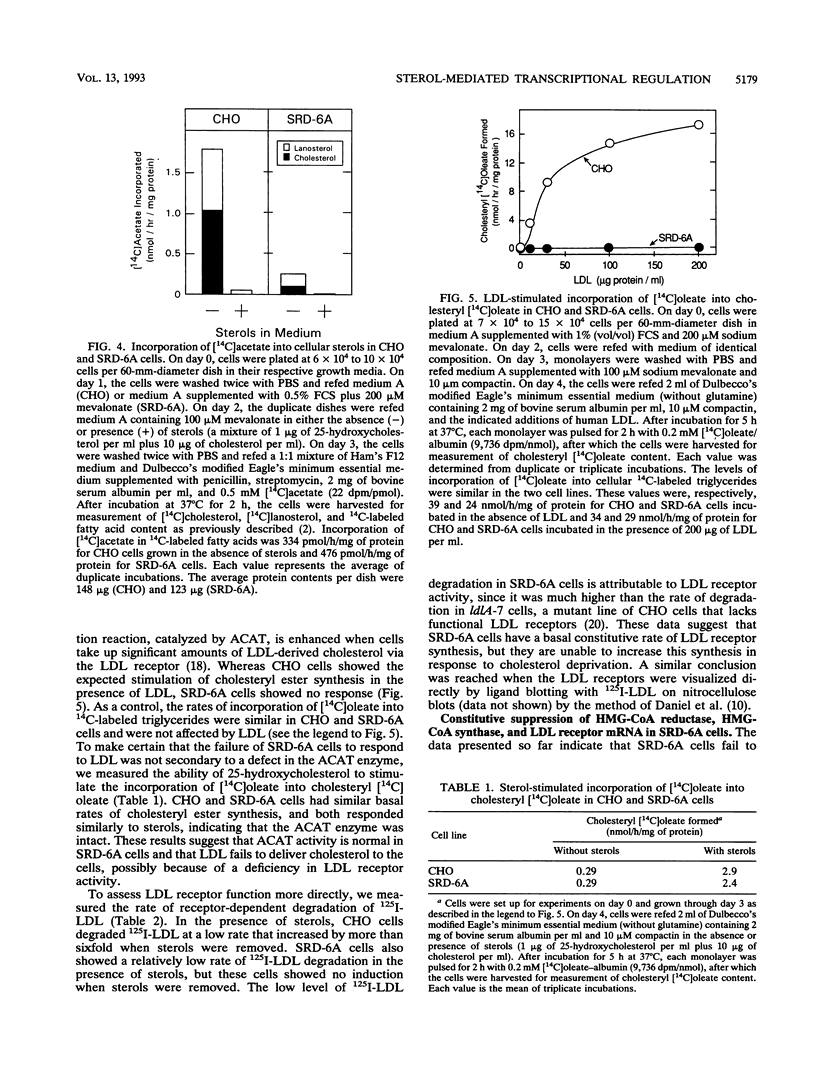
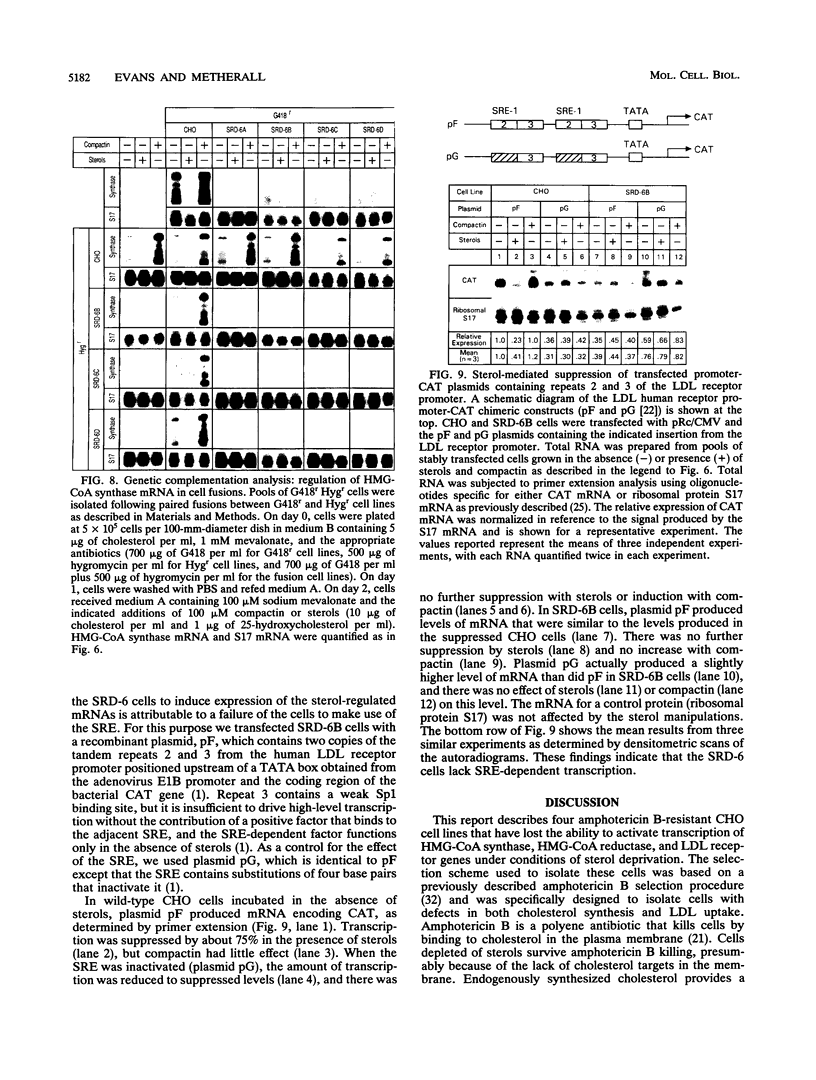
Abstract
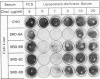
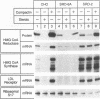
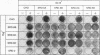
Cholesterol biosynthesis and uptake are controlled by a classic end product-feedback mechanism whereby elevated cellular sterol levels suppress transcription of the genes encoding 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) synthase, HMG-CoA reductase, and the low-density lipoprotein receptor. The 5'-flanking region of each gene contains a common cis-acting element, designated the sterol regulatory element (SRE), that is required for transcriptional regulation. In this report, we describe mutant Chinese hamster ovary (CHO) cell lines that lack SRE-dependent transcription. Mutant cell lines were isolated on the basis of their ability to survive treatment with amphotericin B, a polyene antibiotic that kills cells by interacting with cholesterol in the plasma membrane. Four mutant lines (SRD-6A, -B, -C, and -D) were found to be cholesterol auxotrophs and demonstrated constitutively low levels of mRNA for all three sterol-regulated genes even under conditions of sterol deprivation. The mutant cell lines were found to be genetically recessive, and all four lines belonged to the same complementation group. When transfected with a plasmid containing a sterol-regulated promoter fused to a bacterial reporter gene, SRD-6B cells demonstrated constitutively low levels of transcription, in contrast to wild-type CHO cells, which increased transcription under conditions of sterol deprivation. Mutation of the SREs in this plasmid prior to transfection reduced the level of expression in wild-type CHO cells deprived of sterols to the level of expression found in SRD-6B cells. The defect in SRD-6 cells is limited to transcriptional regulation, since posttranscriptional mechanisms of sterol-mediated regulation were intact: the cells retained the ability to posttranscriptionally suppress HMG-CoA reductase activity and to stimulate acyl-CoA:cholesterol acyltransferase activity. These results suggest that SRD-6 cells lack a factor required for SRE-dependent transcriptional activation. We contrast these cells with a previously isolated oxysterol-resistant cell line (SRD-2) that lacks a factor required for SRE-dependent transcriptional suppression and propose a model for the role of these genetically defined factors in sterol-mediated transcriptional regulation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown M. S., Faust J. R., Goldstein J. L., Kaneko I., Endo A. Induction of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in human fibroblasts incubated with compactin (ML-236B), a competitive inhibitor of the reductase. J Biol Chem. 1978 Feb 25;253(4):1121–1128. [PubMed] [Google Scholar]
- Chang T. Y., Chang C. C. Revertants of a Chinese hamster ovary cell mutant resistant to suppression by an analogue of cholesterol: isolation and partial biochemical characterization. Biochemistry. 1982 Oct 12;21(21):5316–5323. doi: 10.1021/bi00264a030. [DOI] [PubMed] [Google Scholar]
- Chang T. Y., Limanek J. S. Regulation of cytosolic acetoacetyl coenzyme A thiolase, 3-hydroxy-3-methylglutaryl coenzyme A synthase, 3-hydroxy-3-methylglutaryl coenzyme A reductase, and mevalonate kinase by low density lipoprotein and by 25-hydroxycholesterol in Chinese hamster ovary cells. J Biol Chem. 1980 Aug 25;255(16):7787–7795. [PubMed] [Google Scholar]
- Chang T. Y., Telakowski C., Heuvel W. V., Alberts A. W., Vagelos P. R. Isolation and partial characterization of a cholesterol-requiring mutant of Chinese hamster ovary cells. Proc Natl Acad Sci U S A. 1977 Mar;74(3):832–836. doi: 10.1073/pnas.74.3.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. W., Cavenee W. K., Kandutsch A. A. Sterol synthesis in variant Chinese hamster lung cells selected for resistance to 25-hydroxycholesterol. Cross-resistance to 7-ketocholesterol, 20alpha-hydroxycholesterol, and serum. J Biol Chem. 1979 Feb 10;254(3):715–720. [PubMed] [Google Scholar]
- Chin J., Chang T. Y. Evidence for coordinate expression of 3-hydroxy-3-methylglutaryl coenzyme A reductase ad low density lipoprotein binding activity. J Biol Chem. 1981 Jun 25;256(12):6304–6310. [PubMed] [Google Scholar]
- Clarke C. F., Tanaka R. D., Svenson K., Wamsley M., Fogelman A. M., Edwards P. A. Molecular cloning and sequence of a cholesterol-repressible enzyme related to prenyltransferase in the isoprene biosynthetic pathway. Mol Cell Biol. 1987 Sep;7(9):3138–3146. doi: 10.1128/mcb.7.9.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel T. O., Schneider W. J., Goldstein J. L., Brown M. S. Visualization of lipoprotein receptors by ligand blotting. J Biol Chem. 1983 Apr 10;258(7):4606–4611. [PubMed] [Google Scholar]
- Dawson P. A., Hofmann S. L., van der Westhuyzen D. R., Südhof T. C., Brown M. S., Goldstein J. L. Sterol-dependent repression of low density lipoprotein receptor promoter mediated by 16-base pair sequence adjacent to binding site for transcription factor Sp1. J Biol Chem. 1988 Mar 5;263(7):3372–3379. [PubMed] [Google Scholar]
- Dawson P. A., Metherall J. E., Ridgway N. D., Brown M. S., Goldstein J. L. Genetic distinction between sterol-mediated transcriptional and posttranscriptional control of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 1991 May 15;266(14):9128–9134. [PubMed] [Google Scholar]
- Faust J. R., Goldstein J. L., Brown M. S. Squalene synthetase activity in human fibroblasts: regulation via the low density lipoprotein receptor. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5018–5022. doi: 10.1073/pnas.76.10.5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil G., Faust J. R., Chin D. J., Goldstein J. L., Brown M. S. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985 May;41(1):249–258. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Basu S. K., Brown M. S. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Brown M. S. Regulation of the mevalonate pathway. Nature. 1990 Feb 1;343(6257):425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Brown M. S. The LDL pathway in human fibroblasts: a receptor-mediated mechanism for the regulation of cholesterol metabolism. Curr Top Cell Regul. 1976;11:147–181. doi: 10.1016/b978-0-12-152811-9.50011-0. [DOI] [PubMed] [Google Scholar]
- Goldstein J. L., Dana S. E., Brown M. S. Esterification of low density lipoprotein cholesterol in human fibroblasts and its absence in homozygous familial hypercholesterolemia. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4288–4292. doi: 10.1073/pnas.71.11.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka K., Endo H., Akiyama S., Kuwano M. Isolation and characterization of amphotericin B-resistant cell lines in Chinese hamster cells. Cell. 1978 Jun;14(2):415–421. doi: 10.1016/0092-8674(78)90126-5. [DOI] [PubMed] [Google Scholar]
- Kingsley D. M., Krieger M. Receptor-mediated endocytosis of low density lipoprotein: somatic cell mutants define multiple genes required for expression of surface-receptor activity. Proc Natl Acad Sci U S A. 1984 Sep;81(17):5454–5458. doi: 10.1073/pnas.81.17.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsky S. C. Antibiotic interaction with model membranes. Annu Rev Pharmacol. 1970;10:119–142. doi: 10.1146/annurev.pa.10.040170.001003. [DOI] [PubMed] [Google Scholar]
- Krieger M., Martin J., Segal M., Kingsley D. Amphotericin B selection of mutant Chinese hamster cells with defects in the receptor-mediated endocytosis of low density lipoprotein and cholesterol biosynthesis. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5607–5611. doi: 10.1073/pnas.80.18.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limanek J. S., Chin J., Chang T. Y. Mammalian cell mutant requiring cholesterol and unsaturated fatty acid for growth. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5452–5456. doi: 10.1073/pnas.75.11.5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum L., Luskey K. L., Chin D. J., Ho Y. K., Goldstein J. L., Brown M. S. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase and its mRNA in rat liver as studied with a monoclonal antibody and a cDNA probe. J Biol Chem. 1983 Jul 10;258(13):8450–8455. [PubMed] [Google Scholar]
- Metherall J. E., Goldstein J. L., Luskey K. L., Brown M. S. Loss of transcriptional repression of three sterol-regulated genes in mutant hamster cells. J Biol Chem. 1989 Sep 15;264(26):15634–15641. [PubMed] [Google Scholar]
- Metherall J. E., Ridgway N. D., Dawson P. A., Goldstein J. L., Brown M. S. A 25-hydroxycholesterol-resistant cell line deficient in acyl-CoA: cholesterol acyltransferase. J Biol Chem. 1991 Jul 5;266(19):12734–12740. [PubMed] [Google Scholar]
- Mosley S. T., Brown M. S., Anderson R. G., Goldstein J. L. Mutant clone of Chinese hamster ovary cells lacking 3-hydroxy-3 -methylglutaryl coenzyme A reductase. J Biol Chem. 1983 Nov 25;258(22):13875–13881. [PubMed] [Google Scholar]
- Nakanishi M., Goldstein J. L., Brown M. S. Multivalent control of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mevalonate-derived product inhibits translation of mRNA and accelerates degradation of enzyme. J Biol Chem. 1988 Jun 25;263(18):8929–8937. [PubMed] [Google Scholar]
- Osborne T. F., Bennett M., Rhee K. Red 25, a protein that binds specifically to the sterol regulatory region in the promoter for 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 1992 Sep 15;267(26):18973–18982. [PubMed] [Google Scholar]
- Osborne T. F. Single nucleotide resolution of sterol regulatory region in promoter for 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Biol Chem. 1991 Jul 25;266(21):13947–13951. [PubMed] [Google Scholar]
- Peffley D., Miyake J., Leonard S., von Gunten C., Sinensky M. Further characterization of a somatic cell mutant defective in regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase. Somat Cell Mol Genet. 1988 Nov;14(6):527–539. doi: 10.1007/BF01535308. [DOI] [PubMed] [Google Scholar]
- Saito Y., Chou S. M., Silbert D. F. Animal cell mutants defective in sterol metabolism: a specific selection procedure and partial characterization of defects. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3730–3734. doi: 10.1073/pnas.74.9.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer-Polokoff R., von Gunten C., Logel J., Torget R., Sinensky M. Isolation and characterization of a mammalian cell mutant defective in 3-hydroxy-3-methylglutaryl coenzyme A synthase. J Biol Chem. 1982 Jan 10;257(1):472–476. [PubMed] [Google Scholar]
- Sinensky M., Armagast S., Mueller G., Torget R. Somatic cell genetic analysis of regulation of expression of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6621–6623. doi: 10.1073/pnas.77.11.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinensky M., Logel J., Torget R. Complementary recessive 25-hydroxycholesterol-resistant somatic cell mutants--assay of 25-hydroxycholesterol binding activity. J Cell Physiol. 1982 Nov;113(2):314–319. doi: 10.1002/jcp.1041130220. [DOI] [PubMed] [Google Scholar]
- Sinensky M., Torget R., Edwards P. A. Radioimmune precipitation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from Chinese hamster fibroblasts. Effect of 25-hydroxycholesterol. J Biol Chem. 1981 Nov 25;256(22):11774–11779. [PubMed] [Google Scholar]
- Smith J. R., Osborne T. F., Goldstein J. L., Brown M. S. Identification of nucleotides responsible for enhancer activity of sterol regulatory element in low density lipoprotein receptor gene. J Biol Chem. 1990 Feb 5;265(4):2306–2310. [PubMed] [Google Scholar]
- Tanaka R. D., Lee L. Y., Schafer B. L., Kratunis V. J., Mohler W. A., Robinson G. W., Mosley S. T. Molecular cloning of mevalonate kinase and regulation of its mRNA levels in rat liver. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2872–2876. doi: 10.1073/pnas.87.8.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]