Abstract
The aetiology of childhood asthma is complex. An early dysfunction in the immunological development of the innate immune system in combination with environmental factors possibly triggers asthma. CD14 and toll-like receptors are important components of the innate immune system. The aim of this systematic review was to obtain a better insight into the relation between CD14 and toll-like receptors and childhood asthma in Caucasians. We searched PubMed and EMBASE for relevant articles. In total, 44 articles were included. The quality of the selected studies was independently assessed by the first two authors using the Newcastle-Ottawa quality assessment scale. Toll-like receptor 2, toll-like receptor 6, toll-like receptor 9, and toll-like receptor 10 appear to have some association with childhood asthma in Caucasians. The evidence for a relation of CD14 with childhood asthma is limited. In conclusion, there is no convincing evidence yet for a role of CD14 and toll-like receptors in relation to childhood asthma. Future studies should include haplotype analysis and take environmental factors into account to further clarify the role of CD14 and toll-like receptors on childhood asthma.
Keywords: Asthma, Caucasian, CD14, Children, Gene expression, Genetic variants, Polymorphisms, TLR, Wheeze
Review
Background
Asthma is a common disease in childhood and is characterized by chronic eosinophilic airway inflammation and airway (hyper) responsiveness [1]. Both, genetic and environmental factors have been associated with asthma. The cumulative effect of these factors is large, though the individual contribution of each factor may be limited [2-6]. Also, genetic and environmental factors can modulate each other. As this modulation can differ with age, it can be time dependent [3]. Hence, the aetiology of childhood asthma is complex.
It is expected that an early dysfunction in the immunological development of the innate immune system, in combination with environmental factors may trigger childhood asthma [7]. The innate immune system makes the first contact with pathogens. Genetic variations in components of the innate immune system can alter the capability to deal with pathogens. The direction of the innate immune system response can depend on presented environmental signals, like endotoxin exposure [8]. The difference in genetic make-up combined with differences in environmental factors leads to variations in the immune response. Important components of the innate immune system are CD14, an adaptor molecule, and a system of pathogen receptors named toll-like receptors (TLRs) [9].
CD14 is a multifunctional high-affinity receptor for endotoxins, lipopolysaccharides and other bacterial wall components. It has been implicated in the development and maturation of the innate immune system [10-13]. Several studies have associated CD14 with determination of the balance between Th1 versus Th2 cytokines [14-16]. It is expressed on the surface of monocytes, macrophages, and neutrophils and occurs as a membrane-bound form and a soluble form [17,18]. Polymorphisms in CD14 and levels of soluble CD14 (sCD14) have been implicated in childhood asthma [10,19-23].
TLRs are evolutionarily conserved receptor complexes in the pathogen-recognition process [24]. Each of the ten TLRs described in humans recognizes a different spectrum of pathogen-associated molecular conformations. After interaction with pathogens, TLRs induce the innate immune response. They are involved in balancing Th1 versus Th2 immune responses, shifting the balance towards a Th1 response [9] TLRs are expressed in immune cells such as antigen-presenting cells, and in epithelial cells [24]. Polymorphisms in TLRs have been linked to childhood asthma [25-28].
In this systematic review we summarize research on CD14 and TLRs in relation to asthma in Caucasian children. By selecting this specific group we expect more clarity in study outcome as conclusions can differ between ethnicities [29]. Consequently, a better insight into the relation between CD14 and TLRs and pathology of childhood asthma in Caucasians would be achieved. Further, we analyzed the effect of environmental factors on the relation between CD14 and TLRs and asthma in Caucasian children.
Methods
Search strategy
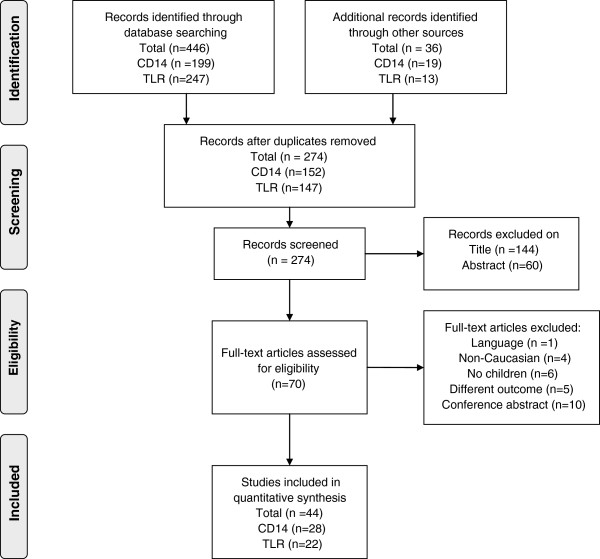
PubMed and EMBASE were searched for relevant articles up to October 2012 by two independent investigators. Primary search terms (CD14, toll-like, TLR) were combined with secondary search terms (asthma*, wheez*) and limitation ‘child’ to find as many relevant publications as possible. Articles were included if they concerned asthma in Caucasian children and investigated the relation with CD14 or TLRs. Articles were excluded if the study population was non-Caucasian (based on area where the study was conducted and stated by the authors) or if the full text was not available in English. Also, conference abstracts and reviews were excluded. The articles were screened by the first two authors for suitability first by title, later by abstract, and finally by full text. In case of disagreement, the senior author was asked to screen the article as well. Additionally, some studies were identified by a manual search of references of included studies. A PRISMA flow diagram of the search is included in Figure 1[30].
Figure 1.
PRISMA flow diagram of search. The flow diagram depicts the flow of information through the different phases of the systematic review. It maps out the number of records identified, included and excluded, and the reasons for exclusions.
Quality assessment
The first two authors independently assessed the quality of the selected studies. Quality assessment was carried out using the Newcastle-Ottawa quality assessment scale which has especially been developed for assessment of observational studies in systematic reviews [31]. In case criteria were met, a “*” was assigned. In case criteria were not met, a “-” was assigned. The overall quality of each article was assessed as better than average (+), average (+/−), or less than average (−). In case of disagreement the senior author was asked to score the article as well. We did not attempt to pool the data.
The categories for cohort studies were as follows: S1: Representativeness of the exposed cohort, S2: Selection of non-exposed cohort, S3: Ascertainment of exposure, S4: Outcome of interest not present at start of study, C1a/C1b: Comparability of cohorts on the basis of the design or analysis, O1: Assessment of outcome, O2: Duration of follow-up, O3: Adequacy of follow up. The categories of case control studies were as follows: S1: Case definition, S2: Representativeness of the cases, S3: Selection of controls, S4: Definition of controls, C1a/C1b Comparability of cases and controls on the basis of the design or analysis, E1: Ascertainment of exposure, E2: Same method of ascertainment for cases and controls, E3: Non-response rate. As no cross-sectional assessment tool was available in the Newcastle-Ottowa quality assessment scale, cross-sectional studies (as identified in the method section of the respective article) were assessed in both the cohort scale and the case control scale. Nested case control studies were scored as case control studies. Polymorphisms, gene expression and/or levels were analyzed by a standard method in all studies. Therefore ascertainment of exposure (S3 for cohorts, E1 and E2 for case control studies) was only scored in case of exposure to environmental factors. Also, we assigned a “*” for E3 in case control studies if the non-response rate was mentioned. Five articles could not be scored as they had neither cohort nor case control or cross-sectional design [19,32-34].
Results
Of the 274 collected records, a total of 44 articles were included in this study (Figure 1). Only one article was excluded based on language [35]. Characteristics of included studies are provided in Additional file 1 (see Additional file 1). The results of the quality scoring can be viewed in Table 1 for cohort studies, and in Table 2 for case control studies.
Table 1.
Quality scoring based on the Newcastle-Ottawa quality assessment scale: cohort studies
| First author | Year | S1 | S2 | S3 | S4 | C1a | C1b | O1 | O2 | O3 | OQ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bieli † [10] |
2007 |
* |
* |
* |
- |
* |
* |
- |
* |
- |
+/− |
| Bottema [26] |
2010 |
* |
* |
NA |
* |
- |
- |
- |
* |
- |
+/− |
| Custovic [62] |
2011 |
* |
* |
- |
* |
* |
* |
- |
* |
* |
+ |
| Eder † [25] |
2004 |
* |
* |
* |
- |
* |
* |
- |
* |
- |
+/− |
| Ege † [22] |
2007 |
* |
* |
* |
- |
* |
* |
- |
* |
- |
+/− |
| Fageras Bottcher † [39] |
2004 |
- |
* |
* |
- |
* |
* |
* |
* |
- |
+/− |
| Guerra [11] |
2004 |
* |
* |
- |
* |
* |
* |
- |
- |
- |
+/− |
| Jones [48] |
2002 |
- |
* |
NA |
* |
- |
- |
- |
- |
- |
– |
| Kabesch † [13] |
2004 |
* |
* |
NA |
* |
- |
- |
- |
* |
- |
+/− |
| Kerkhof [56] |
2010 |
* |
* |
* |
* |
* |
* |
- |
* |
- |
+ |
| Lange [63] |
2011 |
* |
* |
NA |
- |
* |
* |
* |
* |
- |
+ |
| Lodrup Carlsen [51] |
2010 |
* |
* |
NA |
* |
* |
* |
- |
* |
* |
+ |
| O’Donnell [36] |
2004 |
* |
* |
NA |
- |
* |
* |
- |
* |
* |
+ |
| Reijmerink [41] |
2010 |
* |
* |
NA |
* |
- |
- |
- |
* |
- |
+/− |
| Rothenbacher [53] |
2005 |
* |
* |
* |
* |
* |
* |
- |
- |
* |
+ |
| Simpson [8] |
2006 |
* |
* |
* |
* |
* |
* |
- |
* |
* |
+ |
| Snijders † [52] |
2006 |
* |
* |
* |
* |
* |
* |
- |
- |
* |
+ |
| Soferman [54] | 2004 | - | * | NA | * | * | * | * | * | * | + |
S1: Representativeness of the exposed cohort, S2: Selection of non-exposed cohort, S3: Ascertainment of exposure, S4: Outcome of interest not present at start of study, C1a/C1b: Comparability of cohorts on the basis of the design or analysis, O1: Assessment of outcome, O2: Duration of follow-up, O3: Adequacy of follow up (*: criteria were met; -: criteria were not met); OQ: Overall quality (+ better than average quality, +/− average quality, – less than average quality); NA = not applicable; † cross-sectional study.
Table 2.
Quality scoring based on the Newcastle-Ottawa quality assessment scale: case control studies
| First author | Year | S1 | S2 | S3 | S4 | C1a | C1b | E1 | E2 | E3 | OQ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bieli † [10] |
2007 |
- |
* |
* |
- |
* |
* |
* |
* |
* |
+ |
| Bjornvold [38] |
2009 |
*/- |
* |
* |
* |
- |
- |
NA |
NA |
- |
+/− |
| Choudhry [40] |
2005 |
- |
- |
* |
- |
* |
* |
- |
* |
- |
+/− |
| Daley [21] |
2009 |
- |
*/- |
* |
- |
- |
- |
NA |
NA |
- |
+/− |
| Eder † [25] |
2004 |
- |
* |
* |
- |
* |
* |
* |
* |
- |
+/− |
| Ege † [22] |
2007 |
- |
* |
* |
- |
* |
* |
* |
* |
* |
+ |
| Fageras Bottcher † [39] |
2004 |
* |
- |
- |
- |
* |
* |
* |
* |
- |
+/− |
| Genuneit [46] |
2009 |
- |
* |
* |
*/- |
* |
- |
NA |
NA |
- |
+/− |
| Heinzmann [12] |
2003 |
* |
* |
* |
- |
- |
- |
NA |
NA |
- |
+/− |
| Heinzmann [58] |
2010 |
* |
- |
* |
- |
- |
- |
NA |
NA |
- |
+/− |
| Hoffjan [61] |
2005 |
- |
- |
- |
* |
- |
- |
NA |
NA |
- |
– |
| Hussein [59] |
2012 |
* |
* |
- |
* |
- |
- |
NA |
NA |
- |
+/− |
| Jackola [23] |
2006 |
* |
- |
* |
* |
* |
- |
NA |
NA |
- |
+/− |
| Kabesch † [13] |
2004 |
- |
* |
* |
- |
- |
- |
NA |
NA |
- |
+/− |
| Kormann§ [27] |
2008 |
* |
* |
* |
* |
* |
- |
NA |
NA |
- |
+ |
| Kurowski [44] |
2011 |
- |
- |
* |
- |
- |
- |
* |
* |
- |
– |
| Marcos [50] |
2010 |
* |
- |
- |
* |
- |
- |
* |
- |
- |
– |
| Miedema§ [45] |
2012 |
- |
- |
* |
* |
- |
- |
NA |
NA |
- |
+/− |
| Moller-Larsen [28] |
2008 |
* |
- |
* |
* |
* |
- |
NA |
NA |
- |
+/− |
| Perin [42] |
2011 |
* |
- |
- |
* |
- |
- |
NA |
NA |
- |
+/− |
| Puthothu [57] |
2006 |
* |
- |
* |
- |
- |
- |
NA |
NA |
- |
+/− |
| Schubert [60] |
2006 |
* |
- |
* |
- |
- |
- |
NA |
NA |
- |
+/− |
| Sengler § [37] |
2003 |
- |
* |
* |
* |
* |
- |
NA |
NA |
- |
+/− |
| Snijders † [52] |
2006 |
- |
* |
* |
* |
* |
* |
* |
* |
- |
+ |
| Sorensen § [47] |
2009 |
- |
* |
* |
* |
* |
* |
* |
* |
* |
+ |
| Tremblay [20] |
2008 |
- |
*/- |
* |
- |
- |
- |
NA |
NA |
- |
+/− |
| Yang [55] |
2004 |
- |
- |
* |
* |
* |
* |
NA |
NA |
- |
+/− |
| Zhang [43] | 2011 | * | * | * | * | * | * | * | * | - | + |
S1: Case definition; S2: Representativeness of the cases; S3: Selection of controls; S4: Definition of controls; C1a/C1b Comparability of cases and controls on the basis of the design or analysis; E1: Ascertainment of exposure; E2: Same method of ascertainment for cases and controls; E3: Non-response rate (*: criteria were met; -: criteria were not met); OQ: Overall quality (+: better than average quality, +/− average quality, – less than average quality); † cross-sectional study; § nested case control study; NA = not applicable.
Limited evidence for a relation between polymorphisms of CD14 and childhood asthma
The relation between childhood asthma and polymorphisms in CD14 was studied in 17 studies [10,12,13,19-21,34,36-45]. In most studies asthma was taken as outcome. In two studies asthma severity was taken as outcome [19,34]. Age ranged from 0 up to 19 years of age. The number of cases varied from 10 to 644, and controls from 115 to 1858 subjects. CD14 rs2569190 polymorphism, also known as C-260T or C-159T, was studied in 16 studies [10,12,13,19-21,34,36-40,42-45]. Only one study found an association of this polymorphism with childhood asthma, showing an association of the T allele with decreased asthma severity (Table 3) [19]. For the rs4914 polymorphism an association was indicated for decreased asthma incidence in one of the two children cohorts (Study of Asthma Genetics and Environment Cohort) included in one study (Table 3) [20,21]. However, another study investigating rs4914 found no association [38]. All other polymorphisms studied did not show any association with asthma [10,12,20,21,38,40,41,43-45]. Partial duplication in studied population and studied single nucleotide polymorphisms was present between two studies [20,21].
Table 3.
Single nucleotide polymorphisms with association with asthma
| Gene | rs-number | Alleles | Number of associations | Minor allel related to | Association ref. | No association ref. |
|---|---|---|---|---|---|---|
|
CD14 |
rs2569190 |
C > T |
1 out of 15 (duplication [20,21]) |
decreased asthma severity |
[19] |
[10,12,13,20,21,34,36-40,42-45] |
|
CD14 |
rs4914 |
G > C |
1 out of 2 (duplication [20,21]) |
decreased asthma incidence |
[20,21] |
[38] |
|
TLR1 |
rs5743595 |
T > C |
1 out of 1 |
atopic asthma |
[27] |
NA |
|
TLR1 |
rs4833095 |
A > G |
1 out of 1 |
atopic asthma |
[27] |
NA |
|
TLR2 |
rs4696480 |
A > T |
3 out of 5 (partial duplication [26,56]) |
asthma/current asthma symptoms |
[25,26,56] |
[27,45] |
|
TLR2 |
rs3804099 |
T > C |
1 out of 6 (partial duplication [26,56]) |
nonatopic asthma (inverse) |
[27] |
[25,26,38,45,56] |
|
TLR2 |
rs3804100 |
T > C |
1 out of 6 (partial duplication [26,56]) |
allergic asthma (inverse) |
[38] |
[21,25,26,45,56] |
|
TLR2 |
rs1898830 |
A > G |
1 out of 4 |
asthma |
[26] |
[21,27,56] |
|
TLR2 |
rs5743708 |
G > A |
1 out of 2 |
asthma severity in atopic children |
[59] |
[43] |
|
TLR4 |
rs2737190 |
A > G |
1 out of 1 |
(non-atopic) asthma |
[27] |
NA |
|
TLR4 |
rs4986790 |
A > G |
3 out of 10 |
asthma, atopic asthma and asthma severity in atopic children |
[34,39,59] |
[41,43-45,55,56,60] |
|
TLR4 |
rs4986791 |
C > T |
1 out of 4 |
mild atopic asthma |
[34] |
[27,56,60] |
|
TLR6 |
rs6531666 |
T > C |
1 out of 1 |
asthma |
[45] |
NA |
|
TLR6 |
rs5743789 |
T > A |
1 out of 1 |
atopic asthma |
[27] |
NA |
|
TLR6 |
rs5743798 |
C > T |
1 out of 1 |
asthma |
[45] |
NA |
|
TLR6 |
rs5743810 |
C > T |
2 out of 5 |
(atopic) asthma |
[27,61] |
[21,45,57] |
|
TLR7 |
rs179008 |
A > T |
1 out of 2 |
asthma |
[28] |
[27] |
|
TLR8 |
rs2407992 |
G > C |
1 out of 1 |
asthma |
[28] |
NA |
|
TLR9 |
rs187084 |
T > C |
1 out of 3 |
asthma |
[27] |
[21,45] |
|
TLR10 |
rs4129009 |
A > G |
1 out of 2 |
atopic asthma inverse |
[27] |
[45] |
| TLR10 | rs11096957 | A > C | 1 out of 3 | asthma | [21] | [45,57] |
Ref: reference, TLR: Toll-Like Receptor, NA: not applicable.
In three studies the association between rs2569190 in CD14 and wheeze was addressed [8,46,47]. The age of the population studied varied from 18 months up to 13 years. One study investigated two additional polymorphisms (rs5744441 and rs5744455) [46]. None of these studies found a relationship between CD14 polymorphisms and childhood wheeze.
The relation between polymorphisms of CD14 and childhood asthma can be influenced by environmental exposures
The protective effect of farm milk consumption was increased in carriers of one or, even more so, two major alleles of rs2915863 [10]. Also, in individuals exposed to tobacco smoke, a significant association was found between rs2569190 and rs3776138 polymorphisms and asthma severity [40]. Eastern (Russian) or Western (Finnish) environment/lifestyles did not influence the relationship of rs2569190 and rs5744455 on asthma ever in childhood [43]. Neither did age, breastfeeding, allergies to mold, house dust mites or pollens, and exposure to cats and dogs or other large animals in combination with rs5744454, rs2569190 and rs3776138 and asthma-related traits [40], nor dust or living area (rural or urban) with rs2569190 or rs3138078 and asthma [44]. Finally, a dose-dependent relation of endotoxin with non-atopic wheeze for individuals with homozygosity of the major allele of rs2569190 was shown [8]. The association between smoking or pet keeping and the risk of recurrent wheeze was not affected by the rs2569190 polymorphism [47].
Polymorphisms in CD14 lead to increased levels of sCD14
Several studies have shown that sCD14 levels are higher in the presence of one or two minor allele(s) of rs2569190 in chronic and also acute asthma in children [8,13,19,33]. In addition, during asthma attacks an inverse correlation was shown between asthma severity scores and the log value of sCD14 for the major allele of rs2569190 [19]. No significant association was found between rs574441, rs2569193, and rs2569190 and gene expression of CD14 [10].
Higher levels of sCD14 are related to childhood asthma depending on disease status and time of measurement
Levels of sCD14 in cord blood were significantly higher in children with transient-atopy (positive skin prick test at two years of age but not at five years of age and without asthma), compared with never-atopic or persistent-atopic children. However, no changes between transient-atopy, never-atopic or persistant-atopic children could be shown in plasma sCD14 at six months, one year, and five years of age [48]. Also, levels of sCD14 were significantly higher during acute asthma attacks than at recovery in children with status asthmaticus [32]. However, these results might be biased by the administration of corticosteroids during the acute stage which might have caused suppression of cell activation, thereby reducing sCD14 levels in serum [49]. Multiple studies showed no association of sCD14 levels with asthma as measured in cord blood, serum or bronchoalveolar lavage fluid [23,50,51]. In one study no association of sCD14 levels in breast milk with wheeze was found [52].
(Expression) levels of (s)CD14 can be related to environmental exposures leading to a different disease risk
One study showed that of the protective farm related factors that were associated with asthma (pig farming, farm milk consumption, frequent stay in animal sheds, and child’s involvement in haying), only being a farm child in general was associated with higher expression levels of CD14 [22]. Breastfed children who received breast milk with high sCD14 levels had less chance to develop asthma, reaching statistical significance in children of mothers with no history of atopic disease [53]. Another study showed that age or gender did not influence sCD14 production in asthmatic families [23].
CD14 serum levels in infants without recurrent wheezing was significantly higher than in wheezing infants, twelve months after hospitalization due to RSV-induced bronchiolitis. Also, in hospitalized patients sCD14 levels increased with age of admission [54].
Overall, results suggest that some environmental exposures like being a farm child or breast milk with high sCD14 levels protect against development of asthma, however, the number of studies is limited.
Some evidence for a relation between polymorphisms of TLRs (2, 6, 9, 10) and childhood asthma
Childhood asthma in relation to polymorphisms in TLRs was addressed in 19 studies [21,25-28,34,38,39,41,43-45,55-61]. The age of the population in these studies ranged from 0 to 18 years with 31 to 644 cases and 184 to 2927 controls. Outcome was defined as atopic/allergic asthma, non-atopic asthma, bronchial asthma, current asthma symptoms or asthma in general.
TLR4 is by far the most studied of all TLRs in relation to asthma in children, but only some studies showed an association (Table 3). Rs4986791 was statistically more frequent in mild atopic asthmatics [34], whereas rs2737190 showed an association with asthma in general and also non-atopic asthma [27] Rs4986790 showed no association with asthma in seven studies [41,43-45,55,56,60], but was associated with symptoms of (atopic) asthma in two other studies [34,39]. Also, rs4986790 was associated with increased asthma severity in atopic children [59]. However, in most studies polymorphisms of TLR4 did not show any association with asthma [21,25,27,38,41,44,45,55],[56,59,60]. Some duplication between the studies of Reijmerink et al. and Kerkhof et al. was present as the same population was used for single nucleotide polymorphism analysis [41,56]. Studied polymorphisms in the study of Reijmerink were not specified [41].
TLR2, TLR6, TLR9, and TLR10 have been associated with asthma in several studies. For TLR2, rs4696480, rs3804099, rs3804100, and rs1898830 showed association with childhood asthma in a number of studies, but not in others (Table 3) [21,25-27,38,43,56,62]. Moreover, rs5743708 was associated with increased asthma severity in atopic children (Table 3) [59]. Finally, rs13150331, rs4696483, rs5743704, rs7656411, rs1339, rs2289318, and rs5743708 showed no association with childhood asthma [21,43,59]. Two studies had an overlap in data [26,56]. For TLR6, rs6531666, rs5743789, rs5743798, and rs5743810 significant associations with asthma and atopic asthma were found, but not with asthma in other studies (Table 3) [21,27,45,57,61]. Haplotype spanning of polymorphisms in TLR6 and TLR10 (see TLR10) revealed an association with childhood asthma, indicating no individual but a combined influence of these polymorphisms on asthma in children [21,57]. Other TLR6 polymorphisms were not associated with asthma in children [21,27,45]. For TLR9, rs187084 showed a positive association with childhood asthma (Table 3) [27], but not in all studies [21,45]. Other polymorphisms revealed no association with childhood asthma [21,27,44,45,60,63]. For TLR10, out of the eight studied polymorphisms, rs4129009 and rs11096957 showed association with (atopic) asthma in some studies (Table 3) [21,27,45,57]. The rs11096957 polymorphism only demonstrated association in one of the two children cohorts (Study of Asthma Genetics and Environment Cohort) studied by Daley et al [21]. Several polymorphisms (rs4274855, rs10856839, rs11096957, rs11096956, rs11096955, rs11466657, rs4219009) showed no individual association with bronchial asthma, however, haplotype spanning together with polymorphisms in TLR6 (rs5743794, and rs5743810) did [57]. Also rs4274855, rs10856839, rs11096957, and rs4129009 demonstrated a haplotype-based association [58].
TLR7 and TLR8 were only studied in two studies and TLR1, TLR3, and TLR5 in one study: For TLR7, associations with asthma phenotypes for rs179008 separately, and rs179008, rs5743781, rs864058 haplotypes were observed in one study, however, rs179008 showed no association in another study (Table 3) [27,28]. For TLR8, association with asthma phenotype in childhood was shown for rs2407992 separately, and rs5741883, rs3764879, rs3764880, rs5744077, rs2159377, and rs2407992 haplotypes, but not rs3761624 (Table 3) [27,28]. For TLR1, rs5743595, rs4833095, and not rs5743594 showed significant inverse effects with atopic asthma in childhood (Table 3) [27]. For TLR3, rs3775291 showed no association with childhood asthma [27]. Also, rs5744168, rs2072493 and rs5744174 of TLR5 showed no significant association with childhood asthma [27].
One study with 1105 cases and 3137 controls aged 8 to 12 years used wheeze in the past 12 months as outcome variable [46]. For TLR4, rs11536896 showed association, but three other polymorphisms did not. In TLR2, rs1898830 showed association with more frequent wheeze. For TLR9, out of the three studied polymorphisms, rs187084 showed positive association with wheeze.
The relation between polymorphisms of TLRs and childhood asthma is dependent on environmental influences
Four studies addressed the relation between TLR polymorphisms and environmental exposure on childhood asthma [25,43,44,56]. The polymorphisms rs4696480 and rs1898830 in TLR2 and the polymorphisms rs2770150, rs10759931, rs6478317, rs10759932, and rs1927911 in TLR4 increased the effect of fine particulate matter exposure on the prevalence of doctor-diagnosed asthma from birth up to eight years of age. Polymorphism rs10759931 in TLR4 showed an interaction between childhood asthma symptoms and air pollutant levels [56]. High or low endotoxin exposure revealed no interaction with rs4696480, rs3804099, and rs3804100 in TLR2 for asthma diagnosis or current asthma symptoms in children [25]. There was no relation between TLR2 rs5743708 or TLR4 rs4986790 on asthma ever when Eastern or Western environment was taken into account [43]. A relationship neither existed for TLR4 rs4986790 or TLR9 rs352140 with asthma when dust or area (rural/urban) was taken into account [44].
Also, for polymorphism rs4696480 in TLR2 a significant interaction was shown with day care attendance on the development of atopic wheeze in a longitudinal analysis from birth until the age of five years [62].
Polymorphisms in TLRs relate to an increased gene expression of TLRs
Minor alleles of TLR1 rs5743595, TLR6 rs5743789, and TLR10 rs4129009 polymorphisms were associated with increased mRNA expression of the respective TLRs in children [27]. In asthmatic children, farm related factors in combination with TLR polymorphisms were related to higher expression levels of respective TLRs in children [22].
Discussion
We reviewed the literature on the relationship between CD14 and TLRs (polymorphisms) and asthma in Caucasian children. Although a number of studies suggest such a relation, the reviewed literature does not provide convincing evidence for a significant direct role of CD14. TLR2, TLR6, TLR9, and TLR10 did show moderate association with asthma in childhood, but TLR4 did not.
CD14 rs2569190 may be exemplary for the relevance of polymorphisms in relation to asthma. This polymorphism has been analysed in two meta-analysis [64,65]. No significant overall association was detected between rs2569190 and asthma. However, when analysis was stratified by atopy, there was an association indicating a protective effect of the T-allele of rs2569190 in relation to atopic asthma [65]. This was also the case when the analysis was stratified by atopy and restricted to children [64]. However, these results should be interpreted with caution because of the exclusion of studies and the small sample sizes. Also, as the current review was restricted to Caucasian children, comparison is difficult. The lack of similarity in the set-up and populations of these studies makes deduction of conclusions difficult, even more so if the complexity of the disease is taken in account.
Although some TLRs showed associations with asthma in childhood, the most investigated TLR4 did not. A possible explanation is that TLR4, contrary to TLR2, TLR6, TLR9 and TLR10, mainly plays a role in immune response to gram-negative bacteria [25,61]. As the other TLRs are involved in the immune response to multiple microbial products, it can be expected that their role in heterogeneous symptoms and/or diseases is easier to establish than TLR4 [25,27,56,61].
Since CD14 and TLRs are the first mediators in the response of the innate immune system, it is plausible that their contribution to asthma development is influenced by environmental conditions. An increasing number of association studies have included such conditions. However, up to now, the evidence for an interplay between environment and polymorphisms of the mentioned genes is scanty and based on few observations only. If important environmental factors are not included, associations might be overlooked or misinterpreted. For example, it has been demonstrated that the direction of the relation between a CD14 polymorphism and asthma was dependent on endotoxin exposure [8]. Also interesting is the fact that sCD14 in serum seems to be increased at specific time points, which might indicate a time window at which children are more vulnerable for exposure [54]. Consequently, increasing the number of populations studied, without inclusion of environmental influences, will not guarantee more valid results. Indeed, one may argue that studies not taking environmental influences into account underestimate the complexity of the disease.
Another important development in recent years is the focus on haplotype analysis instead of individual polymorphisms [66-68]. It has been shown that the combined analysis of several polymorphisms by haplotypes increases the power to detect associations. In this review, two studies clearly show the strength of this approach, since in these studies individual polymorphisms did not provide an association, whereas haplotype spanning did [57,58]. Consequently, our insight into the role of CD14 and TLRs in relation to asthma would benefit from haplotype analysis. Application of this method will not demand a major investment since previous collected study populations can be reused.
Our review has several strengths. Firstly, by systematic evaluation of the available literature, our review provides an efficient and comprehensive summary of CD14 and TLRs polymorphisms in relation to childhood asthma. Secondly, to increase the number of articles included, we choose to include studies with various outcome measures of asthma. Thirdly, independent searching and scoring of articles was performed to come to a balanced opinion. Also, no articles were rejected based on assigned score, as there is no general agreement on evaluation of observational studies [69]. Fourthly, we decided to focus on asthma as outcome measure as it is a more established entity than for example wheeze. While some studies only used parent reported diagnosis, other studies used more objective methods like doctor diagnosis and lung function [10,12,23,36,38,56]. However, as wheeze is a good predictor in older age for presence of asthma we included studies with outcome wheeze in our review as well. Though, we did separate the used definitions as the underlying mechanisms can differ. Finally, contrary to most reviews, the population included is restricted to Caucasian children.
Some limitations need to be mentioned. Firstly, the number of articles on this subject is limited. Secondly, the wide scatter in age within and between studies may lead to difficulties in interpretation of results due to such factors as behaviour and environment. Thirdly, as is the case for most reviews, the comparability between the studies was diminished by methodological differences between the studies.
Conclusions
All studies taken together, no clear role for CD14 and TLRs in childhood asthma can be determined based on this review. This is probably due to unknown underlying confounding or mediating environmental factors. Another reason can be the fact that analysis has mostly been performed on individual polymorphisms instead of haplotype spanning. Therefore, planning future studies should include haplotype analysis and take environmental factors into account. Also, to be able to identify and confirm associations and gene-gene and gene-environmental interactions with sufficient power, large populations will be necessary. If the above mentioned suggestions are taken into consideration, future studies on the role of CD14 and TLRs in childhood asthma may be more successful.
Abbreviations
sCD14: Soluble CD14; TLR: Toll-like receptor.
Competing interests
The authors declare that they have no competing interest.
Authors’ contributions
EK and BT contributed to the data-acquisition and writing of the manuscript, GE and ED were mainly involved in revision of the manuscript. QJ supervised the project. All authors substantially contributed to the design of the review, interpretation of data and revision of article. All authors read and approved the final manuscript.
Supplementary Material
Characteristics of included studies. Characteristics of included studies are provided: First author, year of publication, type of study, parameters studied, genetic variants included, outcome, environmental exposures, sample size, age of population, study setting, analysis technique, statistics used consistency with the Hardy-Weinberg equilibrium and genotyping success rates. Only data relevant for the current review are provided. Abbreviations in alphabetical order: ALEX: Allergy and Endotoxin, ANOVA: analysis of variance, CAMP: Childhood Asthma Management Program, CAPPS: Canadian Asthma Primary Prevention Study, Cc: case control design, Cs: cross-sectional design, Co: cohort design, DNBC: The Danish National Birth Cohort, ECA: Environment and Childhood Asthma, ELISA: Enzyme-Linked ImmunoSorbent Assay, FACS: Fluorescence-Activated Cell Sorting, FBAT: Family-Based Association Test, GEE: Generalized estimating equations, HWE: Hardy Weinberg Equilibrium, HPLC: High-Performance Liquid Chromatography, irt: in relation to, ISAAC: International Study of Asthma and Allergies in Childhood, ISS: Infant Immune Study, KOALA: Kind, Ouders en gezondheid: Aandacht voor Leefstijl en Aanleg, prospective birth cohort study in the Netherlands, LR: logistic regression, MAAS: Manchester Asthma and Allergy Study, MAS:Multicentre Allergy Study, MWU: Mann–Whitney U, NA: not applicable, NS: not stated, PARSIFAL: Prevention of Allergy - Risk Factors for Sensitisation in Children Related to Farming and Anthroposophic Lifestyle, PCR: Polymerase Chain Reaction, PIAMA: Prevention and Incidence of Asthma and Mite Allergy, PREVASC: Prevention of asthma in genetically susceptible children, RFLP: restriction fragment length polymorphism, RSV: Respiratory Syncytial Virus, SAGE: Study of Asthma Genes and Environment, sCD14: soluble CD14, SNP: Single Nucleotide Polymorphism, SPT: skin prick test, SSCP: single strand conformation polymorphism, TDT: transmission disequilibrium test, TLR: Toll-like receptor, USA: United States of America vs: versus, yrs: years, X2: Chi-square.
Contributor Information
Ester MM Klaassen, Email: e.klaassen@maastrichtuniversity.nl.
Brenda EJT Thönissen, Email: Brenda.thonissen@gmail.com.
Guillaume van Eys, Email: vanEys@MOLCELB.unimaas.nl.
Edward Dompeling, Email: edward.dompeling@mumc.nl.
Quirijn Jöbsis, Email: r.jobsis@mumc.nl.
References
- Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59:469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- Le Souef PN. Gene-environmental interaction in the development of atopic asthma: new developments. Curr Opin Allergy Clin Immunol. 2009;9:123–127. doi: 10.1097/ACI.0b013e3283292283. [DOI] [PubMed] [Google Scholar]
- Bartlett NW, McLean GR, Chang YS, Johnston SL. Genetics and epidemiology: asthma and infection. Curr Opin Allergy Clin Immunol. 2009;9:395–400. doi: 10.1097/ACI.0b013e32833066fa. [DOI] [PubMed] [Google Scholar]
- Prescott SL. Effects of early cigarette smoke exposure on early immune development and respiratory disease. Paediatr Respir Rev. 2008;9:3–9. doi: 10.1016/j.prrv.2007.11.004. quiz 10. [DOI] [PubMed] [Google Scholar]
- Pinto LA, Stein RT, Kabesch M. Impact of genetics in childhood asthma. J Pediatr (Rio J) 2008;84:S68–S75. doi: 10.1590/S0021-75572008000500010. [DOI] [PubMed] [Google Scholar]
- Thomsen SF, van der Sluis S, Kyvik KO, Skytthe A, Skadhauge LR, Backer V. Increase in the heritability of asthma from 1994 to 2003 among adolescent twins. Respir Med. 2011;105:1147–1152. doi: 10.1016/j.rmed.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Halfhide C, Smyth RL. Innate immune response and bronchiolitis and preschool recurrent wheeze. Paediatr Respir Rev. 2008;9:251–262. doi: 10.1016/j.prrv.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson A, John SL, Jury F, Niven R, Woodcock A, Ollier WE, Custovic A. Endotoxin exposure, CD14, and allergic disease: an interaction between genes and the environment. Am J Respir Crit Care Med. 2006;174:386–392. doi: 10.1164/rccm.200509-1380OC. [DOI] [PubMed] [Google Scholar]
- Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- Bieli C, Eder W, Frei R, Braun-Fahrlander C, Klimecki W, Waser M, Riedler J, von Mutius E, Scheynius A, Pershagen G. A polymorphism in CD14 modifies the effect of farm milk consumption on allergic diseases and CD14 gene expression. J Allergy Clin Immunol. 2007;120:1308–1315. doi: 10.1016/j.jaci.2007.07.034. [DOI] [PubMed] [Google Scholar]
- Guerra S, Lohman IC, Halonen M, Martinez FD, Wright AL. Reduced interferon gamma production and soluble CD14 levels in early life predict recurrent wheezing by 1 year of age. Am J Respir Crit Care Med. 2004;169:70–76. doi: 10.1164/rccm.200304-499OC. [DOI] [PubMed] [Google Scholar]
- Heinzmann A, Dietrich H, Jerkic SP, Kurz T, Deichmann KA. Promoter polymorphisms of the CD14 gene are not associated with bronchial asthma in Caucasian children. Eur J Immunogenet. 2003;30:345–348. doi: 10.1046/j.1365-2370.2003.00414.x. [DOI] [PubMed] [Google Scholar]
- Kabesch M, Hasemann K, Schickinger V, Tzotcheva I, Bohnert A, Carr D, Baldini M, Hackstein H, Leupold W, Weiland SK. A promoter polymorphism in the CD14 gene is associated with elevated levels of soluble CD14 but not with IgE or atopic diseases. Allergy. 2004;59:520–525. doi: 10.1111/j.1398-9995.2004.00439.x. [DOI] [PubMed] [Google Scholar]
- Holt PG, Sly PD, Bjorksten B. Atopic versus infectious diseases in childhood: a question of balance? Pediatr Allergy Immunol. 1997;8:53–58. doi: 10.1111/j.1399-3038.1997.tb00145.x. [DOI] [PubMed] [Google Scholar]
- Baldini M, Lohman IC, Halonen M, Erickson RP, Holt PG, Martinez FD. A Polymorphism* in the 5′ flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol. 1999;20:976–983. doi: 10.1165/ajrcmb.20.5.3494. [DOI] [PubMed] [Google Scholar]
- Vercelli D, Baldini M, Stern D, Lohman IC, Halonen M, Martinez F. CD14: a bridge between innate immunity and adaptive IgE responses. J Endotoxin Res. 2001;7:45–48. [PubMed] [Google Scholar]
- Haziot A, Chen S, Ferrero E, Low MG, Silber R, Goyert SM. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. J Immunol. 1988;141:547–552. [PubMed] [Google Scholar]
- Ulevitch RJ, Tobias PS. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu Rev Immunol. 1995;13:437–457. doi: 10.1146/annurev.iy.13.040195.002253. [DOI] [PubMed] [Google Scholar]
- Martin AC, Laing IA, Khoo SK, Zhang G, Rueter K, Teoh L, Taheri S, Hayden CM, Geelhoed GC, Goldblatt J, LeSouef PN. Acute asthma in children: Relationships among CD14 and CC16 genotypes, plasma levels, and severity. Am J Respir Crit Care Med. 2006;173:617–622. doi: 10.1164/rccm.200509-1367OC. [DOI] [PubMed] [Google Scholar]
- Tremblay K, Daley D, Chamberland A, Lemire M, Montpetit A, Laviolette M, Musk AW, James AL, Chan-Yeung M, Becker A. Genetic variation in immune signaling genes differentially expressed in asthmatic lung tissues. J Allergy Clin Immunol. 2008;122:529–536. doi: 10.1016/j.jaci.2008.05.049. e517. [DOI] [PubMed] [Google Scholar]
- Daley D, Lemire M, Akhabir L, Chan-Yeung M, Qing He J, McDonald T, Sandford AJ, Stefanowicz D, Tripp B, Zamar D. Analyses of associations with asthma in four asthma population samples from Canada and Austrialia. Hum Genet. 2009;125:445–459. doi: 10.1007/s00439-009-0643-8. [DOI] [PubMed] [Google Scholar]
- Ege MJ, Frei R, Bieli C, Schram-Bijkerk D, Waser M, Benz MR, Weiss G, Nyberg F, van Hage M, Pershagen G. Not all farming environments protect against the development of asthma and wheeze in children. J Allergy Clin Immunol. 2007;119:1140–1147. doi: 10.1016/j.jaci.2007.01.037. [DOI] [PubMed] [Google Scholar]
- Jackola DR, Basu S, Liebeler CL, Willaert R, Luah SS, Oetting W, King RA, Blumenthal MN. CD14 promoter polymorphisms in atopic families: implications for modulated allergen-specific immunoglobulin E and G1 responses. Int Arch Allergy Immunol. 2006;139:217–224. doi: 10.1159/000091167. [DOI] [PubMed] [Google Scholar]
- Imler JL, Hoffmann JA. Toll receptors in innate immunity. Trends Cell Biol. 2001;11:304–311. doi: 10.1016/S0962-8924(01)02004-9. [DOI] [PubMed] [Google Scholar]
- Eder W, Klimecki W, Yu L, von Mutius E, Riedler J, Braun-Fahrlander C, Nowak D, Martinez FD. Toll-like receptor 2 as a major gene for asthma in children of European farmers. J Allergy Clin Immunol. 2004;113:482–488. doi: 10.1016/j.jaci.2003.12.374. [DOI] [PubMed] [Google Scholar]
- Bottema RW, Kerkhof M, Reijmerink NE, Thijs C, Smit HA, van Schayck CP, Brunekreef B, van Oosterhout AJ, Postma DS, Koppelman GH. Gene-gene interaction in regulatory T-cell function in atopy and asthma development in childhood. J Allergy Clin Immunol. 2010;126:338–346. doi: 10.1016/j.jaci.2010.04.024. 346 e331-310. [DOI] [PubMed] [Google Scholar]
- Kormann MS, Depner M, Hartl D, Klopp N, Illig T, Adamski J, Vogelberg C, Weiland SK, von Mutius E, Kabesch M. Toll-like receptor heterodimer variants protect from childhood asthma. J Allergy Clin Immunol. 2008;122:86–92. doi: 10.1016/j.jaci.2008.04.039. 92 e81-88. [DOI] [PubMed] [Google Scholar]
- Moller-Larsen S, Nyegaard M, Haagerup A, Vestbo J, Kruse TA, Borglum AD. Association analysis identifies TLR7 and TLR8 as novel risk genes in asthma and related disorders. Thorax. 2008;63:1064–1069. doi: 10.1136/thx.2007.094128. [DOI] [PubMed] [Google Scholar]
- Zhang G, Goldblatt J, LeSouef PN. Does the relationship between IgE and the CD14 gene depend on ethnicity? Allergy. 2008;63:1411–1417. doi: 10.1111/j.1398-9995.2008.01804.x. [DOI] [PubMed] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62:1006–1012. doi: 10.1016/j.jclinepi.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Wells GA, Shea B, O'Connell D, Peterson J, Welch V, Losos M, Tugwell P. Quality assessment scales for observational studies. Ottowa Health Research Institute; 2004. www.ohri.ca/programs/clinical_epidemiology/oxford.htm. [Google Scholar]
- Garty BZ, Monselise Y, Nitzan M. Soluble CD14 in children with status asthmaticus. Isr Med Assoc J. 2000;2:104–107. [PubMed] [Google Scholar]
- Keskin O, Birben E, Sackesen C, Soyer OU, Alyamac E, Karaaslan C, Tokol N, Ercan H, Kalayci O. The effect of CD14-c159T genotypes on the cytokine response to endotoxin by peripheral blood mononuclear cells from asthmatic children. Ann Allergy Asthma Immunol. 2006;97:321–328. doi: 10.1016/S1081-1206(10)60796-X. [DOI] [PubMed] [Google Scholar]
- Sackesen C, Karaaslan C, Keskin O, Tokol N, Tahan F, Civelek E, Soyer OU, Adalioglu G, Tuncer A, Birben E. The effect of polymorphisms at the CD14 promoter and the TLR4 gene on asthma phenotypes in Turkish children with asthma. Allergy. 2005;60:1485–1492. doi: 10.1111/j.1398-9995.2005.00874.x. [DOI] [PubMed] [Google Scholar]
- Lis G, Kostyk E, Sanak M, Pietrzyk JJ. Molecular studies in a population of children with bronchial asthma. I. Polymorphism in the promotor region of gene CD14. Pneumonol Alergol Pol. 2001;69:265–272. [PubMed] [Google Scholar]
- O’Donnell AR, Toelle BG, Marks GB, Hayden CM, Laing IA, Peat JK, Goldblatt J, Le Souef PN. Age-specific relationship between CD14 and atopy in a cohort assessed from age 8 to 25 years. Am J Respir Crit Care Med. 2004;169:615–622. doi: 10.1164/rccm.200302-278OC. [DOI] [PubMed] [Google Scholar]
- Sengler C, Haider A, Sommerfeld C, Lau S, Baldini M, Martinez F, Wahn U, Nickel R. Evaluation of the CD14 C-159 T polymorphism in the German Multicenter Allergy Study cohort. Clin Exp Allergy. 2003;33:166–169. doi: 10.1046/j.1365-2222.2003.01549.x. [DOI] [PubMed] [Google Scholar]
- Bjornvold M, Munthe-Kaas MC, Egeland T, Joner G, Dahl-Jorgensen K, Njolstad PR, Akselsen HE, Gervin K, Carlsen KC, Carlsen KH, Undlien DE. A TLR2 polymorphism is associated with type 1 diabetes and allergic asthma. Genes Immun. 2009;10:181–187. doi: 10.1038/gene.2008.100. [DOI] [PubMed] [Google Scholar]
- Fageras Bottcher M, Hmani-Aifa M, Lindstrom A, Jenmalm MC, Mai XM, Nilsson L, Zdolsek HA, Bjorksten B, Soderkvist P, Vaarala O. A TLR4 polymorphism is associated with asthma and reduced lipopolysaccharide-induced interleukin-12(p70) responses in Swedish children. J Allergy Clin Immunol. 2004;114:561–567. doi: 10.1016/j.jaci.2004.04.050. [DOI] [PubMed] [Google Scholar]
- Choudhry S, Avila PC, Nazario S, Ung N, Kho J, Rodriguez-Santana JR, Casal J, Tsai HJ, Torres A, Ziv E. CD14 tobacco gene-environment interaction modifies asthma severity and immunoglobulin E levels in Latinos with asthma. Am J Respir Crit Care Med. 2005;172:173–182. doi: 10.1164/rccm.200409-1232OC. [DOI] [PubMed] [Google Scholar]
- Reijmerink NE, Bottema RW, Kerkhof M, Gerritsen J, Stelma FF, Thijs C, van Schayck CP, Smit HA, Brunekreef B, Koppelman GH, Postma DS. TLR-related pathway analysis: novel gene-gene interactions in the development of asthma and atopy. Allergy. 2009;65:199–207. doi: 10.1111/j.1398-9995.2009.02111.x. [DOI] [PubMed] [Google Scholar]
- Perin P, Berce V, Potocnik U. CD14 gene polymorphism is not associated with asthma but rather with bronchial obstruction and hyperreactivity in Slovenian children with non-atopic asthma. Respir Med. 2011;105(Suppl 1):S54–S59. doi: 10.1016/S0954-6111(11)70012-9. [DOI] [PubMed] [Google Scholar]
- Zhang G, Candelaria P, Makela JM, Khoo SK, Hayden MC, von Hertzen L, Laatikainen T, Vartiainen E, Goldblatt J, Haahtela T, LeSouef NP. Disparity of innate immunity-related gene effects on asthma and allergy on Karelia. Pediatr Allergy Immunol. 2011;22:621–630. doi: 10.1111/j.1399-3038.2011.01186.x. [DOI] [PubMed] [Google Scholar]
- Kurowski M, Majkowska-Wojciechowska B, Wardzynska A, Kowalski ML. Associations of allergic sensitization and clinical phenotypes with innate immune response genes polymorphisms are modified by house dust mite allergen exposure. Arch Med Sci. 2011;7:1029–1036. doi: 10.5114/aoms.2011.26616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedema KG, Tissing WJ, Te Poele EM, Kamps WA, Alizadeh BZ, Kerkhof M, de Jongste JC, Smit HA, de Pagter AP, Bierings M. Polymorphisms in the TLR6 gene associated with the inverse association between childhood acute lymphoblastic leukemia and atopic disease. Leukemia. 2012;26:1203–1210. doi: 10.1038/leu.2011.341. [DOI] [PubMed] [Google Scholar]
- Genuneit J, Cantelmo JL, Weinmayr G, Wong GW, Cooper PJ, Riikjarv MA, Gotua M, Kabesch M, von Mutius E, Forastiere F. A multi-centre study of candidate genes for wheeze and allergy: the International Study of Asthma and Allergies in Childhood Phase 2. Clin Exp Allergy. 2009;39:1875–1888. doi: 10.1111/j.1365-2222.2009.03364.x. [DOI] [PubMed] [Google Scholar]
- Sorensen M, Allermann L, Vogel U, Andersen PS, Jespersgaard C, Loft S, Raaschou-Nielsen O. Polymorphisms in inflammation genes, tobacco smoke and furred pets and wheeze in children. Pediatr Allergy Immunol. 2009;20:614–623. doi: 10.1111/j.1399-3038.2009.00855.x. [DOI] [PubMed] [Google Scholar]
- Jones CA, Holloway JA, Popplewell EJ, Diaper ND, Holloway JW, Vance GH, Warner JA, Warner JO. Reduced soluble CD14 levels in amniotic fluid and breast milk are associated with the subsequent development of atopy, eczema, or both. J Allergy Clin Immunol. 2002;109:858–866. doi: 10.1067/mai.2002.123535. [DOI] [PubMed] [Google Scholar]
- Djukanovic R, Roche WR, Wilson JW, Beasley CR, Twentyman OP, Howarth RH, Holgate ST. Mucosal inflammation in asthma. Am Rev Respir Dis. 1990;142:434–457. doi: 10.1164/ajrccm/142.2.434. [DOI] [PubMed] [Google Scholar]
- Marcos V, Latzin P, Hector A, Sonanini S, Hoffmann F, Lacher M, Koller B, Bufler P, Nicolai T, Hartl D, Griese M. Expression, regulation and clinical significance of soluble and membrane CD14 receptors in pediatric inflammatory lung diseases. Respir Res. 2010;11:32. doi: 10.1186/1465-9921-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodrup Carlsen KC, Mowinckel K, Granum B, Carlsen KH. Can childhood asthma be predicted at birth? Clin Exp Allergy. 2010;40:1767–1775. doi: 10.1111/j.1365-2222.2010.03620.x. [DOI] [PubMed] [Google Scholar]
- Snijders BE, Damoiseaux JG, Penders J, Kummeling I, Stelma FF, van Ree R, van den Brandt PA, Thijs C. Cytokines and soluble CD14 in breast milk in relation with atopic manifestations in mother and infant (KOALA Study) Clin Exp Allergy. 2006;36:1609–1615. doi: 10.1111/j.1365-2222.2006.02613.x. [DOI] [PubMed] [Google Scholar]
- Rothenbacher D, Weyermann M, Beermann C, Brenner H. Breastfeeding, soluble CD14 concentration in breast milk and risk of atopic dermatitis and asthma in early childhood: birth cohort study. Clin Exp Allergy. 2005;35:1014–1021. doi: 10.1111/j.1365-2222.2005.02298.x. [DOI] [PubMed] [Google Scholar]
- Soferman R, Bar-Zohar D, Jurgenson U, Fireman E. Soluble CD14 as a predictor of subsequent development of recurrent wheezing in hospitalized young children with respiratory syncytial virus-induced bronchiolitis. Ann Allergy Asthma Immunol. 2004;92:545–548. doi: 10.1016/S1081-1206(10)61762-0. [DOI] [PubMed] [Google Scholar]
- Yang IA, Barton SJ, Rorke S, Cakebread JA, Keith TP, Clough JB, Holgate ST, Holloway JW. Toll-like receptor 4 polymorphism and severity of atopy in asthmatics. Genes Immun. 2004;5:41–45. doi: 10.1038/sj.gene.6364037. [DOI] [PubMed] [Google Scholar]
- Kerkhof M, Postma DS, Brunekreef B, Reijmerink NE, Wijga AH, de Jongste JC, Gehring U, Koppelman GH. Toll-like receptor 2 and 4 genes influence susceptibility to adverse effects of traffic-related air pollution on childhood asthma. Thorax. 2010;65:690–697. doi: 10.1136/thx.2009.119636. [DOI] [PubMed] [Google Scholar]
- Puthothu B, Heinzmann A. Is toll-like receptor 6 or toll-like receptor 10 involved in asthma genetics–or both? Allergy. 2006;61:649–650. doi: 10.1111/j.1398-9995.2006.01098.x. [DOI] [PubMed] [Google Scholar]
- Heinzmann A, Brugger M, Bierbaum S, Mailaparambil B, Kopp MV, Strauch K. Joint influences of Acidic-Mammalian-Chitinase with Interleukin-4 and Toll-like receptor-10 with Interleukin-13 in the genetics of asthma. Pediatr Allergy Immunol. 2010;21:e679–e686. doi: 10.1111/j.1399-3038.2010.01053.x. [DOI] [PubMed] [Google Scholar]
- Hussein YM, Awad HA, Shalaby SM, Ali AS, Alzahrani SS. Toll-like receptor 2 and Toll-like receptor 4 polymorphisms and susceptibility to asthma and allergic rhinitis: a case–control analysis. Cell Immunol. 2012;274:34–38. doi: 10.1016/j.cellimm.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Schubert K, von Bonnsdorf H, Burke M, Ahlert I, Braun S, Berner R, Deichmann KA, Heinzmann A. A comprehensive candidate gene study on bronchial asthma and juvenile idiopathic arthritis. Dis Markers. 2006;22:127–132. doi: 10.1155/2006/373620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffjan S, Stemmler S, Parwez Q, Petrasch-Parwez E, Arinir U, Rohde G, Reinitz-Rademacher K, Schultze-Werninghaus G, Bufe A, Epplen JT. Evaluation of the toll-like receptor 6 Ser249Pro polymorphism in patients with asthma, atopic dermatitis and chronic obstructive pulmonary disease. BMC Med Genet. 2005;6:34. doi: 10.1186/1471-2350-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custovic A, Rothers J, Stern D, Simpson A, Woodcock A, Wright AL, Nicolaou NC, Hankinson J, Halonen M, Martinez FD. Effect of day care attendance on sensitization and atopic wheezing differs by Toll-like receptor 2 genotype in 2 population-based birth cohort studies. J Allergy Clin Immunol. 2011;127:390–397. doi: 10.1016/j.jaci.2010.10.050. e391-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange NE, Zhou X, Lasky-Su J, Himes BE, Lazarus R, Soto-Quiros M, Avila L, Celedon JC, Hawrylowicz CM, Raby BA, Litonjua AA. Comprehensive genetic assessment of a functional TLR9 promoter polymorphism: no replicable association with asthma or asthma-related phenotypes. BMC Med Genet. 2011;12:26. doi: 10.1186/1471-2350-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tian C, Zhang J, Li X, Wan H, He C, Guo L, Meilang Q, Peng C, Duo L. The -159C/T polymorphism in the CD14 gene and the risk of asthma: a meta-analysis. Immunogenetics. 2011;63:23–32. doi: 10.1007/s00251-010-0489-1. [DOI] [PubMed] [Google Scholar]
- Zhao L, Bracken MB. Association of CD14–260 (−159) C > T and asthma: a systematic review and meta-analysis. BMC Med Genet. 2011;12:93. doi: 10.1186/1471-2350-12-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Haplotype phasing: existing methods and new developments. Nat Rev Genet. 2011;12:703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele EJ, Williamson JF, Lester S, Stewart BJ, Millman JA, Carnegie P, Lindley RA, Pain GN, Dawkins RL. Genesis of ancestral haplotypes: RNA modifications and reverse transcription-mediated polymorphisms. Hum Immunol. 2011;72:283–293. doi: 10.1016/j.humimm.2010.12.005. e281. [DOI] [PubMed] [Google Scholar]
- Stauffer D, Cebrat S. Haplotype complementarity under mutational pressure. Front Biosci (Schol Ed) 2011;3:408–415. doi: 10.2741/s160. [DOI] [PubMed] [Google Scholar]
- Sanderson S, Tatt ID, Higgins JP. Tools for assessing quality and susceptibility to bias in observational studies in epidemiology: a systematic review and annotated bibliography. Int J Epidemiol. 2007;36:666–676. doi: 10.1093/ije/dym018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of included studies. Characteristics of included studies are provided: First author, year of publication, type of study, parameters studied, genetic variants included, outcome, environmental exposures, sample size, age of population, study setting, analysis technique, statistics used consistency with the Hardy-Weinberg equilibrium and genotyping success rates. Only data relevant for the current review are provided. Abbreviations in alphabetical order: ALEX: Allergy and Endotoxin, ANOVA: analysis of variance, CAMP: Childhood Asthma Management Program, CAPPS: Canadian Asthma Primary Prevention Study, Cc: case control design, Cs: cross-sectional design, Co: cohort design, DNBC: The Danish National Birth Cohort, ECA: Environment and Childhood Asthma, ELISA: Enzyme-Linked ImmunoSorbent Assay, FACS: Fluorescence-Activated Cell Sorting, FBAT: Family-Based Association Test, GEE: Generalized estimating equations, HWE: Hardy Weinberg Equilibrium, HPLC: High-Performance Liquid Chromatography, irt: in relation to, ISAAC: International Study of Asthma and Allergies in Childhood, ISS: Infant Immune Study, KOALA: Kind, Ouders en gezondheid: Aandacht voor Leefstijl en Aanleg, prospective birth cohort study in the Netherlands, LR: logistic regression, MAAS: Manchester Asthma and Allergy Study, MAS:Multicentre Allergy Study, MWU: Mann–Whitney U, NA: not applicable, NS: not stated, PARSIFAL: Prevention of Allergy - Risk Factors for Sensitisation in Children Related to Farming and Anthroposophic Lifestyle, PCR: Polymerase Chain Reaction, PIAMA: Prevention and Incidence of Asthma and Mite Allergy, PREVASC: Prevention of asthma in genetically susceptible children, RFLP: restriction fragment length polymorphism, RSV: Respiratory Syncytial Virus, SAGE: Study of Asthma Genes and Environment, sCD14: soluble CD14, SNP: Single Nucleotide Polymorphism, SPT: skin prick test, SSCP: single strand conformation polymorphism, TDT: transmission disequilibrium test, TLR: Toll-like receptor, USA: United States of America vs: versus, yrs: years, X2: Chi-square.