy metabolism | oxidoreductase - regulation of oxidoreductase activity | 5 | 5 | 100 | 0 | 0 |
| Energy Metabolism | oxygen and reactive oxygen species metabolic process | 8 | 0 | 0 | 8 | 100 |
| Energy metabolism | proton transport | 12 | 2 | 17 | 10 | 83 |
| Energy metabolism | release of cytochrome c from mitochondria | 5 | 2 | 40 | 3 | 60 |
| Energy metabolism | respiratory electron transport chain | 12 | 1 | 8 | 11 | 92 |
| Energy Metabolism | Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. | 23 | 6 | 26 | 17 | 74 |
| Energy metabolism | respiratory gaseous exchange | 6 | 4 | 67 | 2 | 33 |
| Energy Metabolism | The citric acid (TCA) cycle and respiratory electron transport | 27 | 7 | 26 | 20 | 74 |
| Energy Metabolism | Transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds | 4 | 0 | 0 | 4 | 100 |
| Glycan Biosynthesis and Metabolism | hexose metabolic process | 24 | 8 | 33 | 16 | 67 |
| Glycan Biosynthesis and Metabolism | monosaccharide biosynthetic process | 9 | 5 | 56 | 4 | 44 |
| Glycan Biosynthesis and Metabolism | monosaccharide metabolic process | 28 | 11 | 39 | 17 | 61 |
| Glycan Biosynthesis and Metabolism | Other glycan degradation | 5 | 5 | 100 | 0 | 0 |
| Homeostasis | catalytic activity - negative regulation of catalytic activity | 27 | 15 | 56 | 12 | 44 |
| Homeostasis | catalytic activity - positive regulation of catalytic activity | 32 | 16 | 50 | 16 | 50 |
| Homeostasis | homeostasis - calcium ion homeostasis | 15 | 10 | 67 | 5 | 33 |
| Homeostasis | homeostasis - cation homeostasis | 22 | 11 | 50 | 11 | 50 |
| Homeostasis | homeostasis - cellular homeostasis | 45 | 24 | 53 | 21 | 47 |
| Homeostasis | homeostasis - cellular ion homeostasis | 30 | 17 | 57 | 13 | 43 |
| Homeostasis | homeostasis - chemical homeostasis | 37 | 19 | 51 | 18 | 49 |
| Homeostasis | homeostasis - di-, tri-valent inorganic cation homeostasis | 21 | 11 | 52 | 10 | 48 |
| Homeostasis | homeostasis - homeostatic process | 61 | 31 | 51 | 30 | 49 |
| Homeostasis | homeostasis - ion homeostasis | 31 | 18 | 58 | 13 | 42 |
| Homeostasis | homeostasis - iron ion homeostasis | 5 | 1 | 20 | 4 | 80 |
| Homeostasis | homeostasis - multicellular organismal homeostasis | 8 | 3 | 38 | 5 | 63 |
| Homeostasis | hydrolase - negative regulation of hydrolase activity | 8 | 5 | 63 | 3 | 38 |
| Homeostasis | hydrolase - regulation of hydrolase activity | 15 | 12 | 80 | 3 | 20 |
| Homeostasis | molecular function - negative regulation of molecular function | 34 | 19 | 56 | 15 | 44 |
| Homeostasis | molecular function - positive regulation of molecular function | 38 | 20 | 53 | 18 | 47 |
| Homeostasis | phosphate metabolic process | 58 | 25 | 43 | 33 | 57 |
| Homeostasis | phosphorus metabolic process - negative regulation of phosphorus metabolic process | 6 | 6 | 100 | 0 | 0 |
| Lipid Metabolism | Arachidonic acid metabolism | 5 | 3 | 60 | 2 | 40 |
| Lipid Metabolism | carboxylic acid biosynthetic process | 12 | 6 | 50 | 6 | 50 |
| Lipid Metabolism | Fatty acid, triacylglycerol, and ketone body metabolism | 8 | 8 | 100 | 0 | 0 |
| Lipid Metabolism | Glycerophospholipid metabolism | 5 | 2 | 40 | 3 | 60 |
| Lipid Metabolism | Lipid - fatty acid biosynthetic process | 8 | 5 | 63 | 3 | 38 |
| Lipid Metabolism | Lipid - negative regulation of lipid metabolic process | 5 | 4 | 80 | 1 | 20 |
| Lipid Metabolism | Lipid - Regulation of Lipid Metabolism by Peroxisome 2proliferator-activated receptor alpha (PPARalpha) | 5 | 5 | 100 | 0 | 0 |
| Lipid Metabolism | Lipid - Sphingolipid metabolism | 4 | 4 | 100 | 0 | 0 |
| Lipid Metabolism | Lipid - unsaturated fatty acid biosynthetic process | 8 | 6 | 75 | 2 | 25 |
| Lipid Metabolism | lipid localization | 12 | 8 | 67 | 4 | 33 |
| Lipid Metabolism | lipid storage | 6 | 6 | 100 | 0 | 0 |
| Lipid Metabolism | Metabolism of lipids and lipoproteins | 21 | 15 | 71 | 6 | 29 |
| Lipid metabolism | prostaglandin metabolic process | 5 | 4 | 80 | 1 | 20 |
| Lipid Metabolism | Response to elevated platelet cytosolic Ca2+ | 16 | 7 | 44 | 9 | 56 |
| Lipid Metabolism | steroid biosynthetic - regulation of steroid biosynthetic process | 4 | 3 | 75 | 1 | 25 |
| Metabolism of Cofactors and Vitamins | coenzyme metabolic process | 14 | 5 | 36 | 9 | 64 |
| Metabolism of Cofactors and Vitamins | cofactor metabolic process | 16 | 6 | 38 | 10 | 63 |
| Metabolism of Cofactors and Vitamins | Metabolism of vitamins and cofactors | 7 | 3 | 43 | 4 | 57 |
| Mineral Metabolism | Iron uptake and transport | 9 | 1 | 11 | 8 | 89 |
| Nucleotides Metabolism | Metabolism of nucleotides | 8 | 4 | 50 | 4 | 50 |
| Nucleotide Metabolism | nucleoside triphosphate catabolic process | 4 | 2 | 50 | 2 | 50 |
| Nucleotide Metabolism | Purine metabolism | 7 | 4 | 57 | 3 | 43 |
| Nucleotide Metabolism | purine nucleoside triphosphate biosynthetic process | 14 | 4 | 29 | 10 | 71 |
| Nucleotide Metabolism | purine nucleotide biosynthetic process | 16 | 6 | 38 | 10 | 63 |
| Nucleotide Metabolism | purine nucleotide metabolic process | 19 | 7 | 37 | 12 | 63 |
| Nucleotide Metabolism | purine ribonucleotide biosynthetic process | 15 | 5 | 33 | 10 | 67 |
| Nucleotide Metabolism | purine ribonucleotide metabolic process | 17 | 5 | 29 | 12 | 71 |
| Nucleotide Metabolism | pyridine nucleotide metabolic process | 9 | 3 | 33 | 6 | 67 |
| Nucleotide Metabolism | Pyrimidine metabolism | 5 | 3 | 60 | 2 | 40 |
| Overview | cellular biosynthetic - positive regulation of cellular biosynthetic process | 49 | 31 | 63 | 18 | 37 |
| Prostanoid Metabolism | Prostanoid metabolism | 4 | 3 | 75 | 1 | 25 |
| Protein Metabolism | Metabolism of proteins | 59 | 15 | 25 | 44 | 75 |
| Protein metabolism | peptidase - negative regulation of peptidase activity | 6 | 3 | 50 | 3 | 50 |
| Protein metabolism | peptidase - regulation of peptidase activity | 5 | 5 | 100 | 0 | 0 |
| Protein metabolism | peptide metabolic process | 6 | 1 | 17 | 5 | 83 |
| Protein metabolism | protein catabolic - regulation of protein catabolic process | 7 | 6 | 86 | 1 | 14 |
| Protein metabolism | protein kinase - positive regulation of protein kinase cascade | 16 | 12 | 75 | 4 | 25 |
| Protein metabolism | protein kinase - regulation of protein kinase cascade | 6 | 3 | 50 | 3 | 50 |
| Protein metabolism | protein metabolic - negative regulation of protein metabolic process | 13 | 10 | 77 | 3 | 23 |
| Protein metabolism | protein metabolic - positive regulation of protein metabolic process | 12 | 7 | 58 | 5 | 42 |
| Protein Metabolism | protein metabolic - regulation of cellular protein metabolic process | 2 | 0 | 0 | 2 | 100 |
| Protein Metabolism | protein metabolic - regulation of protein metabolic process | 19 | 8 | 42 | 11 | 58 |
| Protein metabolism | protein modification - negative regulation of protein modification process | 5 | 4 | 80 | 1 | 20 |
| Protein metabolism | protein modification - positive regulation of protein modification process | 5 | 4 | 80 | 1 | 20 |
| Protein metabolism | protein modification - regulation of protein modification process | 14 | 3 | 21 | 11 | 79 |
| 1715 | 770 | 45 | 945 | 55 | ||
| Organismal Systems | ||||||
| Circulatory System | angiogenesis | 16 | 9 | 56 | 7 | 44 |
| Circulatory System | angiogenesis - positive regulation of angiogenesis | 5 | 5 | 100 | 0 | 0 |
| Circulatory System | blood pressure - regulation of blood pressure | 10 | 5 | 50 | 5 | 50 |
| Circulatory System | blood vessel development | 20 | 12 | 60 | 8 | 40 |
| Circulatory System | Cardiac muscle contraction | 15 | 6 | 40 | 9 | 60 |
| Circulatory System | circulatory system process | 15 | 7 | 47 | 8 | 53 |
| Circulatory System | erythrocyte differentiation | 6 | 1 | 17 | 5 | 83 |
| Circulatory System | erythrocyte homeostasis | 8 | 2 | 25 | 6 | 75 |
| Circulatory System | Factors involved in megakaryocyte development and platelet production | 9 | 8 | 89 | 1 | 11 |
| Circulatory System | hemopoiesis | 20 | 10 | 50 | 10 | 50 |
| Circulatory System | Hemostasis | 44 | 28 | 64 | 16 | 36 |
| Circulatory System | Integrin cell surface interactions | 6 | 5 | 83 | 1 | 17 |
| Circulatory System | Muscle contraction | 5 | 2 | 40 | 3 | 60 |
| Circulatory System | myeloid cell differentiation | 11 | 5 | 45 | 6 | 55 |
| Circulatory System | myeloid cell differentiation - negative regulation of myeloid cell differentiation | 5 | 4 | 80 | 1 | 20 |
| Circulatory System | myeloid cell differentiation - regulation of myeloid cell differentiation | 4 | 3 | 75 | 1 | 25 |
| Circulatory System | myeloid leukocyte differentiation - regulation of myeloid leukocyte differentiation | 7 | 5 | 71 | 2 | 29 |
| Circulatory System | Platelet activation, signaling and aggregation | 24 | 12 | 50 | 12 | 50 |
| Circulatory System | Vascular smooth muscle contraction | 8 | 5 | 63 | 3 | 38 |
| Circulatory System | vasoconstriction - regulation of vasoconstriction | 5 | 4 | 80 | 1 | 20 |
| Development | Axon guidance | 20 | 14 | 70 | 6 | 30 |
| Development | cell differentiation - negative regulation of cell differentiation | 14 | 10 | 71 | 4 | 29 |
| Development | cell differentiation - positive regulation of cell differentiation | 12 | 9 | 75 | 3 | 25 |
| Development | cell maturation | 8 | 3 | 38 | 5 | 63 |
| Development | development - positive regulation of developmental process | 22 | 17 | 77 | 5 | 23 |
| Development | Developmental Biology | 23 | 17 | 74 | 6 | 26 |
| Development | developmental growth | 9 | 6 | 67 | 3 | 33 |
| Development | developmental maturation | 9 | 4 | 44 | 5 | 56 |
| Development | mesoderm development | 8 | 4 | 50 | 4 | 50 |
| Development | Osteoclast differentiation | 16 | 13 | 81 | 3 | 19 |
| Development | osteoclast differentiation - regulation of osteoclast differentiation | 4 | 3 | 75 | 1 | 25 |
| Development | Semaphorin interactions | 7 | 5 | 71 | 2 | 29 |
| Development | vasculature development | 21 | 13 | 62 | 8 | 38 |
| Digestive System | Gastric acid secretion | 5 | 5 | 100 | 0 | 0 |
| Digestive System | Mineral absorption | 6 | 3 | 50 | 3 | 50 |
| Digestive System | Pancreatic secretion | 6 | 4 | 67 | 2 | 33 |
| Digestive System | Salivary secretion | 5 | 5 | 100 | 0 | 0 |
| Endocrine System | Adipocytokine signaling pathway | 5 | 5 | 100 | 0 | 0 |
| Endocrine System | Progesterone-mediated oocyte maturation | 5 | 3 | 60 | 2 | 40 |
| Endocrine System | Signaling by GnRH | 8 | 5 | 63 | 3 | 38 |
| Endocrine System | Signaling by insulin | 10 | 4 | 40 | 6 | 60 |
| Endocrine System | Signaling by Insulin receptor | 10 | 1 | 10 | 9 | 90 |
| Endocrine System | Signaling by PPAR | 7 | 5 | 71 | 2 | 29 |
| Environmental Adaptation | hydrogen peroxide metabolic process | 5 | 0 | 0 | 5 | 100 |
| Environmental Adaptation | response to abiotic stimulus | 24 | 16 | 67 | 8 | 33 |
| Environmental Adaptation | response to acid | 5 | 1 | 20 | 4 | 80 |
| Environmental Adaptation | response to amino acid stimulus | 4 | 1 | 25 | 3 | 75 |
| Environmental Adaptation | response to drug | 19 | 12 | 63 | 7 | 37 |
| Environmental Adaptation | response to dsRNA | 5 | 3 | 60 | 2 | 40 |
| Environmental Adaptation | response to endogenous stimulus | 34 | 22 | 65 | 12 | 35 |
| Environmental Adaptation | response to endoplasmic reticulum stress | 6 | 4 | 67 | 2 | 33 |
| Environmental Adaptation | response to ethanol | 7 | 3 | 43 | 4 | 57 |
| Environmental Adaptation | response to external stimulus - positive regulation of response to external stimulus | 9 | 4 | 44 | 5 | 56 |
| Environmental Adaptation | response to external stimulus - regulation of response to external stimulus | 4 | 3 | 75 | 1 | 25 |
| Environmental Adaptation | response to extracellular stimulus | 23 | 13 | 57 | 10 | 43 |
| Environmental Adaptation | response to glucocorticoid stimulus | 12 | 10 | 83 | 2 | 17 |
| Environmental Adaptation | response to heat | 6 | 4 | 67 | 2 | 33 |
| Environmental Adaptation | response to hormone stimulus | 30 | 20 | 67 | 10 | 33 |
| Environmental Adaptation | response to hydrogen peroxide | 9 | 4 | 44 | 5 | 56 |
| Environmental Adaptation | response to hypoxia | 17 | 8 | 47 | 9 | 53 |
| Environmental Adaptation | response to inorganic substance | 21 | 11 | 52 | 10 | 48 |
| Environmental Adaptation | response to insulin stimulus | 10 | 6 | 60 | 4 | 40 |
| Environmental Adaptation | response to mechanical stimulus | 7 | 5 | 71 | 2 | 29 |
| Environmental Adaptation | response to metal ion | 12 | 5 | 42 | 7 | 58 |
| Environmental Adaptation | response to nutrient | 14 | 8 | 57 | 6 | 43 |
| Environmental Adaptation | response to nutrient levels | 19 | 11 | 58 | 8 | 42 |
| Environmental Adaptation | response to organic cyclic substance | 10 | 10 | 100 | 0 | 0 |
| Environmental Adaptation | response to organic nitrogen | 7 | 4 | 57 | 3 | 43 |
| Environmental Adaptation | response to organic substance | 68 | 46 | 68 | 22 | 32 |
| Environmental Adaptation | response to oxidative stress | 22 | 10 | 45 | 12 | 55 |
| Environmental Adaptation | response to oxygen levels | 19 | 9 | 47 | 10 | 53 |
| Environmental Adaptation | response to oxygen radical | 4 | 1 | 25 | 3 | 75 |
| Environmental Adaptation | response to peptide hormone stimulus | 13 | 9 | 69 | 4 | 31 |
| Environmental Adaptation | response to protein stimulus | 19 | 16 | 84 | 3 | 16 |
| Environmental Adaptation | response to reactive oxygen species | 11 | 6 | 55 | 5 | 45 |
| Environmental Adaptation | response to steroid hormone stimulus | 17 | 12 | 71 | 5 | 29 |
| Environmental Adaptation | response to stimulus - positive regulation of response to stimulus | 18 | 11 | 61 | 7 | 39 |
| Environmental Adaptation | response to stress | 33 | 15 | 45 | 18 | 55 |
| Environmental Adaptation | response to temperature stimulus | 9 | 7 | 78 | 2 | 22 |
| Environmental Adaptation | response to unfolded protein | 22 | 17 | 77 | 5 | 23 |
| Environmental Adaptation | response to vitamin | 8 | 5 | 63 | 3 | 38 |
| Excretory System | Collecting duct acid secretion | 6 | 1 | 17 | 5 | 83 |
| Excretory System | Vasopressin-regulated water reabsorption | 5 | 2 | 40 | 3 | 60 |
| Immune System | adaptive immune system | 59 | 38 | 64 | 21 | 36 |
| Immune System | adaptive immune system - positive regulation of adaptive immune response | 6 | 2 | 33 | 4 | 67 |
| Immune System | Cytokine Signaling in Immune system | 38 | 24 | 63 | 14 | 37 |
| Immune System | Cytosolic DNA-sensing pathway | 5 | 3 | 60 | 2 | 40 |
| Immune System | defense response | 63 | 34 | 54 | 29 | 46 |
| Immune System | defense response - positive regulation of defense response | 8 | 4 | 50 | 4 | 50 |
| Immune System | Hematopoietic cell lineage | 13 | 10 | 77 | 3 | 23 |
| Immune System | humoral immune response | 11 | 7 | 64 | 4 | 36 |
| Immune System | IFN - Antiviral mechanism by IFN-stimulated genes | 9 | 4 | 44 | 5 | 56 |
| Immune System | IFN - RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways | 9 | 6 | 67 | 3 | 33 |
| Immune System | IFN - RLR (RIG-like receptor) mediated induction of IFN alpha/beta | 5 | 4 | 80 | 1 | 20 |
| Immune System | immune effector - regulation of immune effector process | 11 | 7 | 64 | 4 | 36 |
| Immune System | immune effector process | 14 | 10 | 71 | 4 | 29 |
| Immune System | Immune System - positive regulation of immune response | 23 | 16 | 70 | 7 | 30 |
| Immune System | immune system development | 24 | 13 | 54 | 11 | 46 |
| Immune System | Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell | 9 | 6 | 67 | 3 | 33 |
| Immune System | inflammatory response | 42 | 26 | 62 | 16 | 38 |
| Immune System | inflammatory response - acute inflammatory response | 11 | 9 | 82 | 2 | 18 |
| Immune System | inflammatory response - positive regulation of inflammatory response | 7 | 3 | 43 | 4 | 57 |
| Immune System | inflammatory response - regulation of inflammatory response to antigenic stimulus | 4 | 1 | 25 | 3 | 75 |
| Immune System | Innate Immune System | 31 | 18 | 58 | 13 | 42 |
| Immune System | Interferon alpha/beta signaling | 10 | 2 | 20 | 8 | 80 |
| Immune System | Interferon gamma signaling | 15 | 11 | 73 | 4 | 27 |
| Immune System | Interferon Signaling | 27 | 16 | 59 | 11 | 41 |
| Immune System | Interleukin signaling | 14 | 11 | 79 | 3 | 21 |
| Immune System | Intestinal immune network for IgA production | 8 | 8 | 100 | 0 | 0 |
| Immune System | ISG15 antiviral mechanism | 9 | 4 | 44 | 5 | 56 |
| Immune System | L1CAM interactions | 12 | 8 | 67 | 4 | 33 |
| Immune System | leukocyte activation - regulation of leukocyte activation | 14 | 9 | 64 | 5 | 36 |
| Immune System | leukocyte adhesion | 7 | 6 | 86 | 1 | 14 |
| Immune System | leukocyte chemotaxis | 5 | 2 | 40 | 3 | 60 |
| Immune System | leukocyte mediated immunity | 10 | 7 | 70 | 3 | 30 |
| Immune System | leukocyte mediated immunity - positive regulation of leukocyte mediated immunity | 5 | 2 | 40 | 3 | 60 |
| Immune System | leukocyte mediated immunity - regulation of leukocyte mediated immunity | 2 | 2 | 100 | 0 | 0 |
| Immune System | leukocyte migration | 19 | 10 | 53 | 9 | 47 |
| Immune System | leukocyte proliferation - positive regulation of leukocyte proliferation | 6 | 5 | 83 | 1 | 17 |
| Immune System | Leukocyte transendothelial migration | 12 | 7 | 58 | 5 | 42 |
| Immune System | lymphocyte activation - positive regulation of lymphocyte activation | 10 | 8 | 80 | 2 | 20 |
| Immune System | lymphocyte mediated immunity | 9 | 6 | 67 | 3 | 33 |
| Immune System | lymphocyte mediated immunity - regulation of lymphocyte mediated immunity | 6 | 3 | 50 | 3 | 50 |
| Immune System | MAPK targets/Nuclear events mediated by MAP kinases | 6 | 5 | 83 | 1 | 17 |
| Immune System | MyD88 cascade initiated on plasma membrane | 13 | 11 | 85 | 2 | 15 |
| Immune System | MyD88 dependent cascade initiated on endosome | 12 | 10 | 83 | 2 | 17 |
| Immune System | MyD88:Mal cascade initiated on plasma membrane | 13 | 11 | 85 | 2 | 15 |
| Immune System | MyD88-independent cascade initiated on plasma membrane | 14 | 11 | 79 | 3 | 21 |
| Immune System | Natural killer cell mediated cytotoxicity | 10 | 6 | 60 | 4 | 40 |
| Immune System | nitric oxide - positive regulation of nitric oxide biosynthetic process | 8 | 7 | 88 | 1 | 13 |
| Immune System | phagocytosis | 7 | 3 | 43 | 4 | 57 |
| Immune System | phagocytosis - Fc epsilon RI signaling pathway | 5 | 2 | 40 | 3 | 60 |
| Immune System | phagocytosis - Fc gamma R-mediated phagocytosis | 8 | 3 | 38 | 5 | 63 |
| Immune System | response to bacterium | 22 | 11 | 50 | 11 | 50 |
| Immune System | response to lipopolysaccharide | 14 | 8 | 57 | 6 | 43 |
| Immune System | response to molecule of bacterial origin | 16 | 8 | 50 | 8 | 50 |
| Immune System | response to virus | 11 | 3 | 27 | 8 | 73 |
| Immune System | response to wounding | 58 | 35 | 60 | 23 | 40 |
| Immune System | signaling - Chemokine signaling pathway | 19 | 9 | 47 | 10 | 53 |
| Immune System | Signaling - NOD-like receptor signaling pathway | 13 | 10 | 77 | 3 | 23 |
| Immune System | Signaling - Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways | 6 | 4 | 67 | 2 | 33 |
| Immune System | Signaling - Opioid Signalling | 5 | 2 | 40 | 3 | 60 |
| Immune System | signaling - TRIF mediated TLR3 signaling | 13 | 10 | 77 | 3 | 23 |
| Immune System | Signaling by Interleukins | 14 | 11 | 79 | 3 | 21 |
| Immune System | Signaling by RIG-I-like receptor | 5 | 3 | 60 | 2 | 40 |
| Immune System | Signaling by TCR | 19 | 17 | 89 | 2 | 11 |
| Immune System | Signaling by the B Cell Receptor (BCR) | 17 | 9 | 53 | 8 | 47 |
| Immune System | T cell - Antigen processing and presentation | 50 | 27 | 54 | 23 | 46 |
| Immune System | T cell - Costimulation by the CD28 family - T cell | 9 | 9 | 100 | 0 | 0 |
| Immune System | T cell - positive regulation of T cell activation | 8 | 6 | 75 | 2 | 25 |
| Immune System | TAK1 activates NFkB by phosphorylation and activation of IKKs complex | 5 | 5 | 100 | 0 | 0 |
| Immune System | TLR - Innate immune response mediated by toll like receptors | 11 | 7 | 64 | 4 | 36 |
| Immune System | TLR - MAP kinase activation in TLR cascade | 9 | 7 | 78 | 2 | 22 |
| Immune System | TLR - Toll-like receptor signaling pathway | 25 | 15 | 60 | 10 | 40 |
| Immune System | TLR - Trafficking and processing of endosomal TLR | 6 | 1 | 17 | 5 | 83 |
| multicellular organismal process | multicellular organismal - negative regulation of multicellular organismal process | 10 | 5 | 50 | 5 | 50 |
| multicellular organismal process | multicellular organismal - positive regulation of multicellular organismal process | 12 | 7 | 58 | 5 | 42 |
| Nervous System | Cholinergic synapse | 5 | 3 | 60 | 2 | 40 |
| Nervous System | Dopaminergic synapse | 6 | 5 | 83 | 1 | 17 |
| Nervous System | Long-term potentiation | 5 | 4 | 80 | 1 | 20 |
| Nervous System | neurological system - positive regulation of neurological system process | 6 | 5 | 83 | 1 | 17 |
| Nervous System | Neuronal System | 11 | 5 | 45 | 6 | 55 |
| Nervous System | Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell | 5 | 2 | 40 | 3 | 60 |
| Nervous System | Serotonergic synapse | 7 | 3 | 43 | 4 | 57 |
| Nervous System | Signaling - Neurotrophin signaling pathway | 13 | 11 | 85 | 2 | 15 |
| Nervous System | Signaling - NGF signalling via TRKA from the plasma membrane | 11 | 7 | 64 | 4 | 36 |
| Nervous System | synaptic plasticity - regulation of synaptic plasticity | 7 | 6 | 86 | 1 | 14 |
| Nervous System | synaptic transmission - positive regulation of synaptic transmission | 6 | 5 | 83 | 1 | 17 |
| Nervous System | synaptic transmission - regulation of synaptic transmission | 5 | 4 | 80 | 1 | 20 |
| Nervous System | Synaptic vesicle cycle | 7 | 0 | 0 | 7 | 100 |
| Nervous System | Transmission across Chemical Synapses | 7 | 4 | 57 | 3 | 43 |
| Nervous System | vesicle docking during exocytosis | 4 | 1 | 25 | 3 | 75 |
| Nervous System | vesicle-mediated transport | 43 | 13 | 30 | 30 | 70 |
| 2299 | 1399 | 61 | 900 | 39 | ||
| Human Diseases | ||||||
| Cancers | Bladder cancer | 7 | 2 | 29 | 5 | 71 |
| Cancers | Glioma | 5 | 3 | 60 | 2 | 40 |
| Cancers | myeloid leukemia - Acute myeloid leukemia | 7 | 4 | 57 | 3 | 43 |
| Cancers | myeloid leukemia - Chronic myeloid leukemia | 6 | 4 | 67 | 2 | 33 |
| Cancers | Pancreatic cancer | 6 | 2 | 33 | 4 | 67 |
| Cancers | Pathways in cancer | 22 | 14 | 64 | 8 | 36 |
| Cancers | Prostate cancer | 10 | 8 | 80 | 2 | 20 |
| Cancers | Renal cell carcinoma | 8 | 3 | 38 | 5 | 63 |
| Cancers | Small cell lung cancer | 6 | 5 | 83 | 1 | 17 |
| Cancers | Transcriptional misregulation in cancer | 13 | 10 | 77 | 3 | 23 |
| Cardiovascular Diseases | Arrhythmogenic right ventricular cardiomyopathy (ARVC) | 6 | 6 | 100 | 0 | 0 |
| Cardiovascular Diseases | Dilated cardiomyopathy | 7 | 6 | 86 | 1 | 14 |
| Cardiovascular Diseases | Hypertrophic cardiomyopathy (HCM) | 8 | 6 | 75 | 2 | 25 |
| Cardiovascular Diseases | Viral myocarditis | 11 | 10 | 91 | 1 | 9 |
| Endocrine and Metabolic Diseases | Diabetes pathways | 19 | 16 | 84 | 3 | 16 |
| Immune Diseases | Allograft rejection | 9 | 8 | 89 | 1 | 11 |
| Immune Diseases | Asthma | 9 | 8 | 89 | 1 | 11 |
| Immune Diseases | Autoimmune thyroid disease | 8 | 7 | 88 | 1 | 13 |
| Immune Diseases | Graft-versus-host disease | 11 | 10 | 91 | 1 | 9 |
| Immune Diseases | Rheumatoid arthritis | 32 | 19 | 59 | 13 | 41 |
| Immune Diseases | Systemic lupus erythematosus | 16 | 16 | 100 | 0 | 0 |
| Infectious Diseases | Amoebiasis | 11 | 10 | 91 | 1 | 9 |
| Infectious Diseases | Bacterial invasion of epithelial cells | 7 | 4 | 57 | 3 | 43 |
| Infectious Diseases | Botulinum neurotoxicity | 4 | 1 | 25 | 3 | 75 |
| Infectious Diseases | Chagas disease (American trypanosomiasis) | 17 | 13 | 76 | 4 | 24 |
| Infectious Diseases | Hepatitis C | 11 | 7 | 64 | 4 | 36 |
| Infectious Diseases | Herpes simplex infection | 28 | 20 | 71 | 8 | 29 |
| Infectious Diseases | HIV Infection | 23 | 4 | 17 | 19 | 83 |
| Infectious Diseases | HTLV-I infection | 30 | 22 | 73 | 8 | 27 |
| Infectious Diseases | Influenza infection | 65 | 28 | 43 | 37 | 57 |
| Infectious Diseases | Legionellosis | 18 | 14 | 78 | 4 | 22 |
| Infectious Diseases | Leishmaniasis | 23 | 21 | 91 | 2 | 9 |
| Infectious Diseases | Malaria | 6 | 4 | 67 | 2 | 33 |
| Infectious Diseases | Measles | 16 | 12 | 75 | 4 | 25 |
| Infectious Diseases | Pathogenic Escherichia coli infection | 12 | 8 | 67 | 4 | 33 |
| Infectious Diseases | Pertussis | 19 | 14 | 74 | 5 | 26 |
| Infectious Diseases | Salmonella infection | 17 | 13 | 76 | 4 | 24 |
| Infectious Diseases | Shigellosis | 10 | 6 | 60 | 4 | 40 |
| Infectious Diseases | Signaling - Epithelial cell signaling in Helicobacter pylori infection | 13 | 5 | 38 | 8 | 62 |
| Infectious Diseases | Staphylococcus aureus infection | 13 | 13 | 100 | 0 | 0 |
| Infectious Diseases | Toxoplasmosis | 21 | 18 | 86 | 3 | 14 |
| Infectious Diseases | Tuberculosis | 33 | 24 | 73 | 9 | 27 |
| Infectious Diseases | Vibrio cholerae infection | 10 | 3 | 30 | 7 | 70 |
| Neurodegenerative Diseases | Alzheimer's disease | 30 | 9 | 30 | 21 | 70 |
| Neurodegenerative Diseases | Amyloids | 9 | 6 | 67 | 3 | 33 |
| Neurodegenerative Diseases | Huntington's disease | 33 | 6 | 18 | 27 | 82 |
| Neurodegenerative Diseases | Parkinson's disease | 30 | 7 | 23 | 23 | 77 |
| Neurodegenerative Diseases | Prion diseases | 10 | 7 | 70 | 3 | 30 |
| 745 | 466 | 63 | 279 | 37 |
A total of 24 pathways in various cellular processes had at least 20 DE genes identified in PAMs infected with PRRSV (Table 1). Of them, PRRSV infection down-regulated more than two thirds of the genes in three pathways: actin filament based processes (69.6%), anti-apoptosis (67.7%) and positive regulation of cell communication (66.7%), while it up-regulated more than two thirds of the genes in two other pathways: membrane organization (67.7%) and lysosome activities (68%) at 24 hours post infection (Table 1).
Among 329 DE genes related to cellular processes, SOM analysis assigned 32 (9.7%), 12 (3.6%), 68 (20.7%), 22 (6.7%), 23 (7.0%), 16 (4.9%), 15 (4.6%), 44 (13.4%), 65 (19.8%) and 32 (9.7%) into expression trend clusters A – J, respectively. All 30 of the following DE genes, VEGFA, ACVRL1, GPX1, SOD1, GSN, MAPK1, CAPG, APP, CD24, CAPZB, UBB, LTB, PPP2CA, IL1B, CFL1, CDKN1A, FLNA, TNF, ANG, EDN1, PRKCQ, ITGB1, JAK2, HBEGF, HMOX1, IL1A, NPM1, PLEK, ACTG1, RPS27A were involved in cellular processes and had multiple functions in at least 10 pathways. The last 17 genes (56.7%) in this list above were clustered in H, I and J, respectively. On the other hand, CREG1, HSBP1, H1F0, BRK1, H1FX, CAPG, S100A6, CAPNS1, CD68, CTSH, MBD3, SCARB2, FXYD5, RNF130, TMBIM6, LAPTM4A, TSPAN31, SERPINI1, IER3, SYNE2, CDC42EP3, CRIP1, ARID5A and FMNL3 were exclusively involved in cellular processes: with the first 12 genes (50%) grouped in clusters A, B and C, respectively. A collection of the top 10 up- and bottom 10 down-regulated genes at each time-point post infection made a pool of 19 genes: TNF, HSPA1B, TIMP1, TNFSF13, BAG3, HSPA1A, ANGPTL4, HMOX1, GJA1, CCRL1, HBEGF, CCL3L1, HSPA6, HLA-DOA, MAN2B1, NUDC, HLA-DMB, ENPP1 and PLA2G15 as the most actively down-regulated genes and a pool of 25 genes: RAB7B, IL3RA, LRPAP1, HLA-A, ACVR1, ACE, CD24, MAEA, RAB11A, SOD2, SFTPA1, GPX1, ARPC2, TIAL1, H1FX, H1F0, ATF5, MMP9, BNIP3, LGALS9, CCL2, CCL8, IDO1, S100A6, CXCL6 as the most actively up-regulated genes in cellular processes.
Reactome of PAMs Infected with PRRSV: Genetic Information Processing
PRRSV infection of PAMs triggered reactions in 262 genes handling genetic information processing, including transcription, translation, replication and repair, and protein folding, sorting and degradation (Figure 7). Most of the genes involved in genetic information processing were involved in two or more sub-category pathways. However, there were large clusters of genes that functioned exclusively in folding, sorting, and degradation as well as in transcription. In contrast, there were small clusters of genes that only contributed to replication and repair, and translation. There were a total of 147 DE genes related to transcription processes with pathways in DNA binding and regulation, gene expression and regulation, mRNA stability and regulation, regulation of the I-kappaB kinase/NF-kappaB cascades, NF-kappaB transcription factor activity and NF-kappaB import into nucleus, nonsense-mediated decay, processing of capped intron-containing pre-mRNA, RNA biosynthetic processes, RNA polymerases I, II and III transcription, spliceosome and regulation of transcription factors and their import into the nucleus. Protein folding, sorting, and degradation was affected by 147 DE genes with specific functions in degradation of the extracellular matrix, regulation of endopeptidase activity, asparagine N-linked glycosylation, nucleocytoplasmic transport and regulation, post-Golgi vesicle-mediated transport, proteasomal ubiquitin-dependent protein catabolic processes, protein folding, protein import into nucleus and regulation, protein localization and regulation, protein localization in organelles, protein processing in endoplasmic reticulum, protein targeting, intracellular protein transport and regulation, regulation of protein ubiquitination, and Soluble NSF Attachment Protein Receptor (SNARE) interactions in vesicular transport. A role for SNARE machinery in virion egress has been proposed for cytomegalovirus [18] and may be similarly involved with PRRSV egress from PAMs. Seventy-four genes associated with translation processes were DE in PRRSV-infected PAMs and specific pathways were related to post-translational protein modification, ribosome and ribosome biogenesis, RNA transport, signal recognition particle (SRP)-dependent cotranslational protein targeting to membrane, translation and regulation, translation elongation, translation initiation and regulation, and translation termination. Pathways related to repair, replication and regulation were affected by 21 genes that were DE in PAMs infected with PRRSV. The genetic information processing networks of 262 DE genes in PAMs infected with PRRSV are illustrated in Figure 7 and summarized in Table 1.
Figure 7. DE gene distributions and interactions among functional categories associated with Genetic Information Processing in PAMs infected with PRRSV.
In the genetic information processing systems, we observed 14 pathways with at least 20 DE genes identified in PAMs infected with PRRSV (Table 1). Among them, more than two-thirds of genes were down-regulated in two pathways, while two-thirds of genes were up-regulated in eight pathways at 24 h post infection. Interestingly, the genes that were down-regulated participated in protein folding (78%) and protein processing in endoplasmic reticulum (75%) pathways and belonged to the broader “protein folding, sorting and degradation” category. The eight up-regulated pathways were related to transcription and translation: transcription processes with gene expression (66%) and nonsense-mediated decay (85%) and translation processes with translation (77%), translation elongation (84%), translation initiation (86%), ribosome (89%), translation termination (90%) and SRP-dependent cotranslational protein targeting to membrane (91%), respectively (Table 1).
For 262 DE genes included in the Genetic Information Processing systems, clusters A – J had 29 (11%), 18 (6.9%), 48 (18%), 19 (7.3%), 14 (5.3%), 13 (5.0%), 13 (5.0%), 31 (12%), 49 (19%) and 28 (11%) genes, respectively. The genes RPS6, RPL23, RPS19, RPS5, RPS16, RPS7, UBB, FLNA, TNF, NFKBIA, JAK2 and RPS27A had multiple functions in at least ten pathways with the first six genes (50%) which code for ribosomal proteins clustered in B, C and D, respectively. Twenty one genes: AKAP12, MRPL52, SUMO2, TSFM, MRPL28, UBXN1, YBX1, HELB, HNRNPH2, AHSA1, DSCR3, HNRNPC, NFIL3, PPIC, HNRNPA2B1, PTBP1, DMXL2, HNRNPA1, LMO4, NARS and SYNCRIP had exclusive functions with the last twelve genes (57%) clustered in H, I and J, respectively. The most actively down-regulated genes were TNF, HSPA1B, TIMP1, TNFSF13, BAG3, HSPA1A, DNAJB1, HMOX1, GJA1, C3, NARS, FOS, EGR1, HSPA6, YWHAE, NUDC, ENPP1, RAMP2, JUNB, RPS7 and the most actively up-regulated genes included CSF1, RAB7B, CCNH, LRPAP1, PPIA, TRAPPC2, NME2, MYBPC3, ACVR1, CD24, POLB, VEGFA, RAB11A, YBX1, GPX1, MRPL28, BST2, PSME2, POLR2I, KAP12, WARS, MMP9, BNIP3, LGALS9, TRAPPC4, respectively as they appeared either on the top 10 up- or bottom 10 down-regulated genes at least once in PAMs at the four time points post-PRRSV infection.
Reactome of PAMs Infected with PRRSV: Environmental Information Processing
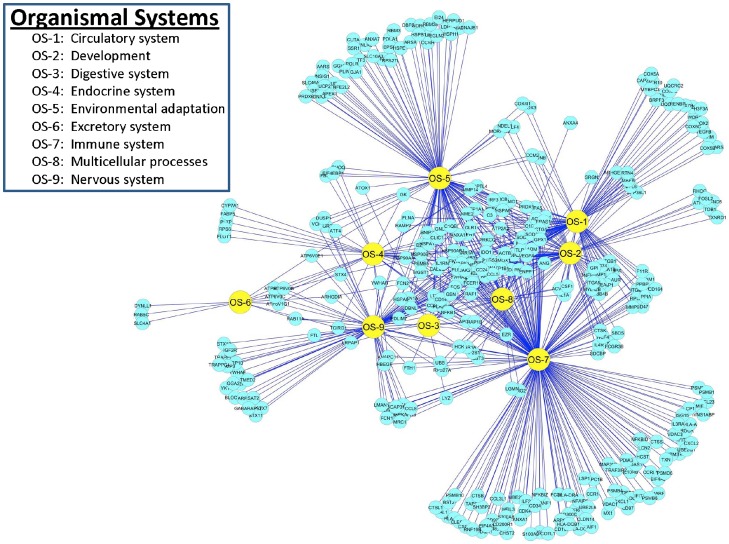
In the environmental information processing systems, a total of 189 genes differentially expressed in PAMs infected with PRRSV were assigned to three functional categories: 1) membrane transport, 2) signal transduction, and 3) signaling molecules and interaction (Figure 8). While there were large clusters of genes that had exclusive pathway functions, many of the genes involved in environmental information processing contributed to each of the three pathways. The GO, KEGG and REACTOME databases mapped 126 DE genes to functions in signal transduction, such as regulation of signal transduction, Ras protein signal transduction, small GTPase mediated signal transduction, calcium signaling, cytokine-mediated signaling, ER-nuclear signaling, platelet-derived growth factor receptor signaling, and signaling by EGFR, ErbB, FGFR, GPCR, Jak-STAT, MAPK, mTOR, NGF, PDGF, SCF-KIT, VEGF and Wnt, respectively. In addition, 66 DE genes functioned as signaling molecules and interactions, such as cell adhesion molecules, regulation of cytokine biosynthetic processes and production, cytokine-cytokine receptor interaction, ECM-receptor interaction and GPCR ligand binding. Furthermore, 53 DE genes were involved with membrane transport and had functions related to Golgi vesicle transport, membrane docking and trafficking, regulation of secretion, aquaporin-mediated transport, SLC-mediated transmembrane transport, and transmembrane transport of small molecules. The 189 DE genes in PAMs infected with PRRSV involved in environmental information processing networks are illustrated in Figure 8 and summarized in Table 1.
Figure 8. DE gene distributions and interactions among functional categories associated with Environmental Information Processing in PAMs infected with PRRSV.
The dominant networks with at least 20 DE genes in the environmental information processing systems included five signal transduction pathways, two signaling molecules and interaction pathways and one membrane transport pathway (Table 1). Among them, at least 65% of the DE genes in PRRSV-infected PAMs at 24 hours post-infection had down-regulation roles in five pathways, including signaling by MAPK (85%), NGF (70%) and GPCR (67%), GPCR ligand binding (67%) and positive regulation of signal transduction (65%). However, genes in only the transmembrane transport of small molecules pathway showed significant up-regulation (68%) by PAMs in response to PRRSV infection 24 hours post-infection (Table 1).
In the environmental information processing systems, expression trend clusters A – J had 22 (12%), 7 (3.7%), 31 (16%), 12 (6.4%), 12 (6.4%), 12 (6.4%), 10 (5.3%), 19 (10%), 44 (23%) and 20 (11%), respectively. The genes RAF1 and MAPK1 were involved in 10 and 12 pathways, respectively. The former gene was member of cluster H, while the latter gene belonged to cluster C. Meanwhile, OR5P3, EMR1, RRAD and HCAR2 were exclusively related to the system and were classified into F, G, I and J clusters, respectively. Compilation of the top 10 up- and bottom 10 down-regulated DE genes in the system each at 6, 12, 16 and 24 hours post infection revealed a pool of 18 genes: TNF, RRAD, HSPA1B, MAP3K8, TNFSF13, HSPA1A, HMOX1, GJA1, CCRL1, C3, HBEGF, CCL3L1, HSPA6, HLA-DOA, HLA-DMB, RAMP2, CD14 and CTSZ as the most actively down-regulated genes, while a pool of 20 genes: RAB7B, CD34, IL3RA, HLA-A, ACVR1, CD24, VEGFA, RAB11A, GPX1, VDAC3, PSME2, SLC16A3, MMP9, LGALS9, PLA2G2D, CCL2, CCL8, TRAPPC4, IDO1 and CXCL6 as the most actively up-regulated genes, respectively.
Reactome of PAMs Infected with PRRSV: Metabolisms
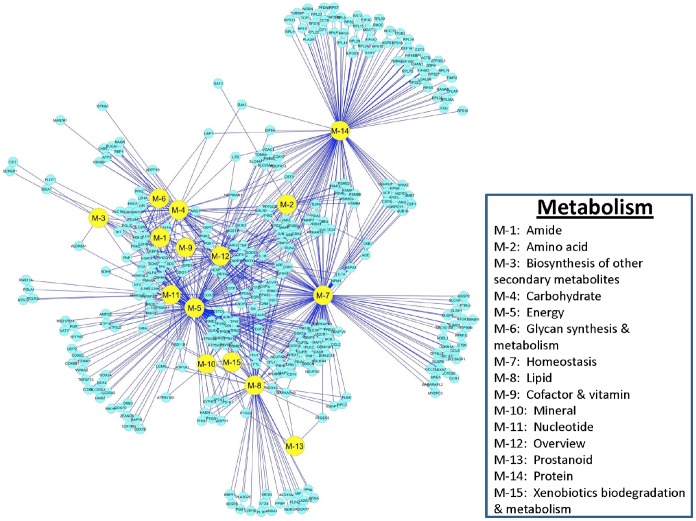
PRRSV infection induced differential expressions of 340 genes in PAMs by 24 hours post-infection that were mainly involved in metabolism of 1) amino acids, 2) carbohydrates, 3) energy, 4) glycans, 5) homeostasis, 6) lipids, 7) cofactors and vitamins, 8) nucleotides and 9) proteins plus a few more functions in amide, secondary metabolites, minerals, prostanoids and cellular biosynthetic processes (Figure 9). The response of the metabolism system of PAMs in response to PRRSV was quite complicated. Small clusters of DE genes identified functioned exclusively in homeostasis, protein metabolism or lipid metabolism. However, the majority of genes were involved in more than two metabolism pathways. More than half (176 genes) of these 340 DE genes were involved in energy metabolism, such as pathways in ATP biosynthetic processes, biological oxidations, cell redox homeostasis, cellular respiration, electron transport chain, energy coupled proton transport, down electrochemical gradient, energy derivation by oxidation of organic compounds, generation of precursor metabolites and energy, integration of energy metabolism, mitochondrial ATP synthesis coupled electron transport, mitochondrial electron transport, NADH to ubiquinone; mitochondrial protein import, mitochondrial transport, mitochondrion organization, regulation of monooxygenase activity, NAD metabolic processes, positive regulation of nitrogen compound metabolic processes, nitrogen compound biosynthetic processes, oxidation reduction, oxidative phosphorylation, regulation of oxidoreductase activity, oxygen and reactive oxygen species metabolic processes, proton transport, release of cytochrome c from mitochondria, respiratory electron transport chain, respiratory electron transport, ATP synthesis by chemiosmotic coupling, heat production by uncoupling proteins, respiratory gaseous exchange, the TCA cycle and respiratory electron transport, and transport of glucose and other sugars, bile salts and organic acids, metal ions and amine compounds. Another group of 151 DE genes participated in homeostasis, such as regulation of catalytic activity, calcium ion homeostasis, cation homeostasis, cellular homeostasis, chemical homeostasis, di- and tri-valent inorganic cation homeostasis, homeostatic processes, ion homeostasis, iron ion homeostasis, multicellular organismal homeostasis, regulation of hydrolase activity, regulation of molecular function and phosphate metabolic processes and regulation. Protein metabolism in PRRSV-infected PAMs was affected by 124 DE genes that were involved in peptidase activity and regulation, peptide metabolic processes and regulation, and regulation of protein kinase cascades, protein metabolic processes and protein modification processes. The data analysis also revealed 64, 48 and 49 DE genes having functions in metabolism of lipids, carbohydrates, and positive regulation of cellular biosynthetic processes, respectively. The remaining metabolic categories had 35 and fewer DE genes involved. The metabolism networks of 340 DE genes in PAMs infected with PRRSV are shown in Figure 9 and summarized in Table 1.
Figure 9. DE gene distributions and interactions among functional categories associated with Metabolism in PAMs infected with PRRSV.
Among pathways involved in energy metabolism, genes related to generation of precursor metabolites and energy; the TCA cycle and respiratory electron transport; respiratory electron transport, ATP synthesis by chemiosmotic coupling and heat production by uncoupling proteins; oxidation reduction; and oxidative phosphorylation accounted for 82%, 74%, 74%, 73% and 68% of the up-regulated DE genes, respectively (Table 1). In homeostasis, 11 pathways had more than 20 DE genes identified, but none of these homeostasis pathways had two-thirds of the DE genes either down- or up-regulated. Other important pathways needing to be mentioned in the metabolism systems include: metabolism of proteins (59 DE genes with 44 (75%) up-regulated), hexose metabolic processes (24 DE genes with 16 genes (67%) up-regulated), metabolism of lipids and lipoproteins (21 genes with 15 (71%) down-regulated) and glucose metabolic processes (21 DE genes with 15 (71%) up-regulated), respectively (Table 1).
Of the 340 DE genes that were involved in the metabolism systems in PRRSV-infected PAMs, 46 (14%) were assigned to cluster A, 25 (7.4%) to B, 74 (22%) to C, 26 (7.7%) to D, 18 (5.3%) to E, 15 (4.4%) to F, 14 (4.1%) to G, 44 (13%) to H, 46 (14%) to I and 32 (9.4%) to J, respectively. The following 55 genes in the metabolism systems had multiple functions in at least 10 pathways: SOD2, SOD1, GPX1, ATP5C1, ATP6V0B, NME2, NDUFB3, NDUFS2, NDUFV2, FTL, NDUFAB1, ATP5J2, PGLS, UQCR10, FTH1, G6PD, ATP6V0E1, TPI1, COX3, MDH2, ATP6V1F, GPI, ATP5G2, NDUFB2, TALDO1, IFI6, UQCR11, CCL2, APP, TCIRG1, ATP6V0C, COX2, NDUFA4, CD24, HEXA, TXNRD1, PNP, ATP5O, APOA5, ATF4, IL1B, GPD1, ENPP1, LDHB, JUN, SDHB, FBP1, TNF, EDN1, JAK2, HEXB, ATP2A2, HMOX1, HERPUD1 and ADM with the first 28 genes (50.91%) clustered in A, B and C, respectively, while the last 15 genes (27.27%) in H and I clusters, respectively. The metabolism systems specific genes were SDS, PGAM1, GBE1, CYP51A1, HSD11B1, ENOPH1, NADH5, PTGR1, DDT, PPM1G, TBC1D1, ATP5J2, PGLS, ISYNA1, PHYH, HSD17B14, MBOAT7, PGS1, TPI1, MDH2, TALDO1, PGK1, COX17, CNDP2, SH3BGRL3, BCKDK, NAGK, POR, GLRX, SLC25A3, ISCA1, AMPD2, ALDH8A1, PTGES3, TXNL1, GSTO1, GPD1, LDHB, CYP4F3, HADH, NT5C2, SCPEP1, CKB, RIOK3, TBC1D10A, TBC1D20 with the first 28 genes (61%, 28/46) grouped into clusters A, B and C, respectively. Two sets of DE genes, one with TNF, TIMP1, MAP3K8, TNFSF13, ANGPTL4, HMOX1, GJA1, NDEL1, PMAIP1, FOS, DUSP6, ANG, EGR1, MGST1, PLAUR, MAN2B1, YWHAE, ENPP1, JUNB, HSPB1, ARSA, PLA2G15 and RPS7 and the other with CSF1, HEXA, LRPAP1, PPIA, MYBPC3, ACVR1, ACE, CD24, MAEA, RAB11A, SOD2, SFTPA1, GPX1, HSD11B1, PSME2, POLR2I, TBC1D1, BCKDK, SLC16A3, BNIP3, LGALS9, PLA2G2D, CCL2, SDS and IDO1 were identified as the most actively down- and up-regulated genes, respectively in the metabolism system.
Reactome of PAMs Infected with PRRSV: Organismal Systems
A total of 346 DE genes identified in PAMs infected with PRRSV impacted organismal systems, shown in Figure 10 and summarized in Table 1. All of the pathways involved in organismal systems were affected by genes that functioned in multiple processes. There were 30 pathways in organismal systems of PAMs infected with PRRSV at 24 hours post infection that had 20 or more DE genes (Table 1). More than two-thirds of DE genes were down-regulated in eight of these pathways - positive regulation of developmental processes (77%), response to unfolded protein (77%), developmental biology (74%), axon guidance (70%), positive regulation of immune response (70%), response to organic substance (68%), response to hormone stimulus (67%) and response to abiotic stimulus (67%), In comparison, more than two-thirds (70%) of the DE genes in the vesicle docking during exocytosis pathway were up-regulated (Table 1).
Figure 10. DE gene distributions and interactions among functional categories associated with Organismal Systems in PAMs infected with PRRSV.
The immune system held the largest group of 297 DE genes that included genes involved in adaptive immunity and regulation, cytokine signaling, cytosolic DNA-sensing pathways, defense response and regulation, hematopoietic cell lineage, humoral immune response, antiviral mechanisms by IFN-stimulated genes, RIG-I/MDA5 mediated induction of IFN-α/β pathways, RLR (RIG-like receptor) mediated induction of IFN-α/β, immune effector processes and regulation, regulation of immune response, immune system development, immunoregulatory interactions between a lymphoid and a non-lymphoid cell, inflammatory response and regulation, acute inflammatory response, regulation of inflammatory response to antigenic stimulus, innate immune system, interferon α/β signaling, IFN-γ signaling, IFN signaling, interleukin signaling, intestinal immune network for IgA production, ISG15 antiviral mechanism, L1CAM interactions, regulation of leukocyte activation, leukocyte adhesion, leukocyte chemotaxis, leukocyte mediated immunity and regulation, regulation of leukocyte mediated immunity, leukocyte migration, positive regulation of leukocyte proliferation, leukocyte transendothelial migration, regulation of lymphocyte activation, lymphocyte mediated immunity and regulation, MAPK targets/nuclear events mediated by MAP kinases, MyD88 cascades initiated on plasma membrane, MyD88 dependent cascades initiated on endosome, MyD88:Mal cascades initiated on plasma membranes, MyD88-independent cascades initiated on plasma membrane, natural killer cell mediated cytotoxicity, regulation of nitric oxide biosynthetic processes, phagocytosis, Fc-ε RI signaling pathway, Fc-γ R-mediated phagocytosis, response to bacterium, response to lipopolysaccharide, response to molecule of bacterial origin, response to virus, response to wounding, chemokine signaling pathway, NOD-like receptor signaling pathway, nucleotide-binding domain, leucine rich repeat containing receptor signaling pathways, opioid signaling, TRIF mediated TLR3 signaling, signaling by interleukins, signaling by RIG-I-like receptor, signaling by TCR, signaling by the B cell receptor, antigen processing and presentation, stimulation by the CD28 family, regulation of T cell activation, TAK1 activation of NFκB by phosphorylation and activation of IKKs complex, innate immune response mediated by toll like receptors, MAP kinase activation in TLR cascades, toll-like receptor signaling pathways and trafficking and processing of endosomal TLR.
Reactome of PAMs Infected with PRRSV: Human Diseases
As a disease in pigs, PRRSV infection of PAMs caused DE of 234 genes that share pathways associated with human diseases, such as 1) cancers, 2) cardiovascular diseases, 3) endocrine and metabolic diseases, 4) immune diseases, 5) infectious diseases and, 6) neurodegenerative diseases (Figure 11; Table 1). Interestingly, most genes had exclusive functions in each disease subcategory. Among them, 169 DE genes are important in human infectious diseases, including pathways for amoebiasis, bacterial invasion of epithelial cells, botulinum neurotoxicity, Chagas disease (American trypanosomiasis), hepatitis C, herpes simplex infection, HIV infection, HTLV-I infection, influenza infection, legionellosis, leishmaniasis, malaria, measles, pathogenic Escherichia coli infection, pertussis, Salmonella infection, shigellosis, Signaling - Epithelial cell signaling in Helicobacter pylori infection, Staphylococcus aureus infection, toxoplasmosis, tuberculosis and Vibrio cholerae infection (Table 1). Fifty-seven genes important in the courses of five human neurodegenerative diseases - Alzheimer's disease, Amyloids, Huntington's disease, Parkinson's disease and prion diseases were also DE in PRRSV-infected PAMs. A total of 42 DE genes expressed in PAMs infected with PRRSV were also involved in human immune diseases, such as allograft rejection, asthma, autoimmune thyroid disease, graft-versus-host disease, rheumatoid arthritis and systemic lupus erythematosus. In addition, KEGG and REACTOME pathway analyses revealed 36, 19 and 18 genes that are related to human cancer diseases, endocrine and metabolic diseases and cardiovascular diseases (Table 1) are also DE in PAMs after infection with PPRSV. The Human Disease networks shared with PRRSV-infected PAMs are illustrated in Figure 11.
Figure 11. DE gene distributions and interactions among functional categories associated with Human Diseases in PAMs infected with PRRSV.
PAMs infected with PRRSV had more than 20 DE genes that are involved in the gene expression pathways of seven human infectious diseases, three neurodegenerative diseases, one immune disease and one pathway in cancer (Table 1). More than two-thirds of the genes associated with leishmaniasis (91%), toxoplasmosis (86%), HTLV-I infection (73%), tuberculosis (73%) and herpes simplex infection (71%), were down-regulated in PAMs infected with PRRSV. On the other hand, PRRSV infection of PAMs up-regulated more than 66% of the genes commonly associated with four human diseases/pathways, including HIV infection (83%), Huntington's disease (82%), Parkinson's disease (77%) and Alzheimer's disease (70%), respectively.
Cluster analysis showed clusters A – J contained 25 (11%), 14 (6.0%), 54 (23%), 23 (10%), 8 (3.4%), 9 (3.9%), 10 (4.3%), 28 (12%), 47 (20%) and 16 (6.8%), respectively, of the 234 genes that were DE in human disease pathways of PRRSV-infected PAMs. Nineteen of the DE genes were involved in at least 10 human disease pathways and included MAPK1, TLR4, IL1B, HLA-DMB, RAF1, JUN, ACTB, TNF, HLA-DRA, HLA-DRB1, HLA-DOA, HLA-DQA2, HLA-DQB1, NFKBIA, ITGB1, IL1A, NFKB1, HLA-DQA1 and ACTG1 with the last 17 (89%) clustered in H, I and J, respectively. Among six systems in human diseases, SLC45A3, CLEC4E and CRTC2 were exclusively involved in human disease network pathways. A set of 18 genes: C1QB, C3, CCL3L1, CD14, DNAJB1, DUSP6, EGR1, FOS, GJA1, HBEGF, HLA-DMB, HLA-DOA, HSPA1A, HSPA1B, HSPA6, RPS7, TNF, TNFSF13 and a set of 25 genes: ACE, ARPC2, BNIP3, CCL2, CCNH, CSF1, CTSK, CXCL6, GPX1, HLA-A, IFIT1, MMP9, MX1, MYBPC3, OAS1, PLA2G2D, POLB, POLR2I, PPIA, PSME2, RAB7B, SFTPA1, SOD2, VDAC3 and VEGFA formed a pool of genes that were predominantly down and up- regulated, respectively in the five categories of human diseases.
Discussion
In the present study, using 91,807 unique tags derived from five SAGE libraries collected from 0 hour mock-infected and 6, 12, 16 and 24 hours PRRSV-infected PAM cells, we identified a total of 699 functionally known genes that showed at least 2.0 fold changes in expression at one of the first three post-infection time points (6, 12 and 16 hours) and were at least 1.5 fold different at the 24 hours post-infection compared to the 0 hour mock infected cells. Our transcriptome profiling represents the largest known set of DE genes of PAMs challenged with PRRSV. The list of DE genes found in our present investigation was extensive, but many were unique as we found that only 8 of 108 (7.4%), 50 of 215 (23%) and 47 of 294 (16%) known coding genes previously reported [19]–[21] were also DE in PRRSV-infected PAMs in our study. Genini and colleagues [19] performed an in vitro study using PAMs obtained from six piglets and challenged with the Lelystad PRRSV strain. The European Lelystad strain of PRRSV has biological similarities but distinct serological properties from the North American VR-2332 isolate [22]. Gene expression was investigated using Affymetrix microarrays, but with very limited annotation available at that time. They detected a total of 1,409 differentially expressed transcripts based on analysis of variance, and found two, five, 25, 16 and 100 transcripts that differed from controls by a minimum of 1.5-fold at 1, 3, 6, 9 and 12 h post-infection, respectively. In addition to three uninfected negative controls, Xiao et al. [20] challenged six conventionally-reared, healthy 6-week-old, crossbred weaned pigs (Landrace×Yorkshire) with the classical North American type PRRSV (N-PRRSV) strain CH 1a. Lung tissues were collected from the control group, three pigs at 96 h (N96) and three pigs at 168 h (N168) post infection and mRNA was extracted. Transcriptome profiling was performed using a Solexa/Illumina next generation sequencing method. Although the authors claimed that there were 5,430 DE genes between all time points (N96/C, N168/C, N168/N96) during infection, they only assigned 215 DE genes to pathways. Using Affymetrix microarrays, Zhou and co-workers [21] reported 294 functionally known genes that were differentially expressed in PAMs derived from three uninfected and three infected 5-week-old Tongcheng pigs at 5 days post infection. The infected groups were challenged with PRRSV-WUH3 by intramuscular inoculation. Here, we performed an in vitro study on PAMs challenged with PRRSV strain VR-2332 and carried out the transcriptome analysis using the SAGE technology. Collectively, these investigations examined responses to infections with different PRRSV strains using either in vitro or in vivo approaches and time courses ranged from 0 to 168 hours. In addition, two different types of tissues, PAMs or lung tissue, were collected for transcriptome profiling using either microarray or tag-based sequencing. Therefore, different experimental designs, transcription profiling formats, time course ranges, virus strains and tissue sources are all reasons that explain the low incidence of common DE genes among the different investigations.
Direct sequencing (whole transcriptome shotgun sequencing or RNA-seq) is likely to yield similar results to SAGE. The Illumina sequencing techniques usually produce sequences with a maximum of 100 bp in length. The major drawbacks of whole transcriptome shotgun sequencing or RNA-seq include insufficient detection of genes/transcripts with low levels of expression, uneven sequencing depth along the length of a transcript and impossible usage of spreadsheet software for data processing due to large file size, (as reviewed by [23]).
Three databases: DAVID (The Database for Annotation, Visualization and Integrated Discovery, v6.7, http://david.abcc.ncifcrf.gov/home.jsp), KEGG (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/pathway.html) and REACTOME (http://www.reactome.org/ReactomeGWT/entrypoint.html) were used in the present study to assign DE genes in PAMs infected with PRRSV to functional pathways. The DAVID Bioinformatics database is owned by the NIH. The team developed a unique single linkage method by which >20 gene identifier types and >40 functional annotation categories from dozens of heterogeneous public databases have been comprehensively integrated in the DAVID Knowledgebase [24]. The KEGG database is described as a resource for understanding high-level functions and utilities of biological systems, such as the cell, the organism and the ecosystem, from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies. The REACTOME tool claims to host a manually curated and peer-reviewed pathway database with cross references to many bioinformatics databases, such as NCBI Entrez Gene, Ensembl and UniProt databases, the UCSC and HapMap Genome Browsers, the KEGG Compound and ChEBI small molecule databases, PubMed, and Gene Ontology. Initially, the DAVID, KEGG and REACTOME databases helped us assign 517, 383 and 369 of 699 DE genes, respectively, to functional pathways. We then combined pathways generated by these three databases and classified them into six systems including 1) cellular processes, 2) genetic information processing, 3) environmental information processing, 4) metabolism, 5) organismal systems and 6) human diseases based on the KEGG classification systems with modifications. Merging the same/similar pathways and editing the overlapping pathways led to functional classification of 573 DE genes. These processes indicate that combining pathway information from different databases helps maximize the coverage of DE genes in pathway analysis.
As shown in Figures 6–11 and Table 1, each of these six systems described above has several functional categories ranging from three in environmental information processing to fifteen in metabolism. The category with the greatest number of DE genes in each of these six systems belongs to the cell growth and death with 191 DE genes identified in cellular processes, the transcription processes with 147 DE genes in the genetic information processing, signal transductions with 126 DE genes in the environmental information processing, energy metabolism with 176 DE genes in metabolism, the immune system with 297 DE genes in organismal systems, and infectious diseases in human disease. These pathway categories with the most DE genes clearly confirmed the basic characteristics of PAMs in pigs responding to PRRSV infection reported by other researchers. For example, Costers and co-workers [25] found that PRRSV stimulates anti-apoptotic pathways in PAMs early in infection and the PRRSV-infected macrophages die by apoptosis late in infection. Gudmundsdottir and Risatti [26] investigated the effect of PRRSV infection on activation of 25 immunomodulatory cellular genes in PAMs at 24 and 48h post-infection and found a regulatory role of PRRSV ORF1A on PAM gene expression. During virus infection, PRRSV modulates the transcription and translation of the host cell to make them survive and propagate [27]. PRRSV infection also suppresses gene expression. Using two-dimensional liquid chromatography-tandem mass spectrometry coupled with isobaric tags for relative and absolute quantification (iTRAQ) labeling approach, Lu and colleagues [28] just recently revealed that signal transduction is one of the differentially expressed proteome components in PAMs infected with PRRSV. Research has shown that viruses and other pathogens usually slow down the host cell’s energy production in order to enhance infection [29], [30]. Resistance response is an expensive activity, which would consume a large amount of energy. Appropriate energy management is important to restrict pathogen propagation or to repair the cells [31]. Innate immunity is critical to the host for defense against various pathogens. PRRSV infection under certain circumstances fails to elicit some components of the innate response [32]. Adaptive immune response is also important to kill viruses. Adaptive immunity depends on antigen presentation, where the MHC (major histocompatibility complex) class II molecule binds antigen to trigger an appropriate adaptive immune response and restrict pathogen growth [33]. We are the first to report DE genes that are common between PRRSV infection and 22 human infectious diseases. In particular, PRRSV infection of PAMs induced DE of 65, 33, 30, 28, 23, 23, and 21 genes that are commonly associated with influenza infection, tuberculosis, HTLV-I infection, Herpes simplex infection HIV Infection, leishmaniasis, and toxoplasmosis, respectively.
A dominant pathway was defined as a pathway with 20 or more DE genes identified in the present study. The numbers of dominant pathways with more than two-thirds of DE genes down- or up-regulated in PAMs infected with PRRSV at 24 h post infection showed some interesting, but different trends among the six systems described above. In the cellular processes and the human diseases systems, the numbers of dominant pathways were relatively even: three down- vs. two up-regulated in the former case and five down- vs. four up-regulated in the latter case. However, in the genetic information processing system there were two down- vs. eight up-regulated pathways and one down- vs. eight up-regulated pathways in the metabolism systems. In contrast, the down- to up-regulated pathway ratio was 5∶1 in the environmental information processing systems and 8∶1 in organismal systems. The dominant pathways with two-thirds of DE genes down-regulated in PAMs infected with PRRSV at 24 h post infection were: actin filament based processes (70%), anti-apoptosis (68%) and positive regulation of cell communication (67%) in cellular processes; protein folding (78%) and protein processing in endoplasmic reticulum (75%) in the genetic information processing systems; signaling by MAPK (85%), NGF (70%) and GPCR (67%), GPCR ligand binding (67%) and positive regulation of signal transduction (65%) in the environmental information processing systems; metabolism of lipids and lipoproteins (71%) in the metabolism systems; positive regulation of developmental processes (77%), response to unfolded protein (77%), developmental biology (74%), axon guidance (70%), positive regulation of immune response (70%), response to organic substances (68%), response to hormone stimulus (67%) and response to abiotic stimulus (67%) in organismal systems; and leishmaniasis (91%), toxoplasmosis (86%), HTLV-I infection (73%), tuberculosis (73%) and Herpes simplex infection (71%) in the human diseases system (Table 1). The dominant pathways with two-thirds of DE genes up-regulated in PAMs infected with PRRSV at 24 h post infection included: membrane organization (68%) and lysosome (68%) in cellular processes; gene expression (66%), nonsense-mediated decay (85%), translation (77%), translation elongation (84%), translation initiation (86%), ribosome (89%), translation termination (90%) and SRP-dependent cotranslational protein targeting to membrane (91%) in the genetic information processing systems; transmembrane transport of small molecules (68%) in the environmental information processing systems; generation of precursor metabolites and energy (82%), the citric acid cycle and respiratory electron transport (74%), respiratory electron transport (74%), ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins (73%), oxidation reduction and oxidative phosphorylation (68%), metabolism of proteins (75%), hexose metabolic processes (67%) and glucose metabolic processes (71%) in the metabolism systems; vesicle docking during exocytosis (70%) in the organismal systems; and HIV Infection (83%), Huntington's disease (82%), Parkinson's disease (77% and Alzheimer's disease (70%) in the human diseases system (Table 1).
As shown in Figure 5, we classified these 699 DE genes into ten clusters based on their expression trends. The abundance of DE genes in cluster A increased from initial infection until 16 h post-infection. Thereafter, gene abundances decreased, but remained up-regulated at 24 h post infection. In comparison, DE gene abundances in Cluster B did not change between 0 h and 6 h post-infection, but rapidly increased by 16 h, and increased slightly at 24 h post infection. The relative abundances of DE genes in cluster C decreased to their lowest levels at 6 h, but gradually increased to reach their highest levels at 24 h post infection. The DE genes in cluster D were up-regulated at 6 h, returned to pre-infection levels between 12 h and 16 h, and were dramatically up-regulated at 24 h post infection. Abundances of DE genes in both clusters H and I decreased dramatically by 6 h post-infection. Expression levels of genes in cluster H slowly returned to pre-infection amounts by 24 h post-infection, while gene abundances in cluster I remained at similar down-regulated levels between 6 h and 24 h post-infection. The DE genes in cluster J were significantly down-regulated by 12 h and remained at similar levels until 24 h post infection. Overall, genes in clusters A, B, C and D are up-regulated, while genes in clusters H, I and J are down-regulated at 24 h post infection. For each system, we identified DE genes that were involved in 10 and more pathways and those that were exclusively included in the systems. In the cellular processes and the organismal systems, we found that more than half of the multi-functional DE genes (17/30 = 57% for the former system and 40/70 = 57% for the latter system) had five point expression patterns clustered in H, I and J, while about half of the exclusively expressed DE genes (12/24 = 50% for the former system and 15/31 = 48% for the latter system) were grouped in clusters A, B and C. In contrast, half of the multi-functional DE genes (6/12 = 50%) in the genetic information processing were clustered in B, C and D, while 12 of 21 (57%) exclusively expressed DE genes in the same system fell into clusters H, I and J. Interestingly, the majority of both multi-functional DE genes (28/55 = 51%) and exclusively expressed DE genes (28/46 = 61%) in the metabolism systems had the same expression trends clustered in A, B and C. A total of 19 DE genes identified in PAMs infected with PRRSV are involved in at least 10 human diseases and most of them (17/19 = 89%) were clustered in H, I and J. System-specific expression patterns were not identified in the environmental information processing and human disease systems because a limited number of multi-functional and exclusively expressed DE genes were identified in these systems.
In addition to those system-based features in PAMs infected with PRRSV, we also observed other specific features related to PRRSV infection. Among 699 DE genes discovered in the present study, 206 were commonly down-regulated genes at different infection time points (Figure 4C) and they were involved in many function processes. Our data showed the signal transduction genes (NFKB1, NFKB2, JUN, JUNB and FOS) that trigger immune and inflammatory responses were significantly decreased. As a result, the proinflammatory cytokine genes (IL-1A, IL-1B and TNF) and chemokine genes (CCL23 and CCL3L1) were down-regulated. The receptors of cytokines and chemokines (IL1RN and CCRL1) were also down-regulated. The combined effects may allow PRRSV to avoid an effective immune response. In addition, complement system activation seemed to be blocked during PRRSV infection. C1QB, C3, C1QC, and FCN2, activators of the lectin pathway of the complement system, were down-regulated [34]. The Heat shock proteins (HSPs) genes (HSP90AA1, HSP90AB1, HSP90B1, HSPA1A, HSPA1B, HSPA5, HSPA6, HSPA8, HSPB1 and HSPH1) were negatively regulated during PRRSV infection. Heat stress proteins interact with viral proteins and enhance development of innate and adaptive immune responses against invading pathogens [35]; therefore, the down-regulation of HSPs in PRRSV-infected PAMs may have resulted in a weakened innate immune response. In addition, a recent study has identified that HSPA5 closely associates with PRRSV nsp2 and as such may be involved in PRRSV replication complexes [36].
The collection of MHC II genes (SLA-DMB, SLA-DOA, SLA-DQA1, SLA-DQA2, SLA-DQB1, SLA-DRA, and SLA-DRB1) and cell surface molecules (CD74 and CD83) that help MHC II bind antigen were all down-regulated. PRRSV inhibits cell death or apoptosis, which allows a prolonged infection [37]. Negative regulation of apoptosis or cell death genes (IER3, CFLAR, YWHAZ, PIM2, ANXA4, HMOX1and BAG3) were also down-regulated. CD163 is a PRRSV receptor that takes part in the internalization and uncoating of the virus. Down-regulation of CD163 could reduce PRRSV replication [38]. On the other hand, only 130 DE genes were commonly up-regulated at all four infected time stages. In particular, the up-regulated genes are involved in regulation of apoptosis and cell death (MAEA, BNIP3, IDO1, GPX1, EI24, and TIAL1) and oxygen metabolism (ACE, VEGFA, BNIP3 and EGLN2) (Figure 4B). Superoxide dismutases (SOD1 and SOD2) involved in the ubiquitin - proteasome pathway were significantly up-regulated, and the genes that encode proteasome subunits such as PSMB1, PSMB2 and PSME2 were also increased. Cyclooxygenase genes (COX1 and COX2) which participate in virus replication and the regulation of inflammatory response following viruses infection [39] were up-regulated at four time points post-infection. The S100 family gene S100A6 is involved in actin and tubulin cytoskeleton organization. Interferon regulatory transcription factor IRF3, which regulates IFN-β expression was activated at all four stages.
PRRSV affected expression of genes in PAMs at critical time points after infection. Interestingly, at 6 h post-infection, most of the affected genes were down-regulated, while many genes were up-regulated at 24 h post infection. At 6 h post infection, the energy metabolism related genes (ATP5G2, COX17, COX5B, GSTP1, NDUFAB1 and NDUFS2) were suppressed. Transcriptional and protein synthesis genes (H1FX and OBFC2B) were also down-regulated. The antiviral gene BST2, which encodes the protein tetherin could directly restrict various viruses; however, BST2 abundance was reduced by PRRSV infection [40], [41]. In addition, PRRSV infection of PAMs decreased the viral replication regulator ANAPC11 to control virus replication. Consistent with a previous report [42], genes in the DUSP family (DUSP1, DUSP2, DUSP5 and DUSP6), which regulate MAPK signaling, were down-regulated at four time points post-infection. At 24 h post infection, when the viral replication is exponential, an increasing amount of protein is needed to assemble whole virions. Consistent with this protein need, PRRSV up-regulated expression of several ribosomal protein genes (RPL15, RPL29, RPL34, RPL36A, RPL41, RPL9, RPLP0, RPLP1, RPS13, RPS16, RPS18, RPS19, RPS27L and RPS7) to make translation more productive. Selected antiviral genes (IFI6, IFIT3 and IFITM3) were also up-regulated, indicating that PAMs were attempting to control the viral infection. S100A9, another member of the S100 family that is involved in induction of the inflammatory response common to many pathogen infections, was also significantly up-regulated. This gene and family was also found to be in the top ten up-regulated genes in the tracheobronchial lymph nodes of pigs infected with highly pathogenic PRRSV rJXwn06 versus control at 14 days post infection [43]. Genes that were down-regulated genes at 24h post infection included inflammatory and anti-pathogen genes (CSF1 and USP2) and the cell apoptosis and death genes (MICB, BUB1B, ITGB2, UBB and BIRC3).
The host-pathogen interaction is another feature to discuss as a virus develops many ways to manipulate host gene expression [44]. During PRRSV infection, PRRSV modulates the transcription and translation of the host cell to enable propagation and survival of the virus [27]. PRRSV infection of PAMS resulted in suppressed expression of IL1α, IL1β and TNF-α, which are important proinflammatory cytokines known to elicit innate immunity to restrict virus replication. Normally, virus infection may enhance IL-1α, IL-1β and TNF-α expression that consequently inhibits virus replication [45]. Lack of early TNF expression may be a method that PRRSV utilizes to evade aspects of the innate host immune response. Therefore, PRRSV evades the early lines of defense by effectively blocking the expression of important innate/inflammatory genes. The abundances of IL1, IL6 and TNF were 10–100 times less in PRRSV single inoculated pigs than PRRSV-LPS inoculated pigs [46]. Similar results were observed in PAMs during PRRSV infection [47]. Our data showed that PRRSV infection restricts expression of these genes from 6 h to 24 h post infection. The complement pathway supports phagocytosis through opsonization and subsequent elimination of pathogens [48]. Down-regulated C3 and other complement pathway genes may weaken antiviral ability of phagocytic cells such as PAMs. Similar results were observed in PAMs during HP-PRRSV infection in vivo [49]. However, Xiao et al. showed that the complement system was activated in lung tissue during PRRSV infection, which may have caused severe lung damage [50], [51]. In the present study, the IFN-stimulated genes (IFIT1, IFIT3, MX1, ISG15, OAS1, and IFI6) were dramatically increased at 16 h and 24 h post infection, which should trigger powerful antiviral functions. This represents a positive signal for a host cell responding to PRRSV infection. Previous studies have shown that very low or negligible levels of IFN-α are produced upon PRRSV infection in pulmonary alveolar macrophages (PAMs) and PRRSV permissive monkey kidney cells (MARC-145) in vitro [52], [53]. IFN-α production in the lungs of pigs acutely infected with PRRSV was either almost undetectable or 100- to 200-fold lower than that induced by porcine respiratory coronavirus (PRCV) [54], [55]. PRRSV has also been found to suppress IFN-α production by transmissible gastroenteritis corona virus (TGEV), a known inducer of IFNs in infected alveolar macrophages [52]. At the same time, externally provided IFN-α or IFN-β have been able to reduce viral replication in cultured alveolar macrophages [52], [56]. PRRSV is thought to suppress type I IFN expression and block its signaling by interfering with STAT1/STAT2 nuclear translocation [57]. The virus was also found to inhibit the dsRNA-mediated up-regulation of IFN-β gene transcription [53]. A microarray analysis of PAMs infected with Lelystad virus (European type PRRSV) showed no significant change in the IFN-α from the control at 12 h post-infection [19]. IRF3, which plays an important role in activating type I interferon was up-regulated at all infected time stages. However, we did not detect differential expression of type I IFN over the course of infection. It is possible that type I IFN might have been induced before 6 h, which was our first PAM collection time. It has been shown the PRRSV can trigger the activation of IRF-3 as well as induce IFN production at 24 h post infection but the activities are much lower than those triggered by Poly(I:C) and PRRSV nsp1 antagonizes IFN production through the TLR3 and RIG-I pathways and down-regulates the protein level of IRF-3 [58]. Pre-treatment of PAMs with LPS down-regulated expression of CD163, a PRRSV receptor involved in PRRSV uncoating, and restricted PRRSV replication [38], [59]. In the present study, CD163 was significantly decreased after PRRSV infection, which indicates that viral evasion methods in PAMs were actively induced.
In conclusion, our current study revealed the largest known set of 699 DE genes in PAMs challenged with PRRSV, which are involved in six biological systems, 60 functional categories and 504 pathways. The major reactomes of PAMs responding to PRRSV infection included cell growth and death, transcription processes, signal transductions, energy metabolism, immune system and infectious diseases. In particular, PRRSV infection dramatically minimized pathway functions involving the actin filament based processes, anti-apoptosis, positive regulation of cell communication, protein folding, protein processing in endoplasmic reticulum, signaling by MAPK, NGF and GPCR, GPCR ligand binding, positive regulation of signal transduction, metabolism of lipids and lipoproteins, positive regulation of developmental processes, response to unfolded protein, developmental biology, axon guidance, positive regulation of immune responses, response to organic substances, response to hormone stimulus and response to abiotic stimulus in PAMs. However, PRRSV invasion maximized pathway functions related to membrane organization, lysosome, gene expression, nonsense-mediated decay, translation, translation elongation, translation initiation, ribosome, translation termination, SRP-dependent cotranslational protein targeting to membrane, transmembrane transport of small molecules, generation of precursor metabolites and energy, the citric acid cycle and respiratory electron transport, respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins, oxidation reduction and oxidative phosphorylation, metabolism of proteins, hexose metabolic processes, glucose metabolic processes and vesicle docking during exocytosis in PAMs. Overall, PRRSV took control of PAMs in the course of a 24 hour infection, but the host started to fight back using its autophagy mechanisms.
Materials and Methods
Ethics Statement
The animal use protocol was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the National Animal Disease Center-USDA-Agricultural Research Service.
Cells, Virus Infection and SAGE Analysis
The experiments were conducted as previously described [60], [61]. In brief, PAM cells were harvested from three clinically healthy, PRRS-negative gilts 6–8 weeks of age and tested free by PCR for both porcine circovirus and Mycoplasma spp. Primary PAM were isolated, cultured, and infected, as previously described [62]. Aliquots of PAMs were then frozen and stored in liquid nitrogen separately for all three pigs. After establishing PAMs in culture, cells were infected with PRRSV strain VR-2332. To achieve a near synchronous infection, flasks containing adherent PAMs were infected at a multiplicity of infection (MOI) of 10 in chilled media and incubated at 4°C for 1 hour to allow for virus binding, but not entry into the cell. Pre-warmed media was added and the cells placed at 37°C, 5% CO2 until collected for RNA isolation. Total cellular RNA was prepared using the Qiagen RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Cell samples were collected from each PRRSV-infected PAM flask at 0, 6, 12, 16 or 24 hours after infection. Total cellular RNA from mock-infected PAMs was collected at 0 and 24 hours. Equimolar amounts of total RNA from the PAMs of each pig at each time point were then pooled to make SAGE (serial analysis of gene expression) libraries using NlaIII as the anchoring enzyme and BsmFI as the tagging enzyme [63]. The SAGE libraries provided the population means of the transcript abundance levels for each time point. SAGE clones were amplified and sequenced using a high-throughput sequencing pipeline with an ABI 3730 automated sequencer and ABI chemistry (Applied Biosystems Inc., Foster City, CA). The SAGE libraries with tag counts were submitted to GenBank GEO and have the accession number GSE10346.
Detection of Differentially Expressed SAGE Tags
We assumed that the counts of the ith SAGE tags,  , followed a Poisson distribution defined as:
, followed a Poisson distribution defined as:
| (1) |
We then let 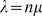 where
where  was interpreted as the “true” frequency that the ith SAGE tag being expressed, among n expressed SAGE tags in total, such that
was interpreted as the “true” frequency that the ith SAGE tag being expressed, among n expressed SAGE tags in total, such that
| (2) |
Here, both 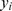 and
and 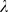 are not comparable between statuses, because the library sizes can vary. We can however, compare
are not comparable between statuses, because the library sizes can vary. We can however, compare 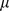 ’s among statuses (i.e., libraries) because they quantify the underlying “true” frequencies of SAGE expression.
’s among statuses (i.e., libraries) because they quantify the underlying “true” frequencies of SAGE expression.
Within the Bayesian framework, we assumed a conjugate Gamma prior distribution, Gamma (a, b), for the parameter 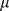 , then, the posterior distribution of
, then, the posterior distribution of 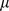 is also a Gamma density with parameters
is also a Gamma density with parameters  and
and  ,
,
| (3) |
Now, we considered 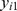 and
and 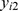 are counts of the i
th SAGE tag at two time points,
are counts of the i
th SAGE tag at two time points, 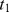 and
and 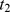 , respectively, and
, respectively, and 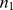 and
and 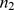 are the total numbers of SAGE tags measured at these two time points. Under the assumption of heterogeneity
are the total numbers of SAGE tags measured at these two time points. Under the assumption of heterogeneity 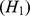 , the means at the two time points are different, that is,
, the means at the two time points are different, that is, 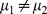 . Then, the likelihood function is
. Then, the likelihood function is
| (4) |
Given a Gamma prior distribution, Gamma (a,b), to 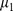 and
and 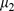 , we showed that their posterior distributions are also Gamma:
, we showed that their posterior distributions are also Gamma:
| (5a) |
| (5b) |
Under the assumption of homogeneity 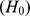 ,
, 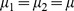 , the likelihood function was
, the likelihood function was
| (6) |
Then the posterior distribution of 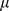 was:
was:
| (7) |
In Bayesian framework, this hypothesis test that contrasts both models (hypotheses) was conducted using the Bayes Factor [64].
Differential gene expression is commonly measured by computing log ratios. In the present study, we similarly computed log ratios (pLR) of posterior frequencies of SAGE tags between each of the treatment time periods (6 h, 12 h, 16 h, or 24 h) and the normal status, as follows:
| (8) |
Alternatively, differential gene expression between two time points can be evaluated by computing the following probability based on posterior samples:
| (9) |
Hence, we numerically constructed the 95% highest posterior density intervals (95% HPD) for 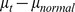 , and differential expression of a SAGE tag was claimed to be true if one of the following criteria held, or false otherwise:
, and differential expression of a SAGE tag was claimed to be true if one of the following criteria held, or false otherwise:
| (10a) |
| (10b) |
In the present study, we employed both criteria, (8) and (10), to identify differentially expressed (DE) SAGE tags.
Assignment of DE Tags to Genes
We downloaded a total of 52,121 porcine mRNA sequences from the GenBank database at National Center for Biotechnology Information (NCBI). A Java program was developed to identify the 3′ most NlaIII cut site for each mRNA sequence and collect the sequence of 10 nucleotides following the anchoring enzyme cut site. The process resulted in 48,988 tags collected from 52,121 mRNA sequences. Interestingly, 95 mRNA sequences had the same AAAAAAAAAA tag and were subsequently deleted from the analysis. We also simply assumed that among the remaining tags, any repeated mRNA sequences with different accession numbers belonged to the same gene. By excluding all repeats, we compiled a list of 26,745 unique tags for different pig mRNAs. The unique mRNA tags were then merged with the DE SAGE tags identified above to determine DE mRNAs of PAMs infected with PRRSV. The mRNA sequences were then annotated for orthologs in the human genome against the Refseq database, as the human genome has been well annotated.
Identification of DE Genes for Pathway Analysis
In order to select DE genes for pathway analysis, we arbitrarily required that each DE gene had at least one time point (6 h, 12 h, 16 h, and 24 h, respectively) that was at least a 2 fold change and 100 tags per million (TPM) different from the 0 h mock-infected control. In addition, we also required that each DE gene was at least 1.5 fold different between the 24 h-infected and the 0 hour mock-infected cells. Generally speaking, we assumed that a cell should express ∼10,000 genes at a given time [65]. As such, each gene should average 100 TPM when a million SAGE tags are sequenced. Therefore, we considered 100 TPM as a minimum requirement to signify a change of functional importance for a DE gene. The associated pathways of all DE genes were identified using DAVID, (http://david.abcc.ncifcrf.gov/home.jsp), KEGG Pathway (http://www.genome.jp/kegg/pathway.html), and Reactome (http://www.reactome.org/ReactomeGWT/entrypoint.html) databases. The DE gene clustering was performed using the self-organizing map (SOM) as described previously [65]. A SOM is a type of artificial neural network that uses unsupervised learning to produce a low-dimensional, discretized representation of the input space of the training samples, called a map. It consists of components called nodes or neurons. Each node is associated with a weight vector of the same dimension (as the input data vectors and a position in the map space). To place a vector from data space onto the map, the method first finds the node with the closest weight vector to the vector taken from data space. Once the closest node is located, it is given the values from the vector taken from the data space.
SOMs operate in two modes: training and mapping. Training builds the map using input examples (data), which features competitive learning, and mapping automatically classifies a new input vector. When a training example is fed to the network, the method computes its Euclidean distance to all weight vectors. The neuron with weight vector that is most similar to this input is called the best matching unit (BMU). The weights of the BMU and neurons close to it in the SOM are adjusted towards the input vector. The magnitude of the change decreases with time and with distance from the BMU. The update formula for a neuron with weight vector W v(t) is as follows:
where α(t) is a monotonically decreasing learning coefficient and D(t) is the input vector. The neighborhood function θ(v, t) depends on the distance between the BMU and neuron v. The neighborhood function shrinks with time. At the beginning when the neighborhood is broad, the self-organizing takes place on the global scale. When the neighborhood has shrunk to just a couple of neurons the weights are converging to local estimates. This process is repeated for each input vector for a number of cycles (denoted as λ, usually in a few hundreds of cycles). Typically, SOMs with a small number of nodes behave similarly to K-means, whereas larger SOMs can rearrange data in a way that is fundamentally topological in character [66].
Supporting Information
PRRSV tags, row counts and TPM detected.
(XLSX)
DE tags, row counts and TPM detected post Bayesian analysis.
(XLSX)
DE genes assigned by SOM analysis to clusters based on their expression trends regardless of their fold-change magnitudes along time points (0 h, 6 h, 12 h, 16 h and 24 h).
(XLSX)
Acknowledgments
We would like to thank W. Boatwright, S. Anderson and R. Steeves for technical assistance, and S. Ohlendorf for secretarial assistance in preparation of the manuscript. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Funding Statement
This work was supported by the National Pork Board #06-122, USDA ARS and PRRS CAP, USDA NIFA Award 2008-55620-19132. Mr. Xiang Zhou is recipient of the China Scholarship Council assistantship for a joint Ph.D. program between Washington State University, USA and Huazhong Agricultural University, China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Goyal SM (1993) Porcine reproductive and respiratory syndrome. J Vet Diagn Invest 5: 656–664. [DOI] [PubMed] [Google Scholar]
- 2. Rossow KD (1998) Porcine reproductive and respiratory syndrome. Vet Pathol 35: 1–20. [DOI] [PubMed] [Google Scholar]
- 3. Albina E, Madec F, Cariolet R, Torrison J (1994) Immune response and persistence of the porcine reproductive and respiratory syndrome virus in infected pigs and farm units. Vet Rec 134: 567–573. [DOI] [PubMed] [Google Scholar]
- 4. Park BK, Joo HS (1997) Induction of dual infections in newborn and three-week-old pigs by use of two plaque size variants of porcine reproductive and respiratory syndrome virus. Am J Vet Res 58: 257–259. [PubMed] [Google Scholar]
- 5. Nelsen CJ, Murtaugh MP, Faaberg KS (1999) Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol 73: 270–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tian K, Yu X, Zhao T, Feng Y, Cao Z, et al. (2007) Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PloS one 2: e526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McOrist S, Khampee K, Guo A (2011) Modern pig farming in the People's Republic of China: growth and veterinary challenges. Revue scientifique et technique 30: 961–968. [DOI] [PubMed] [Google Scholar]
- 8. An TQ, Tian ZJ, Leng CL, Peng JM, Tong GZ (2011) Highly pathogenic porcine reproductive and respiratory syndrome virus, Asia. Emerging infectious diseases 17: 1782–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang YW, Meng XJ (2010) Novel strategies and approaches to develop the next generation of vaccines against porcine reproductive and respiratory syndrome virus (PRRSV). Virus Research 154: 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sang Y, Rowland RR, Blecha F (2011) Interaction between innate immunity and porcine reproductive and respiratory syndrome virus. Animal health research reviews/Conference of Research Workers in Animal Diseases 12: 149–167. [DOI] [PubMed] [Google Scholar]
- 11. Sun XZ, Wertz N, Lager KL, Tobin G, Butler JE (2012) Antibody repertoire development in fetal and neonatal piglets. XXIII: fetal piglets infected with a vaccine strain of PRRS Virus display the same immune dysregulation seen in isolator piglets. Vaccine 30: 3646–3652. [DOI] [PubMed] [Google Scholar]
- 12. Butler JE, Wertz N, Weber P, Lager KM (2008) Porcine reproductive and respiratory syndrome virus subverts repertoire development by proliferation of germline-encoded B cells of all isotypes bearing hydrophobic heavy chain CDR3. Journal of immunology 180: 2347–2356. [DOI] [PubMed] [Google Scholar]
- 13. Butler JE, Lemke CD, Weber P, Sinkora M, Lager KM (2007) Antibody repertoire development in fetal and neonatal piglets: XIX. Undiversified B cells with hydrophobic HCDR3s preferentially proliferate in the porcine reproductive and respiratory syndrome. Journal of immunology 178: 6320–6331. [DOI] [PubMed] [Google Scholar]
- 14. Albina E (1997) [Porcine reproductive and respiratory syndrome: ten years of experience (1986–1996) with this undesirable viral infection]. Vet Res 28: 305–352. [PubMed] [Google Scholar]
- 15. Dokland T (2010) The structural biology of PRRSV. Virus Research 154: 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Breedam W, Delputte PL, Van Gorp H, Misinzo G, Vanderheijden N, et al. (2010) Porcine reproductive and respiratory syndrome virus entry into the porcine macrophage. J Gen Virol 91: 1659–1667. [DOI] [PubMed] [Google Scholar]
- 17. Miller LC, Neill JD, Harhay GP, Lager KM, Laegreid WW, et al. (2010) In-depth global analysis of transcript abundance levels in porcine alveolar macrophages following infection with porcine reproductive and respiratory syndrome virus. Advances in Virology 2010: 864181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu ST, Sharon-Friling R, Ivanova P, Milne SB, Myers DS, et al. (2011) Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proceedings of the National Academy of Sciences of the United States of America 108: 12869–12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Genini S, Delputte PL, Malinverni R, Cecere M, Stella A, et al. (2008) Genome-wide transcriptional response of primary alveolar macrophages following infection with porcine reproductive and respiratory syndrome virus. J Gen Virol 89: 2550–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiao S, Jia J, Mo D, Wang Q, Qin L, et al. (2010) Understanding PRRSV infection in porcine lung based on genome-wide transcriptome response identified by deep sequencing. PloS one 5: e11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou P, Zhai S, Zhou X, Lin P, Jiang T, et al. (2011) Molecular characterization of transcriptome-wide interactions between highly pathogenic porcine reproductive and respiratory syndrome virus and porcine alveolar macrophages in vivo. International journal of biological sciences 7: 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murtaugh MP, Elam MR, Kakach LT (1995) Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch Virol 140: 1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Malone JH, Oliver B (2011) Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol 9: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang da W, Sherman BT, Tan Q, Collins JR, Alvord WG, et al. (2007) The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome biology 8: R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Costers S, Lefebvre DJ, Delputte PL, Nauwynck HJ (2008) Porcine reproductive and respiratory syndrome virus modulates apoptosis during replication in alveolar macrophages. Archives of virology 153: 1453–1465. [DOI] [PubMed] [Google Scholar]
- 26. Gudmundsdottir I, Risatti GR (2009) Infection of porcine alveolar macrophages with recombinant chimeric porcine reproductive and respiratory syndrome virus: effects on cellular gene transcription and virus growth. Virus Research 145: 145–150. [DOI] [PubMed] [Google Scholar]
- 27. Kimman TG, Cornelissen LA, Moormann RJ, Rebel JM, Stockhofe-Zurwieden N (2009) Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine 27: 3704–3718. [DOI] [PubMed] [Google Scholar]
- 28. Lu Q, Bai J, Zhang L, Liu J, Jiang Z, et al. (2012) Two-dimensional liquid chromatography-tandem mass spectrometry coupled with isobaric tags for relative and absolute quantification (iTRAQ) labeling approach revealed first proteome profiles of pulmonary alveolar macrophages infected with porcine reproductive and respiratory syndrome virus. Journal of proteome research 11: 2890–2903. [DOI] [PubMed] [Google Scholar]
- 29. Seo JY, Yaneva R, Hinson ER, Cresswell P (2011) Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science 332: 1093–1097. [DOI] [PubMed] [Google Scholar]
- 30. Dionne MS, Pham LN, Shirasu-Hiza M, Schneider DS (2006) Akt and FOXO dysregulation contribute to infection-induced wasting in Drosophila. Curr Biol 16: 1977–1985. [DOI] [PubMed] [Google Scholar]
- 31. Schneider DS, Ayres JS (2008) Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol 8: 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lunney JK, Chen H (2010) Genetic control of host resistance to porcine reproductive and respiratory syndrome virus (PRRSV) infection. Virus Res 154: 161–169. [DOI] [PubMed] [Google Scholar]
- 33. Yaneva R, Schneeweiss C, Zacharias M, Springer S (2010) Peptide binding to MHC class I and II proteins: new avenues from new methods. Mol Immunol 47: 649–657. [DOI] [PubMed] [Google Scholar]
- 34. Garred P, Honore C, Ma YJ, Munthe-Fog L, Hummelshoj T (2009) MBL2, FCN1, FCN2 and FCN3-The genes behind the initiation of the lectin pathway of complement. Mol Immunol 46: 2737–2744. [DOI] [PubMed] [Google Scholar]
- 35. Oglesbee MJ, Pratt M, Carsillo T (2002) Role for heat shock proteins in the immune response to measles virus infection. Viral Immunol 15: 399–416. [DOI] [PubMed] [Google Scholar]
- 36. Han J, Rutherford MS, Faaberg KS (2010) Proteolytic products of the porcine reproductive and respiratory syndrome virus nsp2 replicase protein. Journal of virology 84: 10102–10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hardwick JM (2001) Apoptosis in viral pathogenesis. Cell Death Differ 8: 109–110. [DOI] [PubMed] [Google Scholar]
- 38. Patton JB, Rowland RR, Yoo D, Chang KO (2009) Modulation of CD163 receptor expression and replication of porcine reproductive and respiratory syndrome virus in porcine macrophages. Virus Res 140: 161–171. [DOI] [PubMed] [Google Scholar]
- 39. Steer SA, Corbett JA (2003) The role and regulation of COX-2 during viral infection. Viral Immunol 16: 447–460. [DOI] [PubMed] [Google Scholar]
- 40. Lopez LA, Yang SJ, Hauser H, Exline CM, Haworth KG, et al. (2010) Ebola virus glycoprotein counteracts BST-2/Tetherin restriction in a sequence-independent manner that does not require tetherin surface removal. J Virol 84: 7243–7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sauter D, Specht A, Kirchhoff F (2010) Tetherin: holding on and letting go. Cell 141: 392–398. [DOI] [PubMed] [Google Scholar]
- 42. Salojin K, Oravecz T (2007) Regulation of innate immunity by MAPK dual-specificity phosphatases: knockout models reveal new tricks of old genes. J Leukoc Biol 81: 860–869. [DOI] [PubMed] [Google Scholar]
- 43. Miller LC, Fleming D, Arbogast A, Bayles DO, Guo B, et al. (2012) Analysis of the swine tracheobronchial lymph node transcriptomic response to infection with a Chinese highly pathogenic strain of porcine reproductive and respiratory syndrome virus. BMC Vet Res 8: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alcami A, Koszinowski UH (2000) Viral mechanisms of immune evasion. Trends Microbiol 8: 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ryan JG, Kastner DL (2008) Fevers, genes, and innate immunity. Curr Top Microbiol Immunol 321: 169–184. [DOI] [PubMed] [Google Scholar]
- 46. Van Gucht S, Labarque G, Van Reeth K (2004) The combination of PRRS virus and bacterial endotoxin as a model for multifactorial respiratory disease in pigs. Vet Immunol Immunopathol 102: 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thanawongnuwech R, Thacker B, Halbur P, Thacker EL (2004) Increased production of proinflammatory cytokines following infection with porcine reproductive and respiratory syndrome virus and Mycoplasma hyopneumoniae. Clin Diagn Lab Immunol 11: 901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu H, Liu B, Frost JL, Hong S, Jin M, et al.. (2012) Complement component C3 and complement receptor type 3 contribute to the phagocytosis and clearance of fibrillar Abeta by microglia. Glia. [DOI] [PMC free article] [PubMed]
- 49. Zhou P, Zhai S, Zhou X, Lin P, Jiang T, et al. (2011) Molecular characterization of transcriptome-wide interactions between highly pathogenic porcine reproductive and respiratory syndrome virus and porcine alveolar macrophages in vivo. Int J Biol Sci 7: 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Lookeren Campagne M, Wiesmann C, Brown EJ (2007) Macrophage complement receptors and pathogen clearance. Cell Microbiol 9: 2095–2102. [DOI] [PubMed] [Google Scholar]
- 51. Xiao S, Mo D, Wang Q, Jia J, Qin L, et al. (2010) Aberrant host immune response induced by highly virulent PRRSV identified by digital gene expression tag profiling. BMC genomics 11: 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Albina E, Carrat C, Charley B (1998) Interferon-alpha response to swine arterivirus (PoAV), the porcine reproductive and respiratory syndrome virus. J Interferon Cytokine Res 18: 485–490. [DOI] [PubMed] [Google Scholar]
- 53. Miller LC, Laegreid WW, Bono JL, Chitko-McKown CG, Fox JM (2004) Interferon type I response in porcine reproductive and respiratory syndrome virus-infected MARC-145 cells. Arch Virol 149: 2453–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Buddaert W, Van Reeth K, Pensaert M (1998) In vivo and in vitro interferon (IFN) studies with the porcine reproductive and respiratory syndrome virus (PRRSV). Adv Exp Med Biol 440: 461–467. [DOI] [PubMed] [Google Scholar]
- 55. Van Reeth K, Labarque G, Nauwynck H, Pensaert M (1999) Differential production of proinflammatory cytokines in the pig lung during different respiratory virus infections: correlations with pathogenicity. Res Vet Sci 67: 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Overend C, Mitchell R, He D, Rompato G, Grubman MJ, et al. (2007) Recombinant swine beta interferon protects swine alveolar macrophages and MARC-145 cells from infection with Porcine reproductive and respiratory syndrome virus. J Gen Virol 88: 925–931. [DOI] [PubMed] [Google Scholar]
- 57. Patel D, Nan Y, Shen M, Ritthipichai K, Zhu X, et al. (2010) Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J Virol 84: 11045–11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shi X, Wang L, Zhi Y, Xing G, Zhao D, et al. (2010) Porcine reproductive and respiratory syndrome virus (PRRSV) could be sensed by professional beta interferon-producing system and had mechanisms to inhibit this action in MARC-145 cells. Virus Res 153: 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Welch SK, Calvert JG (2010) A brief review of CD163 and its role in PRRSV infection. Virus Res 154: 98–103. [DOI] [PubMed] [Google Scholar]
- 60. Miller LC, Harhay GP, Lager KM, Smith TP, Neill JD (2008) Effect of porcine reproductive and respiratory syndrome virus on porcine alveolar macrophage function as determined using serial analysis of gene expression (SAGE). Dev Biol (Basel) 132: 169–174. [DOI] [PubMed] [Google Scholar]
- 61. Miller LC, Neill JD, Harhay GP, Lager KM, Laegreid WW, et al. (2010) In-depth global analysis of transcript abundance levels in porcine alveolar macrophages following infection with porcine reproductive and respiratory syndrome virus. Adv Virol 2010: 864181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chiou MT, Jeng CR, Chueh LL, Cheng CH, Pang VF (2000) Effects of porcine reproductive and respiratory syndrome virus (isolate tw91) on porcine alveolar macrophages in vitro [In Process Citation]. Vet Microbiol 71: 9–25. [DOI] [PubMed] [Google Scholar]
- 63. Velculescu VE, Zhang L, Vogelstein B, Kinzler KW (1995) Serial analysis of gene expression. Science 270: 484–487. [DOI] [PubMed] [Google Scholar]
- 64. Raftery AE (1995) Bayesian model selection in social research (with discussion). Sociological Methodology 25: 111–193. [Google Scholar]
- 65. Wu XL, Griffin KB, Garcia MD, Michal JJ, Xiao Q, et al. (2004) Census of orthologous genes and self-organizing maps of biologically relevant transcriptional patterns in chickens (Gallus gallus). Gene 340: 213–225. [DOI] [PubMed] [Google Scholar]
- 66.Haykin S (1999) Self-organizing maps. Neural networks - A comprehensive foundation. 2nd ed: Prentice-Hall.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRRSV tags, row counts and TPM detected.
(XLSX)
DE tags, row counts and TPM detected post Bayesian analysis.
(XLSX)
DE genes assigned by SOM analysis to clusters based on their expression trends regardless of their fold-change magnitudes along time points (0 h, 6 h, 12 h, 16 h and 24 h).
(XLSX)