Abstract
Background
Gene targeting (GT) provides a powerful tool for the generation of precise genetic alterations in embryonic stem (ES) cells to elucidate gene function and create animal models for human diseases. This technology has, however, been limited to mouse and rat. We have previously established ES cell lines and procedures for gene transfer and selection for homologous recombination (HR) events in the fish medaka (Oryzias latipes).
Methodology and Principal Findings
Here we report HR-mediated GT in this organism. We designed a GT vector to disrupt the tumor suppressor gene p53 (also known as tp53). We show that all the three medaka ES cell lines, MES1∼MES3, are highly proficient for HR, as they produced detectable HR without drug selection. Furthermore, the positive-negative selection (PNS) procedure enhanced HR by ∼12 folds. Out of 39 PNS-resistant colonies analyzed, 19 (48.7%) were positive for GT by PCR genotyping. When 11 of the PCR-positive colonies were further analyzed, 6 (54.5%) were found to be bona fide homologous recombinants by Southern blot analysis, sequencing and fluorescent in situ hybridization. This produces a high efficiency of up to 26.6% for p53 GT under PNS conditions. We show that p53 disruption and long-term propagation under drug selection conditions do not compromise the pluripotency, as p53-targeted ES cells retained stable growth, undifferentiated phenotype, pluripotency gene expression profile and differentiation potential in vitro and in vivo.
Conclusions
Our results demonstrate that medaka ES cells are proficient for HR-mediated GT, offering a first model organism of lower vertebrates towards the development of full ES cell-based GT technology.
Introduction
Gene targeting (GT) in mouse embryonic stem (ES) cells has been used as a powerful tool for analyzing gene function [1]. In this approach, precise alterations are introduced into ES cells at particular loci by gene replacement, and targeted ES cells are introduced into early embryos for the formation of chimeras in which transplanted ES cells contribute to many lineages including the germline. Crossing of germline chimeras lead to the production of animals that are heterozygous or homozygous for the targeted locus. GT in ES cells has been reported also in human [2] and rat [3]. Because of the availability of pluripotent ES cell lines and the possibility of germline chimera formation, however, the production of knockout animals from targeted ES cells has long been limited to mouse [4] before its recent expansion into rat [5].
The effort towards GT has steadily been attempted in non-mammalian species such as lower vertebrates, in particular zebrafish and medaka (Oryzias latipes). In zebrafish, Collodi and his colleagues have reported targeted insertion of a plasmid by HR in zebrafish ES cell cultures [6]. More importantly, zebrafish with targeted gene mutations have been generated by using zinc finger nucleases (ZFNs) [7], [8] and transcription activator-like effector nucleases (TALENs) [9].
We and others use the medaka (Oryzias latipes) as a model organism to develop the GT technology. Like zebrafish, this fish is an excellent model for analyzing vertebrate development [10]. In this organism, we have obtained various stem cell lines of diploid ES cells [11], haploid ES cells [12], [13] and adult germ cells [14], [15], and established procedures for gene transfer in vitro [16], [17], [18], [19] and in vivo [17], [19], [20], positive-negative selection (PNS) to enrich for HR-mediated GT in ES cells [16], and chimera formation [21].
In order to fully develop the GT technology in medaka, we chose the p53 gene as a model. p53 is best known as the “guardian of the genome” and tumor suppressor, as p53 mutations occur in 50% of human cancers [22]. p53 is highly conserved across animal phyla, as its mutation increases the incidence of tumor formation in mouse [22] and fish [23], [24]. In mice, p53 is involved in several other important processes such as senescence and ageing [25]. p53 has been targeted by HR in ES cells of mouse [26] and rat [5]. Previously, we have shown the lack of ultraviolet-light inducibility of the medaka p53 gene [27], and reported a first attempt toward the development of GT in medaka ES cells, in which p53 gene was cloned for the construction of GT vector pGTp53 [28]. In this work, the genuine GT event was not described, thus parameters for PNS and GT efficiency remained speculative. We made use of the pGTp53 vector [28], to continue the effort towards the establishment of GT technology in fish ES cells. pGTp53 was devised to disrupt the medaka p53 by HR on the basis of PNS to allow for enrichment for HR events [29].
This study was aimed at continuing our effort for improving procedures and efficiency for HR-mediated true p53 GT in medaka ES cell lines. We show the effectiveness of PNS procedure in enrichment for the HR event and a high efficiency of GT in medaka ES cells. More importantly, we demonstrate the retention of pluripotency of medaka ES cells after long-term drug selection and targeted p53 disruption.
Results
Gene Transfer and Selection Strategy
For HR-mediated GT experiments, we made use of the GT vector pGTp53 (Figure 1A) that has been described previously [28]. Gene transfer of linearized pGTp53 vector into medaka ES cells was performed by using the GeneJuice reagent (Novagen) [14], [16], [18] in 6-well plates. Each well was considered as a pool for screening of HR-events after gene transfer. Upon HR, the neo would be co-integrated, while the tk would be recombined away, with the homologous recombinant being resistant to G418 and gancyclovir (Gc) owing to the expression of neo and the absence of tk. Upon random integration (RI), both the neo and tk would be co-integrated, with random integrants being resistant to G418 but sensitive to Gc owing to the expression of both neo and tk. Therefore, the PNS by using G418 and Gc is predicted to enrich for HR events by eliminating RI events via their sensitivity to Gc and non-transgenic cells via their sensitivity to G418 (Figure 1B and C).
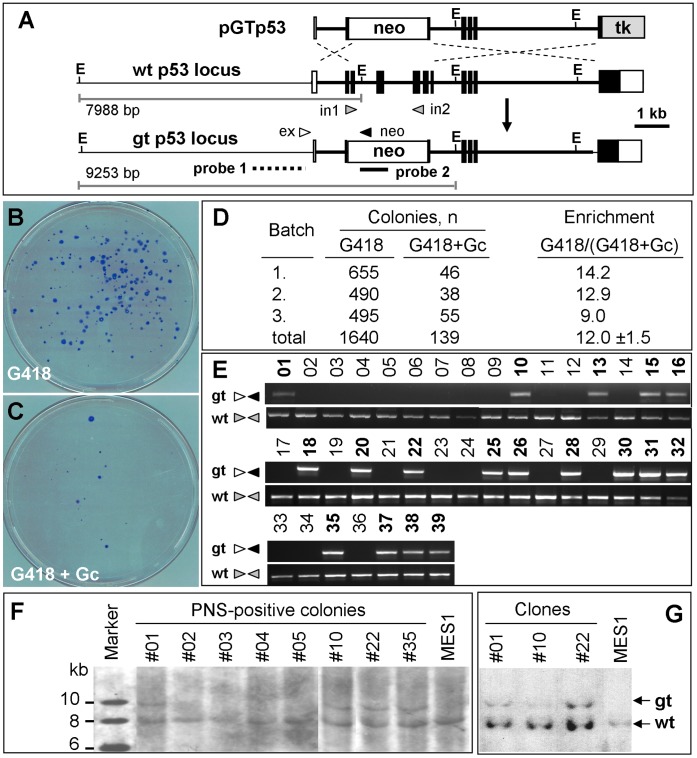
Figure 1. p53 gene targeting in MES1.
(A) Targeting vector pGTp53. gt, gene-targeted; wt, wildtype; filled box, translated exon; open box, untranslated exon; thin line between boxes, intron; thick line, region included in pGTp53; neo, STneo cassette; tk, STk cassette; E, EcoR I site. Arrowheads depict the position and extension direction of primers (in1 and in2, p53 primers whose sequences are absent in pGTp53 and the targeted locus; ex, external primer; neo, neo primer). Probes and EcoR I-digests of predicted sizes for Southern analysis are indicated. (B and C) Colony formation. Colonies formed from pGTp53-transfected MES1 cells in the presence of G418 alone (B) and G418 plus Gc (C) were Giemsa-stained at day 23 of clonal growth. (D) PNS enrichment factor. (E) PCR detection of the GT event. Genomic DNA was isolated from 39 PNS-resistant colonies and subjected to PCR detection by using primers (arrows; for positions see Figure 1A) specific to the targeted (gt) and wildtype (wt) locus. Numbers above lanes, PNS-resistant colonies. Positive colonies are shown in bold. (F) Southern analysis of representative colonies. (G) Southern analysis of representative clones from colonies after ≥20 passages of propagation.
High HR Activity in different Medaka ES Cell Lines
There are three medaka ES cell lines, namely MES1-3 [11]. We first determined whether they possessed reasonable HR activity as a precondition for GT. To this end, the three cell lines were transfected in 12-well plates with pGTp53 and the putative HR event was detected at 5 days post transfection (dpt), by PCR analysis using primers specific to the targeted p53 locus (Figure 1A). This led to the detection of a GT-specific PCR product in all of the six batches of pGTp53-transfected ES cells but not in mock-transfected cells (Figure S1). PNS by using G418 (500 µg/ml) plus Gc (5 µM) increased the yield of the GT-specific PCR product. Importantly, a single round of PCR produced an easily detectable band in MES1 and MES2 but a faint band in MES3 (Figure S1). The band for the targeted allele became more intense after a 2nd round of PCR. Therefore, the three ES cells lines possess high but different levels of cellular HR activity, and MES1 was chosen for subsequent experiments because of its high HR activity and documented ability for chimera formation [21], [30] and retention of pluripotency after gene transfer and long-term drug selection [16].
Efficient p53 Gene Targeting
Compared to RI, HR is a rare event even in mouse ES cells [1]. Previously, we have shown the effectiveness of PNS in model systems of fish cell culture [28], [29]. We wanted to determine the efficiency of PNS for enrichment for the HR event in medaka ES cells. To this, 106 transfectants were seeded in 10-cm dish and grown in the presence of G418 alone or together with Gc, and colony formation was examined after 23–28 days of culture. In a typical experiment, selection with G418 alone produced 100∼300 colonies per dish, while the number of colonies decreased to 5–25 per dish upon PNS (Figure 1B and C). A statistical analysis of three independent experiments led to an enrichment factor of 12.0±1.5 for PNS in pGTp53-transfected MES1 cells (Figure 1D), demonstrating the effectiveness of PNS for enriching for putative homologous recombinants in medaka ES cells.
We utilized four procedures to identify genuine homologous recombinants. A PCR-based genotyping procedure was first used to screen for the targeted p53 locus, which was amenable for PCR amplification by using an external primer and neo primer (Figure 1A). From two batches of experiments, 39 colonies were successfully transferred and expanded, 19 (48.7%) were positive for PCR-genotyping (Figure 1E and Table S2 in File S1). When 11 of the PCR-positive colonies were then subjected to Southern blot analysis, 6 (54.5%) turned out to contain a 9.2-kb band and an 8-kb band without any visible additional bands, whereas normal MES1 cells has only the 8-kb band (Figure 1F and Table S2 in File S1). The 8-kb and 9.2-kb bands conformed to the predicted sizes of genomic EcoR I-digests for the wildtype allele (7988 bp) and targeted allele (9253 bp; Figure 1A; for detail refer to File S2). The targeted allele remained unchanged during ≥20 passages for expansion into individual clones (Figure 1G). Fluorescent in situ hybridization in one of the 6 clones (clone #22) demonstrated the presence of a single neo signal in the cells at different cell cycle phases (Figure S2). Sequencing revealed the predicted junction sequence for the targeted p53 allele (Figure S3). Taken together, p53 GT has efficiently occurred without detectable RI events and the targeted p53 allele is as stable as the wildtype allele during clonal expansion.
Retention of Pluripotency in vitro
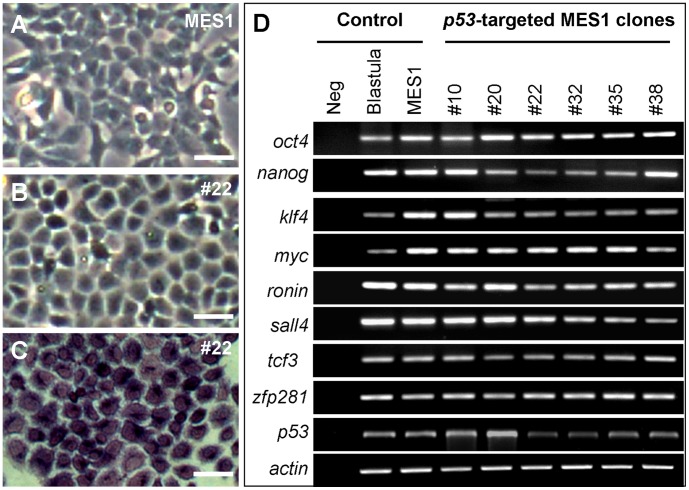
To determine whether the pluripotency was compromised by drug selection and p53 GT, normal MES1 and its p53-targeted counterparts including clone #22 were analyzed for differentiation potential in vitro and chimera competence in vivo. Under the conditions for undifferentiated growth, clone #22 exhibited the ES-cell phenotype such as a small size and round shape similar to that of MES1 (Figure 2A and B), and high alkaline phosphatase activity (Figure 2C), a general marker of fish stem cells [11], [12], [14], [17]. Moreover, all the six p53-targeted clones examined, like normal MES1 cells, did exhibit a high level of expression of 9 pluripotency genes including nanog, oct4 and klf4 (Figure 2D), which have recently been shown to be associated with an undifferentiated state of medaka stem cells in culture [12], [31]. As expected, the p53 transcript was found at a reduced level in 4 out of 6 clones, namely clones 22, 32, 35 and 38, as compared to normal MES1 cells (Figure 2D), in consistence with heterozygosity at the p53 locus in these clones.
Figure 2. Pluripotency in vitro.
(A) Phenotype of parental MES1 cells. (B) Phenotype of p53-targeted clone #22. (C) Alkaline staining of clone #22. (D) Pluripotency gene expression profile in five p53-targeted MES1 clones. neg, negative control without cDNA template. actin was used as a loading control. Blastula embryos and parental MES1 were used as positive controls. Scale bars, 25 µm.
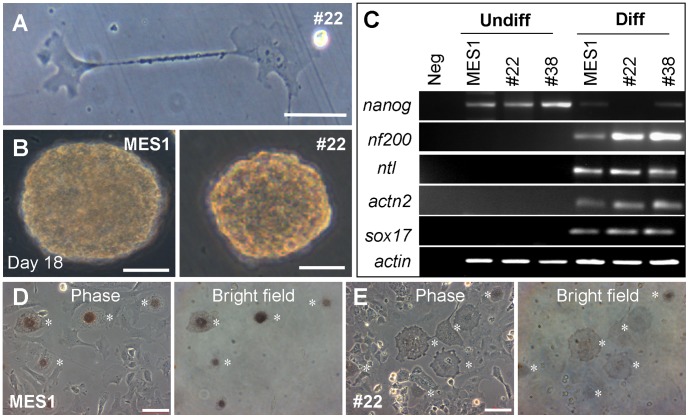
Under the conditions for differentiation induction [11], p53-targeted cells were able to differentiate spontaneously into various specialized cell types including neurons with long and thin cytoplasmic extensions (Figure 3A). They were capable of induced differentiation via embryoid body (EB) formation in suspension culture (Figure 3B), and they lost nanog expression and gained expression of several differentiation genes including the neuroectodermal marker nf200, the mesodermal markers ntl and actinin2 as well as the endodermal marker sox17 (Figure 3C), as have previously been shown for normal medaka ES cells [11], [12], [31], [32]. Normal MES1 cells can be directed for differentiation by forced Mitf (microphthalmia-associated transcription factor) expression into melanocytes [19], [33]. Upon transfection with pXmitf, vector expressing the Xiphophorus Mitf [19], the parental MES1 and p53-targeted counterparts produced pigmented melanocytes at similar efficiencies (Figure 3D and E; Figure S4). Taken together, medaka ES cells after long-term drug selection and p53 GT retain the stem cell phenotype and pluripotency for spontaneous, induced and directed differentiation in vitro.
Figure 3. Differentiation in vitro.
(A) Spontaneous differentiation of p53-targeted MES1 clone #22 into neurons. (B) Embryoid body (EB) formation of MES1 (left) and clone #22 (right). (C) Expression of differentiation genes upon induced differentiation by EB formation. nf200, neurofilament 200; ntl, no tail; actn2, actinin 2. (D and E) Directed differentiation into melanocytes (asterisks) by forced Mitf expression from pXmitf. Scale bars, 50 μm.
Retention of Pluripotency in vivo
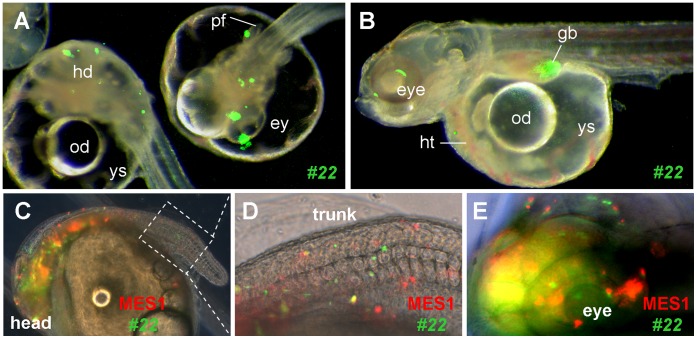
We have previously shown that MES1 cells after stable gene transfer and drug selection were able to give rise to 100% chimera formation and that these donor cells contributed to a wide variety of tissues/organs during chimeric embryogenesis [12], [16], [21], [30], [34]. In order to test whether this property was retained after long-term drug selection plus p53 GT, one of p53-targeted clones, clone #22, was genetically labeled with GFP via transfection by pCVpf and introduced into host blastulae. This generated 100% GFP-positive chimeras in which the GFP-positive donor cells were distributed to many compartments during early embryogenesis and to many tissues/organs at advanced stages (Figure 4A and B). Upon co-transplantation into blastula hosts, GFP-labeled cells of clone #22 were not different from RFP-labeled normal MES1 cells in distribution and differentiation in developing chimeric embryos (Figure 4C-E; Figure S5). Thus, p53-targeted ES cells have retained pluripotency in vivo.
Figure 4. Pluripotency in vivo.
Embryos at the midblastula stage were transplanted with ES cells and photographed at day 3 (A) and 7 post fertilization (B–E). (A and B) Merged micrographs of chimeras, showing GFP-labeled ES cells of p53-targeted clone #22 in many compartments and organ systems including the eye (ey), head (hd), heart (ht), gall bladder (gb) and pectoral fin (pf). od, oil droplet; ys, yolk sac. (C–E) Co-distribution of parental MES1 and p53-targeted clone #22 in the developing chimera at day 7 post fertilization. Following genetic labeling by plasmid transfection, parental MES1 (red) and clone #22 (green) were mixed at 1∶1 ratio and transplanted at approximately 200 cells per blastula. (D and E) Larger magnification of the posterior trunk framed in (C) and anterior end. The embryo is 1 mm in diameter.
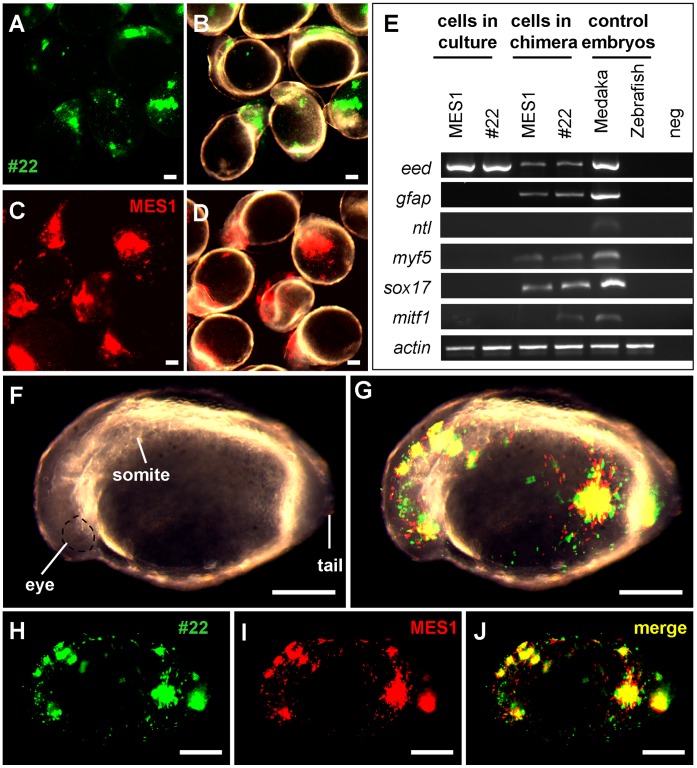
Recently, we have shown that interordinal chimera formation between medaka and zebrafish is a powerful tool for analyzing stem cell differentiation [35]. Specifically, interordinal chimera formation allows for a molecular analysis of ES cell differentiation along multiple cell lineages through RT-PCR assays by using primers that are specific to donor cell cDNAs. We adopted this approach. p53-targeted cells (clone #22) exhibited wide distribution to major compartments of 1-day-old zebrafish host embryos (Figure 5A and B). A similar observation was made also on parental MES1 cells (Figure 5C and D). The detection of medaka three germ layers (ectodermal, eed and gfap; mesodermal, myf5; endomermal, sox17) and neural crest (mitf1) markers in zebrafish host by RT-PCR revealed that in vivo differentiation potential of both parental MES1 cells and p53-targeted ES cells (#22 clone) (Figure 5E). Co-transplantation experiments revealed that MES1 and #22 cells had highly overlapping distribution (Figure 5F–J). Hence, we conclude that, as in mouse ES cells, drug selection and GT do not compromise the pluripotency of medaka ES cells.
Figure 5. Chimeric assay of pluripotency in vivo.
Zebrafish blastulae were used as the host for transplantation of MES1 (red), #22 (green) or both and analyzed at 1 dpf by microscopy and RT-PCR. (A and B) Chimeras by transplantation of #22 cells on fluorescent micrograph (A) and merge between fluorescent and brightfield optics (B). (C and D) Chimeras by transplantation of MES1 cells on fluorescent micrograph (C) and merge between fluorescent and brightfield optics (D). (E) RT-PCR analysis of gene expression. MES1, #22, 1-day-old embryos of medaka and zebrafish were used for comparison. Primers used were specific to medaka cDNAs, except for β-actin primers that amplify the β-actin cDNA of both medaka and zebrafish. (F–J) Chimera by cotransplantation of MES1 and #22 cells on brightfield (F), brightfield-fluorescent merge (G), GFP fluorescence (H), RFP fluorescence (I) and merge between GFP and RFP optics (J). Scale bars, 200 µm.
Discussion
Medaka and zebrafish are excellent twin model organisms for analyzing vertebrate development [10], studying stem cell biology [11], [12], [14], [17], [30], [36], [37] and more importantly, for developing the ES cell technology [16], [28], [29]. As a key step towards the full development of the ES cell technology in this organism, the present work has addressed five important issues. First, we show that all the three medaka ES cell lines possess a significant level of cellular HR activity, allowing for easy detection of HR events as early as just a few days of culture post transfection even in the absence of enrichment by drug selection. Second, we reveal that PNS is effective in enriching for HR events, producing a 12-fold enhancement factor, which is within the range of 2∼100 folds reported for mouse ES cells. Third, we demonstrate that p53 GT in medaka ES cells occurs at a frequency 26% under PNS condition, a proficiency that is 13 times higher than a ∼2% efficiency for p53 GT in ES cells of mouse [38] and rat [5]. With this high proficiency, it is possible to obtain a sufficient number of cell clones with a targeted locus from a few dozens of PNS-resistant colonies in a single transfection experiment. Fourth, our finding that all clones examined in this study are stable in growth and genetic stability indicate that a single copy of wildtype p53 gene is sufficient for ES cell maintenance, in accordance with the observation that p53 heterozygosity is sufficient to support ES cell maintenance in mouse [26] and rat [5] as well as normal development in medaka [24]. Finally and most importantly, we have provided compelling evidence that p53-targeted medaka ES cells retain pluripotency in vitro and in vivo, thus establishing medaka as first lower vertebrate organism to fully develop the HR-based GT technology.
Site-specific gene alterations can also be achieved by using engineered sequence-specific endonucleases such as ZFNs and TALENs. The ZFN or TALEN approach can introduce only minor additions or deletions in an unpredictable manner and are thus limited to gene disruption. Furthermore, this approach may also produce off-target alterations that are difficult to predict and detect. In contrast, HR-based GT generates precisely designed gene replacement, which allows for both gene disruption and correction and essentially alleviate the off-target concern. Therefore, the HR-based GT in ES cells followed by germline transmission still represents the approach of choice to engineer the genome at the best precision [1].
This study has focused on, successes in, HR-based GT in medaka ES cells by using p53 as a model. This success corroborates and extends our previous effort with the same HR vector in MES1 cells [28], and extends the early study by demonstrating the first true success in bona fide GT in MES1 cells and the retention of pluripotency in vitro and in vivo. The next key step is to achieve germline transmission for whole animal production, as has been done in mouse [1] and rat [5]. In zebrafish, blastula cells after short-term culture can form germline chimeras [39]. In medaka, germline chimera formation from ES cell lines has not yet been described, in spite of continuous efforts [21], [30], [34]. Our previous study has indicated that medaka germ cell formation appears to be controlled by a cell-autonomous mechanism [40], raising the possibility that the medaka germline is inaccessible to colonization by cultured ES cells. Recently, we have generated medaka haploid ES cells capable of germline transmission by semicloning [12]. It is anticipated that GT in these haploid ES cells in combination with semicloning will provide an alternative approach for the production of knockout animals in this organism. The procedures and efficiencies established in present study will offer valuable information for GT in medaka haploid ES cells.
Materials and Methods
Fish and Reagents
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Advisory Committee for Laboratory Animal Research in Singapore and approved by this committee (Permit Number: 27/09). Medaka strain af was maintained under an artificial photoperiod of 14-h light to 10-h darkness at 26∼28°C as described [19], [21]. Embryos were maintained at 26∼28°C and staged as described [41]. Unless otherwise indicated, chemicals were purchased from Sigma, enzymes and PCR reagents were from Promega and TaKaRa.
Cell Culture
Maintenance of the medaka ES cell lines MES1-3 were maintained at 28°C under feeder-free culture conditions on gelatin-coated substrata in culture medium ESM4 and staining for alkaline phosphatase activity were performed as previously described [11], [42].
Plasmids
The GT plasmid pGTp53 targeting the medaka p53 gene was described [28]. Vectors pCVpf and pCVpr were used for drug selection and GFP or RFP expression for cell labeling [19]. Plasmid DNA was prepared by using the Qiagen Maxi and Midi preparation kits (Qiagen, Germany).
Cell Transfection and Colony Isolation
Cell transfection with plasmid DNA by using the GeneJuice reagent (Novagen) and drug selection were performed as described [14], [16], [18]. Briefly, cells seeded in 6-well plates were incubated with linearized pGTp53 vector in a serum-free medium for 6 h. After medium change to ESM4, the cells were grown for 3 days and trypsinized into single cells. A quarter of the cells were collected for the PCR analysis (see below), and the remainder were subcultured into 10-cm gelatin-coated Petri dishes (plating) for PNS by using G418 (Gibco) at 500 µg/mL and Gc (Cymevan, Syntex Arzneimittel GmbH, Switzerland) at 5 µM.
After plating, pGTp53-transfected MES1 cells were grown under drug selection for up to 28 days. Dishes with ≤200 colonies were subjected to manual colony isolation. After transferring the PNS-resistant colonies into 96-well plate, a monolayer formed during 10–14 days of culture, with medium being changed every 3–5 days. After 7–12 days of culture, colonies were split into three 96-well plates. Two plates were used for by Southern (plate A) and PCR (plate B) analyses (see below), respectively, where the third was used as master plate (plate C) for cell propagation.
Micro-extraction of DNA
DNA samples from 96-well plates were isolated by using the micro-extraction procedure [43] with minor modifications. Briefly, the DNA was extracted by adding cell lysis buffer (10 mM Tris-HCl, pH7.5, 10 mM EDTA, 10 mM NaCl, 0.5% sarcosyl, and 500 µg/mL proteinase K), incubation at 55°C for 12–16 h and precipitation by NaCl and cold ethanol (1.5 µL of 5 M NaCl to 100 µL of absolute ethanol). After the 70% ethanol wash, 50 µL of autoclaved Milli-Q water was added to each well. DNA was dissolved and restored at 4°C until use.
PCR
PCR-based genotyping was conducted from pooled cell populations or single PNS-resistant colonies by two sequential rounds of PCR. The first round of PCR was run for 35 cycles (94°C for 30 s, 60°C for 30 s and 72°C for 90 s) in a 25-µL volume containing 2 µl of DNA prepared above. The second round of PCR was similarly run for 30 cycles in a 25-µL volume containing 2 µL of the 10 times diluted 1st round PCR product. PCR primers are listed in Table S1 in File S1. The PCR screening of PNS-resistant clones was performed using DNA samples prepared from 96-well plate. PCR products were separated on a 1.0% agarose gel and documented with a bio-imaging system (Synoptics, Cambridge).
For RT-PCR, total RNA was isolated by using the Trizol Reagent (Invitrogen). Synthesis of cDNA templates was primed with oligo (dT)18 by using M-MLV transcriptase (Invitrogen). The cDNA reaction was diluted with water to 10 ng/µL. RT-PCR was run as previously described [12], [14]. PCR primers are listed in Table S1 in File S1.
Southern Analysis
DNA extraction was similarly done as described above but to a larger scale. Ten µg of genomic DNA was digested with EcoR I enzyme at 37°C for 30–40 h. The digested DNA fragments were separated on a 0.8% TBE agarose gel by electrophoresis at 30 V for 16 h at 4°C. After photograph and de-purination treatment with 0.2 M HCl for 10 min, the DNA was transferred to Hybond N+ nylon membrane (GE healthcare, USA) for 5 h by alkaline buffer (0.5 M NaOH, 1.5 M NaCl). Following transfer, the membrane was pre-hybridized with DIG-Easy solution (Roche) containing 100 µg/ml of pre-boiled calf thymus DNA fragments (≤2 kb in size) at 42°C of 12–16 h. The probe was labeled with digoxygenin (a mixture of 100∼1000-bp fragments synthesized from a 1.5-kb 5′-external sequence; for detail see Table S1 in File S1) by random-primed DNA synthesis (Roche), then hybridized to the membrane at 42°C in DIG-easy solution containing 100 µg/ml of calf thymus DNA for 40 h. Non-specific binding was removed by washing in 2 × SSC/0.5% SDS (twice), and 0.1 × SSC/0.5% SDS at 65°C for 10 min each. Followed by an overnight incubation at 4°C with a 1∶5000 diluted anti-DIG-POD antibody (Roche) in 1 × maleic acid solution containing 1.5% blocking reagent (Roche), the hybridization signal was visualized after chemiluminescent reaction by using the ECL reagent (Amersham Biosciences) and developed by exposure for 8 h to films (Kodak).
Sequencing
DNA fragments were subcloned into pGEM-T easy vector (Promega) and sequenced on an ABI3100 automatic sequencer (Applied Biosystems). Sequence analyses were performed by using the Vector NTI package (Invitrogen).
Chromosome Preparation and Fluorescent in situ Hybridization
Chromosome samples were prepared essentially as described [11], [12]. To determine the integrated transgene neo in the genome, the p53GT22 clone was subjected to fluorescent in situ hybridization (FISH) as described [18]. The DNA fragment containing the STneo was obtained from pGTp53, labeled with biotinylated dUTP by using the Biotin-High Prime kit (Roche, Mannheim) and used as a probe for FISH on nuclei and metaphases. Chromosomes were counterstained with propidium iodide (Sigma, USA).
Induced Cell Differentiation
MES1 cells and p53-targeted cell clones were subjected to EB formation in suspension culture in the presence of all-trans retinoic acid (RA; 10 µM, final) for induced differentiation [11], [12], [13]. Cell differentiation was monitored by phenotype and studied by RT-PCR analyses of expression of pluripotency and lineage-specific genes [12], [31].
Cell Labeling
Cells were stably labeled by transfection with pCVpf or pCVpr followed by puromycin selection (1 µg/ml) and clonal expansion of GFP- or RFP-expressing colonies [16], [18]. Alternatively, cells were transiently labeled by staining with fluorescent dyes PKH26 (red; Sigma) or PKH67 (green; Sigma) for 3 min at final concentration of 2 µM in Diluent C, followed by rinses in PBS as described [35].
Chimera Formation
Labeled MES1 cells and p53-targeted cell clones were transplanted alone or together into dechorionated blastula embryos of strain af and donor cells were monitored regularly as described [18], [21], [30]. Transplantation of medaka ES cells into zebrafish blastulae for interordinal chimeric formation was performed as described [35].
Microscopy
Observation and photography on Leica MZFIII stereo microscope, Zeiss Axiovert invert and Axiovert upright microscopes were as described [12], [20], [21], [32], [44].
Statistics
Statistical analyses were calculated by using Origin 6.1 software. Data consolidated were presented as mean ± SD and p values were calculated by using non-parametric student’s t-test.
Supporting Information
Detection of HR activity of medaka ES cell lines. High HR activity of medaka ES cell lines. ES cell lines MES1 to 3 were transfected with pGTp53 (+) or pBluescript as mock transfection control (-), and subjected to PCR detection following 5 days of growth in the absence of drug or 14 days of growth under PNS.
(TIF)
FISH detection of p53 gene targeting. (A-C) Metaphases. (D-F) Nuclei. The FISH signal is green (arrows). Cells of clone #22 were hybridized with the fluorescein-labeled neo probe (green; probe 2 in Figure 1A) and stained for nuclei with propidium iodide (PI, red). The neo signal is highlighted by arrows. Scale bars, 10 µm.
(TIF)
Junction sequence of targeted p53 locus. GT vector and targeted locus. neo, cassette expressing neomycin aminoglycoside phosphotransferase for resistance to G418. Paired arrows depict the position of PCR primers on the targeted p53 locus. Sequences of five p53-targeted ES clones. The p53 sequence included in the GT vector is shown in black. Shown here are only junction sequences. Shown in bold are ATG codon, polyA signal and stop codon introduced immediately downstream of ATG.
(TIF)
Efficiency of Mitf-directed melanocyte differentiation. MES1 and p53-targeted clones were transfected with pXmitf and pCVpf, the former expressing the Xiphophorus melanocyte-specific isoform of microphthalmia-associated transcription factor (mitf-M) and latter expressing pf, a fusion between the puromycin acetyltransferase and GFP. Following co-transfection, cells were pulse-selected with puromycin for 2 days to enrich for transgenic cells. At day 5 post transfection, cell counting was done on merged micrographs of living cells, and percent values of melanocytes and GFP-positive cells were derived by a comparison to the total number of ≥500 cells for MES1 and clones each.
(TIF)
Distribution of MES1 and p53 -targeted clone 22 in developing chimeras. Following genetic labeling by plasmid transfection, parental MES1 (red) clone #22 (green) were mixed at 1∶1 ratio and transplanted at ∼200 cells per blastula. In total, 54 chimeras were daily scored from day 3 to 7 post fertilization.
(TIF)
Table S1 & S2.
(DOC)
(DOC)
Acknowledgments
We thank Jiaorong Deng for breeding fish and Choy Mei Foong for laboratory management.
Funding Statement
This work was supported by the National Research Foundation Singapore (NRF-CRP7-2010-03). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Capecchi MR (2005) Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet 6: 507–512. [DOI] [PubMed] [Google Scholar]
- 2. Zwaka TP, Thomson JA (2003) Homologous recombination in human embryonic stem cells. Nat Biotechnol 21: 319–321. [DOI] [PubMed] [Google Scholar]
- 3. Meek S, Buehr M, Sutherland L, Thomson A, Mullins JJ, et al. (2010) Efficient gene targeting by homologous recombination in rat embryonic stem cells. PLoS One 5: e14225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collins FS, Rossant J, Wurst W (2007) A mouse for all reasons. Cell 128: 9–13. [DOI] [PubMed] [Google Scholar]
- 5. Tong C, Li P, Wu NL, Yan Y, Ying QL (2010) Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature 467: 211–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fan L, Moon J, Crodian J, Collodi P (2006) Homologous recombination in zebrafish ES cells. Transgenic Res 15: 21–30. [DOI] [PubMed] [Google Scholar]
- 7. Ben J, Elworthy S, Ng AS, van Eeden F, Ingham PW (2011) Targeted mutation of the talpid3 gene in zebrafish reveals its conserved requirement for ciliogenesis and Hedgehog signalling across the vertebrates. Development 138: 4969–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, et al. (2008) Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 26: 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang P, Xiao A, Zhou M, Zhu Z, Lin S, et al. (2011) Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 29: 699–700. [DOI] [PubMed] [Google Scholar]
- 10. Wittbrodt J, Shima A, Schartl M (2002) Medaka–a model organism from the far East. Nat Rev Genet 3: 53–64. [DOI] [PubMed] [Google Scholar]
- 11. Hong Y, Winkler C, Schartl M (1996) Pluripotency and differentiation of embryonic stem cell lines from the medakafish (Oryzias latipes). Mech Dev 60: 33–44. [DOI] [PubMed] [Google Scholar]
- 12. Yi M, Hong N, Hong Y (2009) Generation of medaka fish haploid embryonic stem cells. Science 326: 430–433. [DOI] [PubMed] [Google Scholar]
- 13. Yi M, Hong N, Hong Y (2010) Derivation and characterization of haploid embryonic stem cell cultures in medaka fish. Nat Protoc 5: 1418–1430. [DOI] [PubMed] [Google Scholar]
- 14. Hong Y, Liu T, Zhao H, Xu H, Wang W, et al. (2004) Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc Natl Acad Sci U S A 101: 8011–8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakamura S, Kobayashi K, Nishimura T, Higashijima S, Tanaka M (2010) Identification of germline stem cells in the ovary of the teleost medaka. Science 328: 1561–1563. [DOI] [PubMed] [Google Scholar]
- 16. Hong Y, Chen S, Gui J, Schartl M (2004) Retention of the developmental pluripotency in medaka embryonic stem cells after gene transfer and long-term drug selection for gene targeting in fish. Transgenic Res 13: 41–50. [DOI] [PubMed] [Google Scholar]
- 17. Hong Y, Winkler C, Liu T, Chai G, Schartl M (2004) Activation of the mouse Oct4 promoter in medaka embryonic stem cells and its use for ablation of spontaneous differentiation. Mech Dev 121: 933–943. [DOI] [PubMed] [Google Scholar]
- 18. Yan Y, Du J, Chen T, Yi M, Li M, et al. (2009) Establishment of medakafish as a model for stem cell-based gene therapy: efficient gene delivery and potential chromosomal integration by baculoviral vectors. Exp Cell Res 315: 2322–2331. [DOI] [PubMed] [Google Scholar]
- 19. Zhao H, Hong N, Lu W, Zeng H, Song J, et al. (2012) Fusion gene vectors allowing for simultaneous drug selection, cell labeling, and reporter assay in vitro and in vivo. Anal Chem 84: 987–993. [DOI] [PubMed] [Google Scholar]
- 20. Li M, Hong N, Xu H, Yi M, Li C, et al. (2009) Medaka vasa is required for migration but not survival of primordial germ cells. Mech Dev 126: 366–381. [DOI] [PubMed] [Google Scholar]
- 21. Hong N, Li M, Zeng Z, Yi M, Deng J, et al. (2010) Accessibility of host cell lineages to medaka stem cells depends on genetic background and irradiation of recipient embryos. Cell Mol Life Sci 67: 1189–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lane DP (1992) Cancer. p53, guardian of the genome. Nature 358: 15–16. [DOI] [PubMed] [Google Scholar]
- 23. Schartl M, Wilde B, Laisney JA, Taniguchi Y, Takeda S, et al. (2010) A mutated EGFR is sufficient to induce malignant melanoma with genetic background-dependent histopathologies. J Invest Dermatol 130: 249–258. [DOI] [PubMed] [Google Scholar]
- 24. Taniguchi Y, Takeda S, Furutani-Seiki M, Kamei Y, Todo T, et al. (2006) Generation of medaka gene knockout models by target-selected mutagenesis. Genome Biol 7: R116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cao L, Li W, Kim S, Brodie SG, Deng CX (2003) Senescence, aging, and malignant transformation mediated by p53 in mice lacking the Brca1 full-length isoform. Genes Dev 17: 201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA Jr, et al. (1992) Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356: 215–221. [DOI] [PubMed] [Google Scholar]
- 27. Chen S, Hong Y, Scherer SJ, Schartl M (2001) Lack of ultraviolet-light inducibility of the medakafish (Oryzias latipes) tumor suppressor gene p53. Gene 264: 197–203. [DOI] [PubMed] [Google Scholar]
- 28. Chen S, Hong Y, Schartl M (2002) Cloning, structural analysis and construction of homologous recombination vector of p53 gene in medaka fish (Oryzias latipes). Acta Zoologica Sinica 48: 519–526. [Google Scholar]
- 29. Chen S, Hong Y, Schartl M (2002) Development of a positive-negative selection procedure for gene targeting in fish cells. aquaculture 214: 67–79. [Google Scholar]
- 30. Hong Y, Winkler C, Schartl M (1998) Production of medakafish chimeras from a stable embryonic stem cell line. Proc Natl Acad Sci U S A 95: 3679–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang D, Manali D, Wang T, Bhat N, Hong N, et al. (2011) Identification of pluripotency genes in the fish medaka. Int J Biol Sci 7: 440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Z, Bhat N, Manali D, Wang D, Hong N, et al. (2011) Medaka cleavage embryos are capable of generating ES-like cell cultures. Int J Biol Sci 7: 418–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bejar J, Hong Y, Schartl M (2003) Mitf expression is sufficient to direct differentiation of medaka blastula derived stem cells to melanocytes. Development 130: 6545–6553. [DOI] [PubMed] [Google Scholar]
- 34. Hong Y, Winkler C, Schartl M (1998) Efficiency of cell culture derivation from blastula embryos and of chimera formation in the medaka (Oryzias latipes) depends on donor genotype and passage number. Dev Genes Evol 208: 595–602. [DOI] [PubMed] [Google Scholar]
- 35. Hong N, Chen S, Ge R, Song J, Yi M, et al. (2012) Interordinal chimera formation between medaka and zebrafish for analyzing stem cell differentiation. Stem Cells Dev 21: 2333–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yi M, Hong N, Li Z, Yan Y, Wang D, et al. (2010) Medaka fish stem cells and their applications. Sci China Life Sci 53: 426–434. [DOI] [PubMed] [Google Scholar]
- 37. Hong N, Li Z, Hong Y (2011) Fish stem cell cultures. Int J Biol Sci 7: 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mansour SL, Thomas KR, Capecchi MR (1988) Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature 336: 348–352. [DOI] [PubMed] [Google Scholar]
- 39. Ma C, Fan L, Ganassin R, Bols N, Collodi P (2001) Production of zebrafish germ-line chimeras from embryo cell cultures. Proc Natl Acad Sci U S A 98: 2461–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Herpin A, Fischer P, Liedtke D, Kluever N, Neuner C, et al. (2008) Sequential SDF1a and b-induced mobility guides Medaka PGC migration. Dev Biol 320: 319–327. [DOI] [PubMed] [Google Scholar]
- 41. Iwamatsu T (2004) Stages of normal development in the medaka Oryzias latipes. Mech Dev 121: 605–618. [DOI] [PubMed] [Google Scholar]
- 42. Hong Y, Schartl M (2006) Isolation and differentiation of medaka embryonic stem cells. Methods Mol Biol 329: 3–16. [DOI] [PubMed] [Google Scholar]
- 43. Ramirez-Solis R, Rivera-Perez J, Wallace JD, Wims M, Zheng H, et al. (1992) Genomic DNA microextraction: a method to screen numerous samples. Anal Biochem 201: 331–335. [DOI] [PubMed] [Google Scholar]
- 44. Li M, Hong N, Gui J, Hong Y (2012) Medaka piwi is essential for primordial germ cell migration. Curr Mol Med 12: 1040–1049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detection of HR activity of medaka ES cell lines. High HR activity of medaka ES cell lines. ES cell lines MES1 to 3 were transfected with pGTp53 (+) or pBluescript as mock transfection control (-), and subjected to PCR detection following 5 days of growth in the absence of drug or 14 days of growth under PNS.
(TIF)
FISH detection of p53 gene targeting. (A-C) Metaphases. (D-F) Nuclei. The FISH signal is green (arrows). Cells of clone #22 were hybridized with the fluorescein-labeled neo probe (green; probe 2 in Figure 1A) and stained for nuclei with propidium iodide (PI, red). The neo signal is highlighted by arrows. Scale bars, 10 µm.
(TIF)
Junction sequence of targeted p53 locus. GT vector and targeted locus. neo, cassette expressing neomycin aminoglycoside phosphotransferase for resistance to G418. Paired arrows depict the position of PCR primers on the targeted p53 locus. Sequences of five p53-targeted ES clones. The p53 sequence included in the GT vector is shown in black. Shown here are only junction sequences. Shown in bold are ATG codon, polyA signal and stop codon introduced immediately downstream of ATG.
(TIF)
Efficiency of Mitf-directed melanocyte differentiation. MES1 and p53-targeted clones were transfected with pXmitf and pCVpf, the former expressing the Xiphophorus melanocyte-specific isoform of microphthalmia-associated transcription factor (mitf-M) and latter expressing pf, a fusion between the puromycin acetyltransferase and GFP. Following co-transfection, cells were pulse-selected with puromycin for 2 days to enrich for transgenic cells. At day 5 post transfection, cell counting was done on merged micrographs of living cells, and percent values of melanocytes and GFP-positive cells were derived by a comparison to the total number of ≥500 cells for MES1 and clones each.
(TIF)
Distribution of MES1 and p53 -targeted clone 22 in developing chimeras. Following genetic labeling by plasmid transfection, parental MES1 (red) clone #22 (green) were mixed at 1∶1 ratio and transplanted at ∼200 cells per blastula. In total, 54 chimeras were daily scored from day 3 to 7 post fertilization.
(TIF)
Table S1 & S2.
(DOC)
(DOC)