Abstract
The mechanism of action of clinically used Pt-based drugs is through the formation of stable DNA adducts occurring at the nitrogen in position 7 of guanine (N7) and involving one or two spatially closed residues. Nevertheless, proteins can represent alternative targets since in particular sulphur groups, present in cysteine or methionine residues, can efficiently coordinate platinum. Here we have characterized the reactivity profile of cisplatin, transplatin and of two trans-platinum amine derivatives (TPAs) towards three different proteins, bovine α-lactalbumin (α-LA), hen egg lysozyme (LYS) and human serum albumin (HSA).
Our results demonstrate that generally the tested metal complexes react with the selected target causing protein oligomerization, likely through a cross-linking reaction. Interestingly, the extent of such a process is largely modulated by the target protein and by the chemical features of the metal complex, TPAs being the most efficient platinating agents. From a structural point of view the resulting reaction products turned out to be dependenting on the nature of the metal complexes. However, in all instances, a transfer reaction of the metal complex to DNA can also occur, maintaining the relevance of nucleic acids as a biological target. These results can be used to better rationalize the different pharmacological profiles reported for cisplatin and TPAs and can help in designing more predictive SARs within the series.
Keywords: Platinum-based drugs, Nucleic acids, Proteins, Cancer therapy, Adducts formation, Oligomerization
1. Introduction
Platinum complexes represent one of the most successful families of clinically used anticancer drugs. Their mechanism of action is through the formation of stable DNA adducts which are recognized as a DNA damaging event and that, ultimately, result in cell death by apoptosis. From the discovery of cisplatin, research has been largely focused on the characterization of DNA-adducts formation by Pt-based drugs with the aim to provide consistent structure-activity relationships. These studies indicated the nitrogen in position 7 of guanine (N7) as the preferential platination site. Interestingly, according to the nature of the transition metal ion and the stereochemistry of the complex, bifunctional adducts involving two adjacent nucleobases or two residues of the paired DNA strands, can be formed [1].
Nevertheless, nucleobases are not the only biological targets of these drugs. Cysteine or methionine residues in proteins are reactive sites, since platinum can efficiently interact with sulphur groups [2, 3]. Thus, a role of protein as target, reaction intermediate or reservoir system in platinum drug metabolism may be considered. Indeed, the formation of DNA-protein cross-linking has been suggested as part of the mechanism of action of cisplatin as well as to rationalize pharmacokinetics, resistance mechanisms and side effects of this anticancer treatment.
Several studies have focused on the structural characterization of platinated proteins [4]. In particular, plasma proteins such as human serum albumin (HSA) [5, 6], tranferrin [7] and well-known model proteins such as lysozyme (LYS) [8], hemoglobin [9], cytochrome c [10], ubiquitin[11], metallothionein [12] and superoxide dismutase [13] were examined. Additional studies aimed to understand the preferential target of Pt-drugs between protein and nucleic acids. Some of them took advantage of the use of single repetitive units such as nucleotides and amino acids instead of the whole macromolecules. Based on this model, Reedijk and co-workers demonstrated the transition of platinum from S-guanosyl-L-homocysteine to N7 of guanine [14–16]. A common feature emerging from these works is the occurrence of a competition or a transfer mechanism between S- or N-ligands of proteins (Cys, Met and His) and N7 of guanine. Additionally, while interactions with proteins are kinetically promoted, the reaction at guanine-N7 is thermodynamically preferred, thus justifying the nucleic acid as the final platinum acceptor site [17].
Most of the above work used cisplatin due to its clinical efficacy. Although transplatin is clinically inactive, some trans- derivatives showed cytostatic activity profiles comparable to cisplatin. Among them trans-platinum compounds containing at least one planar ammine (TPAs) are as cytotoxic as cisplatin but are not cross-resistant [18]. The effects of leaving groups and of the aromatic heterocyclic planar systems on efficiency, kinetics, and DNA platination mode have been investigated and may partly help to explain the unique cytotoxic profile of TPAs [19, 20]. In addition, different DNA-protein cross-linking efficiency can further contribute to driving distinct cellular responses between cisplatin and TPAs [21].
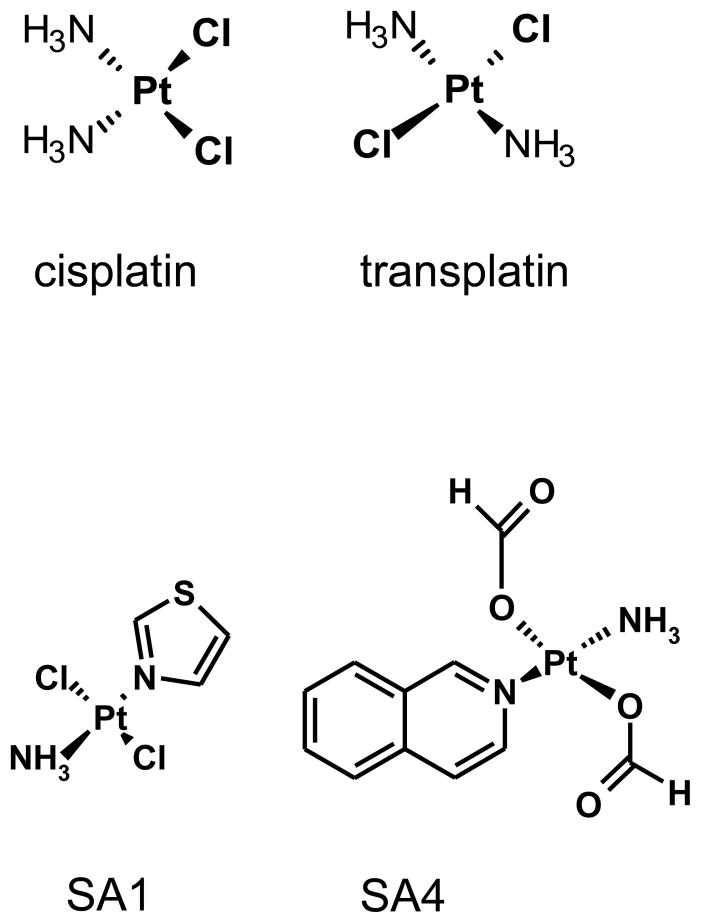
These results prompted us to investigate in more detail the reactivity of these derivatives towards proteins. Thus, in this work, we compared the reactivity of two TPAs (SA1 and SA4) and of the two reference compounds (cisplatin and transplatin, Fig. 1) towards three different proteins. Two of them, bovine α-lactalbumin (α-LA) and hen egg lysozyme (LYS), are small proteins and thus are experimentally useful models whereas human serum albumin (HSA) is the most abundant protein in human plasma thus representing a relevant physiological target. The reactivity of these components was characterized in terms of efficiency, kinetic, nature and conformational features of the resulting reaction products. Additionally, we considered it of importance to better understand the role of proteins in the DNA platination reaction. In order to address this point we performed transfer and competition reactions using both the protein and the nucleic acid as substrates. These studies allow us to propose a more comprehensive model to describe the mechanism of action of Pt-based drugs which may predict some of the toxic side effects specific to this class of compounds.
Fig 1.
Chemical structures of tested Pt-complexes.
2. Materials and methods
2.1. Materials
Proteins (bovine α-LA, hen egg LYS and HSA), cisplatin and transplatin were purchased from Sigma Aldrich (St Louis, MO, USA). TPAs were synthesized and purified according to the reported procedures [22–24]. Stock solutions of Pt-complexes were prepared in Milli Q water, and stored at −20 °C in the dark. Protein solutions were made freshly in MilliQ water.
The DNA oligonucleotides used were the FAM 5′ labelled GGA-TGT-GAG-TGT-GAG-TGT-GAG-G and the corresponding double stranded form obtained by the annealing with its complementary strand. Oligonucleotides were synthesized and purified by HPLC by Eurogentec (Seraing, Belgium).
All other chemicals were of analytical reagent grade and were obtained from Sigma Aldrich.
2.2. Protein-Pt adduct formation
Reactions between proteins and Pt-complexes were performed in 10 mM Tris-HCl, 50 mM KCl, pH 7.4 at 37 °C. A protein concentration of 0.5 mg/ml was added of metal complexes to provide protein:Pt molar ratios corresponding to 1:1 1:3 and 1:10. Aliquots from the reaction mixtures were collected at different incubation time (up to 72 h) and were resolved by SDS-PAGE on polyacrilamide gels according to Laemmli protocol [25] in Trisand 0.1% SDS running buffer, pH 8.3.ample was lyophilized and resuspended in 10 μL of SDS-PAGE loading buffer (4% SDS, 20% glycerol, 125 mM Tris, pH 6.8 and bromophenol blue in traces). Gels were stained with Coomassie Brilliant Blue R-250 and visualized on a Geliance apparatus (Perkin Elmer).
2.3. Pt transfer experiment
The Pt(II) transfer reaction from protein to DNA was performed using the protein-Pt adducts obtained after incubating the protein solution at 37 °C for 24 h in the presence of a ten fold molar excess of metal complexes. Millipore Centricon YM-3 filtration systems were then used to remove non-reacted Pt-complex from the reaction mixture. The eluted material was recovered and the amount of eluted Pt-compound was determined by UV-VIS spectroscopy. The fraction of Pt-complex bound to the protein was calculated by difference from the starting Pt-complex concentration.
Increasing concentrations of single or double stranded 5′-fluorescein labelled oligonucleotide were added to the purified protein-Pt-adducts in order to obtain the following DNA:Pt-complex molar ratios: 1:100, 1:50, 1:25 and 1:10. Samples were further incubated at 37 °C for 24 hours.
Reaction products were finally resolved by loading a constant amount of protein on SDS-PAGE and visualized by Coomassie Brilliant Blue staining. On parallel, aliquots containing constant amounts of DNA were resolved by sequencing denaturating PAGE (20% polyacrylamide gels with 7 M urea in 89 mM Tris, 89 mM boric acid and 2 mM EDTA). Before loading, these samples were lyophilized and finally dissolved in 5 μL of 80% formamide solution containing 0.1% xylene cyanole and 0.1% bromophenol blue. DNA was visualized by recording the fluorescence emission of the fluorescein label in a Storm 840 (Amersham).
2.4. Protein versus DNA competition for Pt adducts formation
To monitor preferential macromolecular platination target, a constant concentration of protein (0.5 mg/ml) and platinum complexes (250 μM) was mixed with increasing concentration of DNA and incubated at 37 °C for 24 hours in 10 mM Tris-HCl, 50 mM KCl pH 7.4 at 37 °C. Additionally, aliquots from the reaction mixtures were collected at different time intervals (up to 48 h) and were resolved by SDS-PAGE or DNA sequencing denaturating gels. Macromolecules were visualized as above described.
2.5. Mass Spectrometry
Mass spectra were acquired by electrospray ionization (ESI) mass spectrometer Q-Tof Micro from Waters Micromass (Manchester, UK) with a Z-spray electrospray ion source. The instrument operated in the positive-ion mode with a source temperature of 80 °C, a capillary voltage of 3 kV and a cone voltage of 35 V. The molecular masses of protein samples and the deconvoluted spectra were estimated using the MassLynx software 4.1 (Micromass).
2.6. Intrinsic protein fluorescence
Fluorescence measurements were performed on a PerkinElmer LS-55 luminescence spectrophotometer using a 1 cm path length quartz cell and an excitation wavelength of 280 nm. Excitation and emission slits were set at 5 nm and the scan speed was fixed at 120 nm/min. Proteins were analyzed at a concentration of 2 μM in 10 mM Tris-HCl, 50 mM KCl pH 7.4 and Pt-complexes were added in a ten fold molar excess. Time drive fluorescence experiments were performed by acquiring spectra at 37 °C for 24 hours with a delay time of 1800 seconds.
2.7. Circular dichroism spectroscopy
CD experiments were performed on a Jasco J-810 spectropolarimeter equipped with a Peltier temperature control system. Quartz cells with pathlength of 1 and 0.1 cm were used for the 250–320 nm and for 197–250 nm regions, respectively. Each CD spectrum was an average of four accumulations scanned at 20 nm min−1. Scans were obtained with bandwidth of 2 nm and response time of 16s. The protein concentration for near-UV CD measurements was of 0.5 mg/mL and for far-UV CD was of 0.1 mg/ml in 10 mM Tris-HCl, 50 mM KCl, pH 7.4. CD spectra were recorded after 24 hours of incubation at 37 °C in the presence/absence of a 10 molar excess of tested drugs. Observed ellipticities were converted to mean residue ellipticity [θ] = deg × cm2 × dmol−1.
3. Results
3.1. Reactivity of platinum complexes towards proteins
To monitor the formation of stable platinum adducts on native proteins, we analyzed the reaction products of cisplatin, transplatin, SA1 and SA4 with α-LA, LYS and HSA by SDS-PAGE.
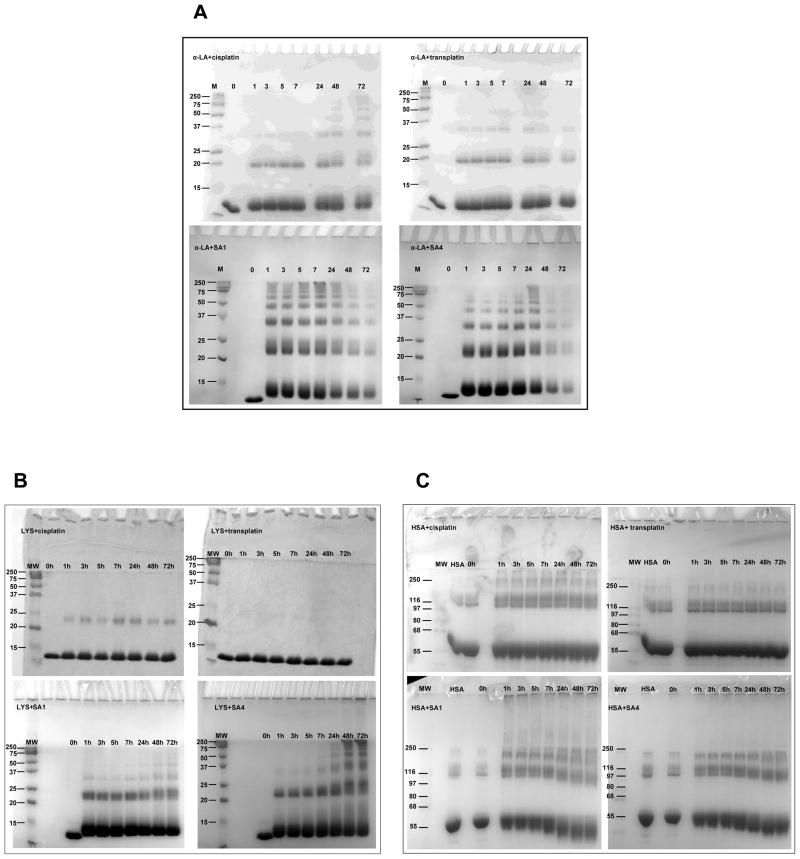
Protein incubation at 37 °C with the tested Pt-complex molar ratio generally showed the formation of SDS-resistant protein products characterized by a discrete increment in molecular weight (Fig. 2). Moreover, experiments performed under the same conditions but with increasing protein amount confirmed that the formation of these products was concentration dependent (data not shown). These data indicated the bands characterized by lower mobility as oligomerization products of protein monomers. Due to their stability in denaturating conditions, they were attributed to cross-linked protein through bifunctional Pt adducts. Protein platination can cause different effects on protein structural organization and among them aggregation and polymerization have been previously evocated [5, 26–28]. Here, our results underline the ability of our metal complexes to preferentially drive an ordinate oligomerization reaction vs an uncontrolled aggregation.
Fig 2.
SDS-PAGE of the reaction products of α-LA (Panel A), LYS (PANEL B) and HSA (PANEL C) incubated with the tested Pt-complexes in 10 mM Tris, 50 mM KCl, pH 7.4 at 37 °C. The reaction at a constant protein:Pt ratio has been followed upon time.
The extent and the kinetics of such a process were a function of protein-Pt molar ratio (Fig. S1). Additionally, they were found to depend directly on both the protein substrate and on the of Pt-complex chemical structure (Fig. 2).
The most striking evidence was a faster and more extensive protein modification in the presence of TPAs. More specifically, a consistent overall reactivity SA4> SA1 ≫ cisplatin > transplatin was confirmed towards all the tested proteins. (Fig S2)
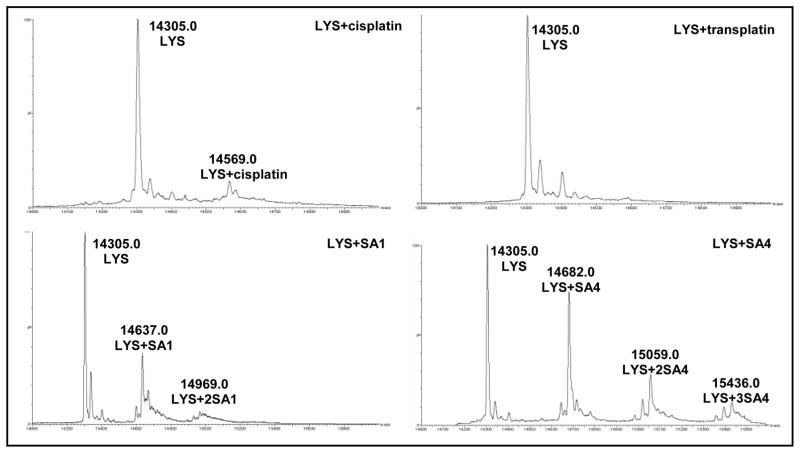
All the tested Pt-complexes promoted protein oligomerization more efficiently on α-LA rather than on LYS. In particular, the reaction on α-LA occurred in the presence of all tested compounds although in the presence of TPAs it appeared at shorter incubation time. Larger modulation was observed in the presence of LYS and HSA where transplatin did not significantly alter the electrophoretic mobility of the proteins. Since protein platination at one single site is not easily identified by SDS-PAGE, this result can reflect a lack of platination or absence of protein cross-linking. To better define this feature, the reaction products of LYS in the presence of a tenfold molar excess of each metal complex (after 24 hours of incubation) were further analyzed by mass spectrometry (Fig. 3). The coordination bonds between Pt and reactive protein residues are sufficiently stable to allow the analysis of the protein-Pt complex in ESI-MS mode. According to our experimental approach, only MS peaks corresponding to the monomeric protein could be resolved. Acquired spectra showed that LYS was not completely platinated since the peak corresponding to the native unmodified protein (14.305 Da) was always detectable.
Fig 3.
Deconvoluted ESI-MS spectra of the reaction products obtained after incubation in 10 mM Tris, 50 mM KCl, pH 7.4 for 24 h at 37 °C of LYS (0.5 mg/ml) with a ten fold molar excess of each tested Pt-complexes.
After reaction with SA1, the mono- (14.637 Da) and bis- platinated (14.969 Da) proteins were identified. In this case, the molecular masses of the most abundant platinated protein forms corresponded to the loss of one chloride group from each metal complex. Mass spectra of the products, deriving from incubation with SA4, showed the formation of protein-Pt adducts with higher metal ions content. Indeed, as reported in Fig. 3, the peaks at 14.682, 15.059 and 15.436 Da correspond to the protein bound to one, two and tree metal complexes, respectively. This correlates with the more prominent protein reactivity exerted by SA4. Here, the mass values indicated the loss of formate groups and eventually their substitution with water.
The deconvoluted spectra of LYS after incubation with cisplatin and transplatin are further in line with electrophoretic results. Upon addition of cisplatin only the peak corresponding to the monoplatinated protein adduct (14.569 Da) appeared, thus confirming a lower reactivity of this metal complex in comparison to TPAs. According to the mass distribution pattern, the loss of one chloride was confirmed. Interestingly, after the addition of the trans- isomer, no adduct formation was evidenced. This suggests that under our conditions the lack of LYS oligomerization in the presence of transplatin is related to a remarkable reduction of platination efficiency. Nevertheless, Casini et al. reported platination of LYS by transplatin, but the reaction occurs to a low extent (15%) and only after long incubation time (72 hours) [8].
Finally, the reaction of HSA with TPAs showed an unusual behavior (Fig. 2C). Indeed, from the analysis of the gels, it was evident that the bands corresponding to the monomeric and the oligomeric species migrate faster and become smeared upon increasing the reaction time. This result suggests the occurrence of intramolecular cross-linking which constrains the protein into a compact structure which is preserved also in denaturating conditions. Such a possibility has been previously taken into account for cisplatin since, in addition to Cys-34 (the preferred platination site), the nitrogen from the imidazole ring of histidine has been found to act as donor ligand for Pt (II) [5]. An analysis of the distribution of the potentially reactive sites along the protein surface supports their random location (Fig. S3A–B). However, several of them are spatially close to one another and are thus suitable to generate intramolecular cross-links.
3.2. Protein structural perturbations upon platination
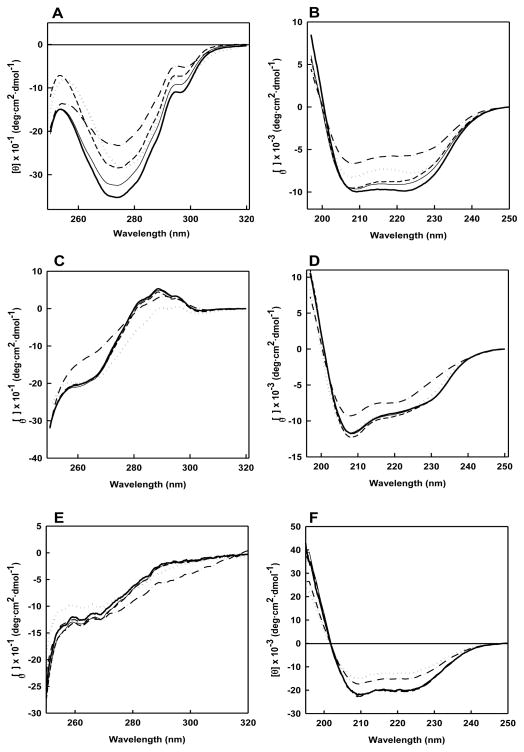
The reaction between protein and Pt-compounds clearly modifies the hydrodynamic properties of the starting material mainly through formation of multimeric adducts. Nevertheless, platination can cause fine protein structural variations. To better characterize this point, we monitored the spectroscopic properties of the tested proteins upon incubation with the metal complexes (Fig. 4). The near-CD spectrum of α-LA presents two minima at 270 and 298 nm, arising from the spectral contribution of aromatic chromophores, whereas in the far UV region the typical CD spectrum of polypeptides in α-helical arrangement with two minima at 208 and 222 nm was recorded (Fig. 4A and 4B). Upon reaction with all the tested Pt-complexes, a gradual decrease of CD signal intensity was observed. Moreover, only after reaction with SA1, the protein spectrum showed a red shift of the negative band at 272 nm. This result is consistent with an increment of the rotation freedom of the aromatic chromophores (loss of the rigid tertiary structure of the protein) and a reduction of secondary structure contents.
Fig 4.
Near and Far UV CD spectra of α-LA (PANELS A and B), LYS (PANELS C and D) and HSA (PANELS E and F) in the absence (bold solid line) and in the presence of a ten fold molar excess of cis-Pt (solid line), trans-Pt (short dash line), SA1 (dotted line) and SA4 (dash-dot line) recorded after 24 h of incubation at 37 °C in 10 mM Tris, 50 mM KCl, pH 7.4.
Native LYS displays a near CD spectrum characterized by a wide negative dichroic signal between 260 and 280 nm and a positive band centered at 293 nm. In particular, the positive signal between 290 and 300 nm is related to the Trp residues that are located in an asymmetric environment (Fig. 4C and 4D) [29, 30]. Both TPAs promoted a reduction of the intensity of this CD feature which appeared as a decreased molar ellipticity of the positive band at 288 nm and of the negative one at 270 nm for SA1 and SA4, respectively. This reflects a reduced organization of the tertiary structure which appears to be a specific consequence of the reaction with these two compounds. Additionally, relevant loss of secondary structure content was induced only by SA4, as evidenced by far UV spectra (Fig. 4D).
With HSA, a consistent reduction of the typical α-helical protein signal at 208 and 222 nm was recorded after incubation with TPAs (Fig. 4E and 4F). Despite this common variation, in the near UV region, SA1 and SA4 altered the Phe signal at 250–270 nm and the Tyr/Trp signals in the 270–300 nm range, respectively.
Distinct from TPAs, cis- and transplatin did not significantly modify the CD spectra of LYS and HSA. Although a lower reactivity of these two compounds has been assessed, this behavior supports the hypothesis that the platinated fraction of the proteins does not undergo substantial conformational rearrangements upon reaction with these metal complexes.
CD data clearly confirmed that the binding of Pt-complexes altered the target proteins conformation by affecting their secondary and tertiary structure arrangements to different extent. To fully characterize these transitions, we monitored the variation of the protein fluorescence spectra upon incubation time in the presence of a ten-fold molar excess of the tested compounds. Examples of the recorded fluorescence spectra are reported in Fig. 5 (see also Supplementary Material Fig. S4 and S5).
Fig 5.
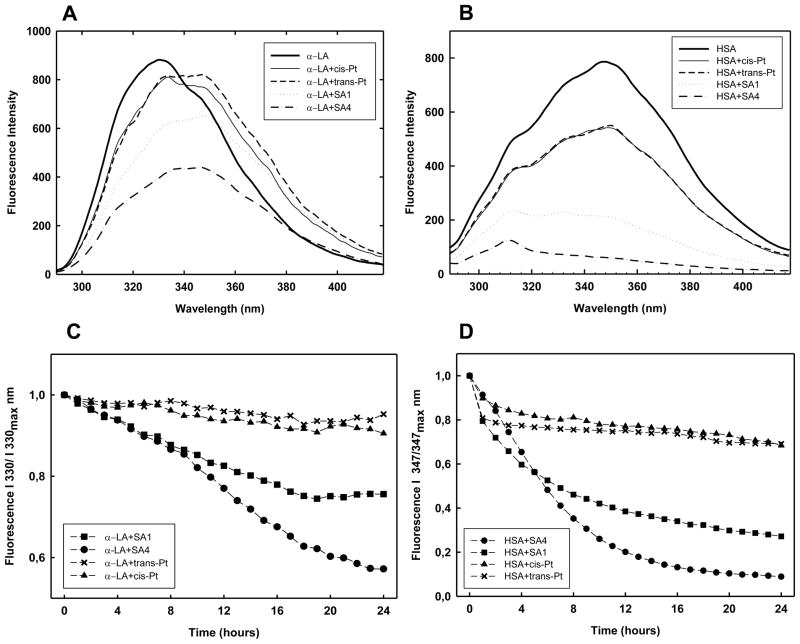
Modification of the fluorescence spectra of α-LA (PANEL A) and HSA (PANEL B) induced by a ten fold molar excess of tested Pt-complexes recorded after 24h incubation in 10 mM Tris, 50 mM KCl, pH 7.4 at 37 °C. Protein concentration was 2 μM. The kinetic variation of the of the fluorescence intensity of α-LA and HSA recorded at 330 and 347 nm, respectively, in the same experimental conditions is reported in PANELS C and D.
At pH 7.4, the four Trp residues of α-LA are buried into the hydrophobic core of the protein, thus resulting in a maximum of fluorescence emission at 330 nm (Fig. 5A) [31]. The reaction with cisplatin and transplatin did not largely affect the signal intensity and only a modest increment of the 350 vs 330 nm contribution was recorded. This indicates that the reaction induces a weak conformational modification in the protein structure. Conversely, in the presence of TPAs, a red shift in protein emission maximum approximately to 350 nm occurred in about 15 h, which is related to the exposition of Trp residues to the aqueous solvent. This transition is paired to a quenching of the 330 nm signal (Fig. 5C) that we cannot ascribe to an energy transfer process between Trp and TPA due to lack of overlapping bands. Thus it should be related to more general mechanisms such as the quenching effect exerted by water or to an increased lifetime of the exited state which occurs in the denaturated forms. The reduced content of folded structure upon platination recorded using CD spectroscopy is clearly in line with this last model. Both quenching and shift of the maximum wavelength emission appeared to take place according to a monophasic process for SA1 (Fig. 5C). Interestingly, SA4 caused a more extensive quenching and the two processes occurred according to two events: the first one, faster (0–8 h), shifts the maximum to 338 nm and it is followed by a second slower step (8–22 h) which results in a spectrum with a low intensity and a maximum at 350 nm.
HSA contains a relevant number of Tyr (18) in comparison to Trp (1) residues. Thus, excitation at 280 nm provides an emission spectrum with a maximum at 347 nm and a shoulder at 312 nm due to Trp and Tyr fluorescence, respectively (Fig. 5B and 5D) [32]. After reaction with TPAs the Trp fluorescence was remarkably quenched with SA4 being the most effective. Although CD spectra indicate loss of protein structure content upon reaction with TPAs, in this case no further shift in the maximum emission wavelength associated to Trp was actually expected. Indeed, the value recorded in the native protein already supports its localization in an hydrophilic environment. Thus, the observed, quenching represents a common feature of the Pt(II) reaction on HSA and α-LA. Interestingly, the tyrosyl fluorescence at 312 nm was actually conserved both in terms of intensity (the ratio I347nm vs I312nm parallels the decay of I347nm, data not shown) and of wavelength of maximal emission, thus suggesting that Tyr contribution is not largely affected. Accordingly to CD results, after cisplatin and transplatin addition, only a modest decrease in Trp emission was detected.
3.3. DNA platination by Pt-protein adducts
Proteins can represent per se a pharmacological target relevant to the mechanism of action of Pt-complexes. At the same time, they can largely modulate the availability of Pt-complexes to form DNA adducts. In this context, the distinct efficiency in protein platination, shown by the tested metal complexes, can represent an additional tool for the modulation of the biological properties of these drugs in the cellular environment. This point was assessed by monitoring the transfer of Pt-complex from protein to nucleic acids using bovine α-LA, which was confirmed to be a good substrate for all the tested metal complexes, and HSA, a reliable physiologically relevant target. In this assay, the reactive species were the platinated proteins, obtained after 24 h incubation with the metal complexes and purified from the unbound reactive drugs. Since a variable degree of platination was achieved according to the protein and the metal complex nature, the oligonucleotide was ultimately treated with the platinated proteins at constant DNA:Pt-complex ratios to provide comparable concentrations of reactive species.
DNA resolution by sequencing gels showed the formation of oligonucleotide species characterized by a reduced electrophoretic mobility upon incubation with platinated α-LA (Figs. 6A–C). Coomassie staining of the gel confirmed the co-localization of DNA and protein in these bands thus indicating them as the cross-linking reaction products between the two macromolecules exerted by platinum.
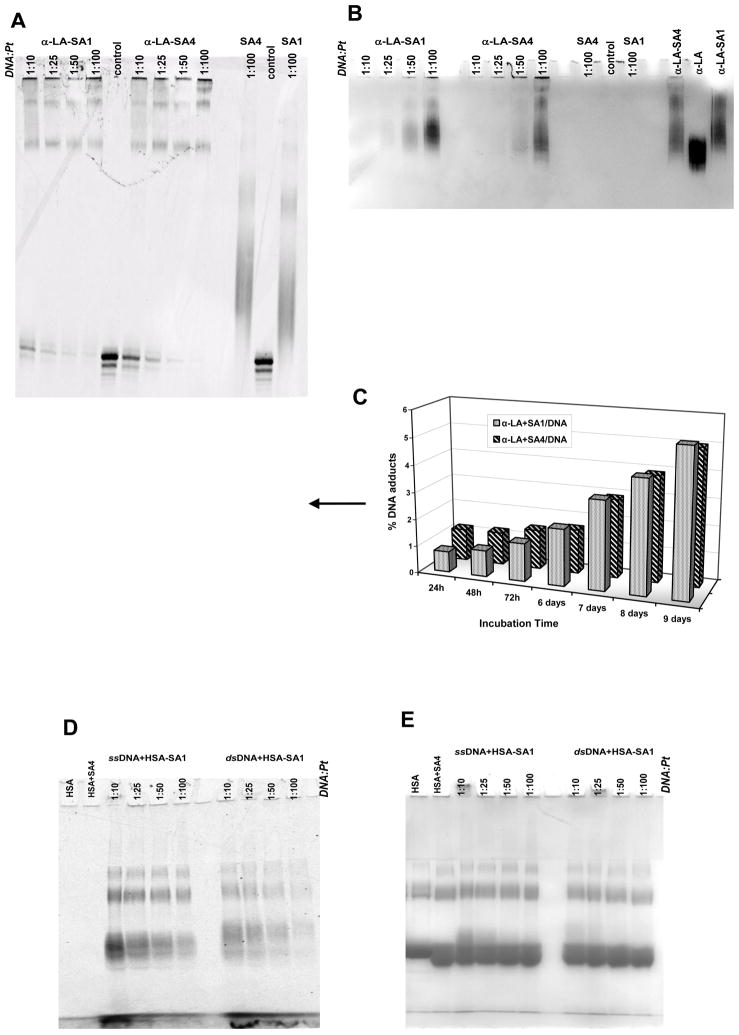
Fig 6.
DNA adduct formation by platinated α-LA (PANEL A and B, sequencing gel) or HSA (PANEL D and E, SDS-PAGE) resolved after 24 hours of incubation at the reported DNA:Pt molar ratio in 10 mM Tris, 50 mM KCl, pH 7.4 at 37 °C. Proteins (0.5 mg/ml) were previously treated with Pt-complexes for 24 h at 37 °C and purified from the unreacted compound.
In PANELS A and D the signal of the fluorescein-labeled DNA was acquired. In PANELS B and E the corresponding gels were stained with Coomassie brilliant blue to visualize the protein.
Proteins incubated in the presence/absence of TPA were used as controls. Additionally, in PANEL A and B, untreated DNA and DNA treated with a one hundred molar excess of SA1 or SA4 in the same experimental conditions were included.
In PANEL C the formation of the DNA-Pt adducts indicated by the arrow is reported as a function of the incubation time.
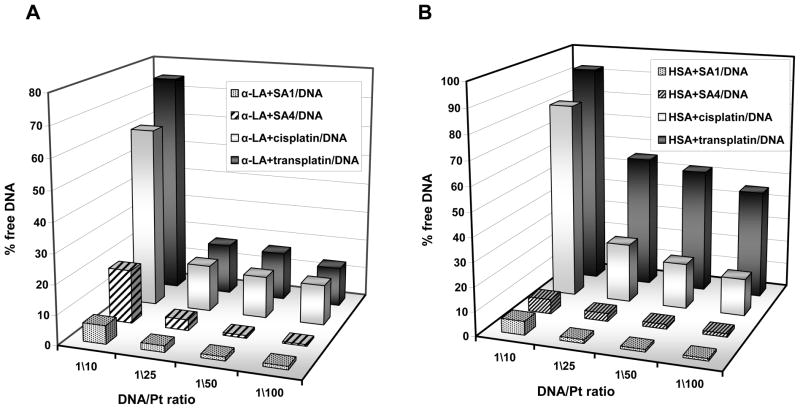
In the presence of TPA-protein adducts, the DNA crosslinking reaction was much more prominent than the one observed with cisplatin or transplatin (Fig. 7A). Moreover, distinct from the above results on protein platination reaction, SA1-protein adducts turned out to be the most efficient DNA platination agents.
Fig 7.
Reduction of unmodified single stranded DNA after 24 hours of incubation in 10 mM Tris, 50 mM KCl, pH 7.4 at 37 °C in the presence of platinated α-LA (PANEL A) or HSA (PANEL B).
Interestingly, DNA-adducts not cross-linked to protein were also observed in the presence of TPA-protein complexes. These correspond mainly to the mono-platinated DNA strand. Since no protein bound Pt-complexes were removed from the reaction mixture before nucleic acid addition, these adducts must derive from protein release upon DNA platination. By monitoring their formation as a function of incubation time (Fig. 6C) we confirmed that the rate of this process was low since it was not completed within 9 days. This feature supports the hypothesis that the transfer of the metal from protein to DNA is not kinetically promoted but represents the thermodynamically favored process.
Comparable experiments performed with the platinated-HSA confirmed the same order of efficiency of the tested compounds in modifying DNA (Fig. 6D and 6E). In this case, the protein-DNA cross-linking products were not easily monitored by DNA sequencing gel due to their high molecular weight and the extensive oligomerization degree of HSA. Thus, a better characterization of these adducts was achieved by resolving them by SDS-PAGE. Interestingly, this allowed the observation that HSA maintains its oligomeric state also upon Pt-mediated cross-linking with the nucleic acid. Indeed, after incubation of platinated protein with increasing oligonucleotide concentration, the Pt-induced oligomeric pattern of HSA was still resolved and was associated to new shifted bands corresponding to protein-Pt-DNA ternary complex.
Finally, although in the presence of transplatin no significant HSA-oligomerization was previously observed, here a reactivity of the platinated protein emerged (Fig. 7B). This suggests that HSA platination occurs but to a limited degree and with no remarkable protein structural changes.
All these data were acquired also using the same oligonucleotide paired to its complementary strand (Fig. 6D and 6E). In all instances a reduction of DNA cross-linking was observed thus suggesting that the flexibility of the single stranded DNA is beneficial for the proper exposition of the reactive site(s).
3.4. Competition between protein and DNA for Pt binding
The above data can help to address the potential role of protein as platinum drugs carriers towards nucleic acid. In addition, in order to evaluate the preferential platinated products in the simultaneous presence of protein and DNA, we performed a competition assay. As target protein we choose α-LA since it forms more usable platination reaction products and for nucleic acid substrate, we included both the more reactive single stranded DNA and the double stranded DNA, the proposed pharmacological target.
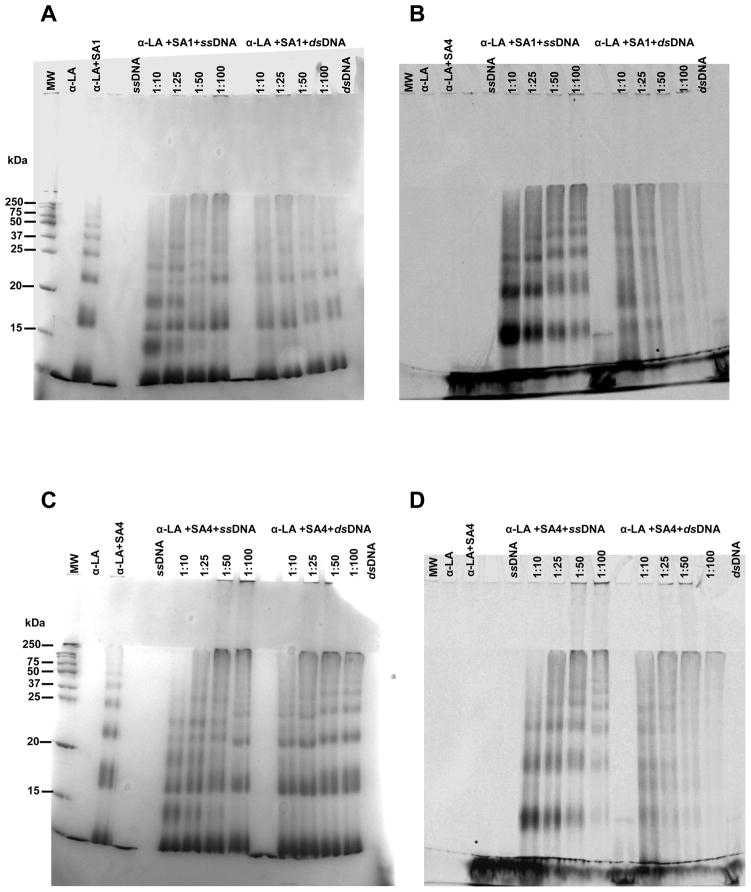
SDS-PAGE of the reaction products (Fig. 8) obtained at low DNA concentration, showed a protein oligomerization pattern superimposable to the one observed in the absence of the nucleic acid, thus identifying the protein as the preferential target. However, by increasing the DNA concentration, a shift of the corresponding bands appeared as the result of protein-DNA cross-linking (as confirmed by co-localization of the fluorescent DNA). Clearly, SA4 was more reactive toward proteins than DNA, promoting the typical oligomerization pattern of α-LA (Fig. 8C and 8D). On the contrary, reactions with SA1 showed a more relevant presence of bands relative to protein-DNA-Pt adducts, indicating an increased DNA reactivity in comparison to the other TPA (Fig. 8A and 8B).
Fig 8.
SDS-PAGE of the reaction products obtained after incubation at 37 °C for 24 hours in 10 mM Tris, 50 mM KCl, pH 7.4 at 37 °C of α-LA (25 μM) with 250 μM of SA1 (PANELS A and B) or SA4 (PANEL C and D) in the presence of single stranded or double stranded DNA at the reported DNA:Pt molar ratio. PANELS A and C show the gels stained with Coomassie Brilliant blue whereas PANEL B and D report the signal of the fluorescein-labeled DNA. MW are the protein molecular weight, the untreated DNA as well as the proteins incubated in the presence/absence of TPA were used as controls.
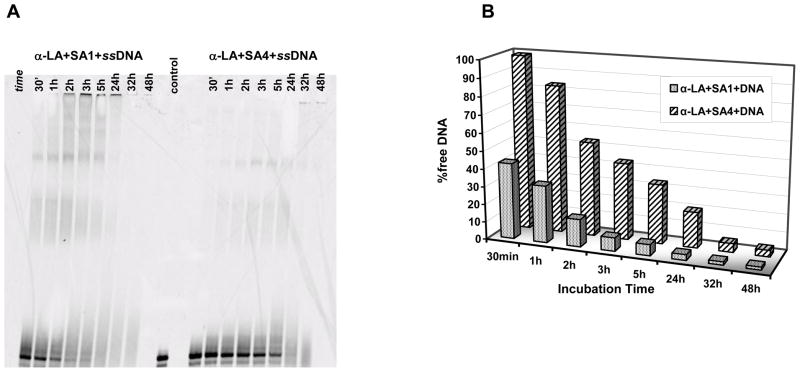
This result was confirmed by a direct comparison of the reaction products obtained in the presence of the two most reactive metal complexes, SA1 and SA4, as a function of incubation time (Fig. 9). Indeed, the quantification of the unreacted DNA indicated a kinetically more efficient modification of the nucleic acid by SA1, which likely reflects its preferential reactivity towards DNA in comparison to protein.
Fig 9.
Sequencing gel resolution of the reaction products obtained after incubation of α-LA (25 μM) and single stranded DNA (25 μM) with 250 μM of SA1 or SA4 (PANEL A) in 10 mM Tris, 50 mM KCl, pH 7.4 at 37 °C. The quantification of the unmodified DNA upon incubation time is reported in PANEL B.
4. Discussion
Data herein presented provide a detailed characterization of protein platination by metal complexes in order to better understand the physiological relevance of this process per se and as an intermediate step in the complex process of DNA-adduct formation.
The extent of such reactivity is expected to be driven by both the reagents. Obviously, we expected to observe different protein platination according to the number of reactive amino acid residues and to their accessibility, which is dictated by the protein tridimensional architecture [33]. In addition the nature of the ligand and leaving groups largely affects the reactivity towards DNA as well as toward single aminoacids [19, 22]. For this reason we thought that the same is likely to occur on full length proteins
These expectations were confirmed. Indeed, a direct comparison of the clinically relevant cisplatin, of its isomer transplatin and of SA1 and SA4, showed that TPAs are more efficient in modifying all the tested proteins. Interestingly, the reactivity order is not the same as the one previously reported for nucleic acid [20]. In particular, SA1 and SA4 preferentially modify DNA and proteins, respectively. Thus, the reduced liability of the carboxylate vs chloride leaving groups, which correlates with the DNA reactivity, appears to be not directly related to the efficiency of protein adduct formation.
This can reflect that the rate limiting step in DNA platination represented by the formation of the acquo species is not necessarily required when an S- substitution can occur as in the presence of S-containing residues. Nevertheless, the same correlation was actually reported for the methionine binding rate in an homologous series of Pt(II) complexes [19]. This suggests that to rationally describe the discrepant reactivity order observed on proteins we cannot take into account only the lability of leaving groups. Indeed, by modulating the nature of the carrier groups protein adducts with distinct coordination profiles are formed and, ultimately, they cause variable alterations of protein conformation.
This is nicely evidenced in Lys (Fig. S3C–D). The native protein has a His exposed to the solvent which is reported to be an efficient platination site. The formation of such an adduct has been reported to not affect the protein folding [8]. In this step the formation rate can favor the Cl-complexes. Indeed, no Cl-derivatives showed significant alteration of the secondary structure content. However, as confirmed by mass spectroscopy results, only in the case of SA4 the reaction efficiently progresses to target two additional sites which can be rationally identified as the two Met residues. They are not easily accessible: likely the chemical features of the aromatic ligand of SA4 can allow only this derivative to react at these positions causing a relevant alteration of the secondary and tertiary protein structure.
In this connection, it is interesting to underline that only with SA4 we were able to observe a biphasic α-LA structural rearrangement. We can easily assume that this represents the access to a first set of easily accessible sites which, upon platination, allow the exposition of a second one. This sequence of events can also fit the conversion of a first non-covalent binding mode into a covalent one.
All the 8 Cys residues of α-LA are involved in disulfur bridges and thus should be poorly reactive towards Pt(II). By mapping the distribution of the other reactive sites (Met and His) on the protein, one His (H68) is located in a very flexible loop letting us to assume that its platination should not largely affect the overall structural content of the protein (Fig. S3E–F). Conversely, the two additional His residues as well as the single Met are inserted in α-helical domains but quite exposed to the solvent. Interestingly, they are clustered in two protein portions closed to Trp residues. Thus, again, the more hysterically hindered SA4 is likely to affect these sites causing a more prominent perturbation of the protein structure.
Thus, although a general oligomerization of proteins is promoted by Pt-protein adduct formation, the structural features of the reaction products are distinct for each Pt-complex. In particular, our results emphasize how the chemical properties of ligands affect the reactivity of Pt-complex although it is not easy to attribute the different behavior to the nature of the leaving group or to other properties such as electronic factor or steric hindrance of the aromatic ligands [19].
From a pharmacological point of view, this prominent protein oligomerization, may be easily related to the nephrotoxicity that characterizes the adverse effects of Pt-based drugs. However, it also represents a modulation of the drug bioavailability at nuclear level. Indeed, some interesting conclusions can be made on the protein-nucleic acid cross-reactivity. First of all DNA-adducts can actually represent the final reaction products. This was confirmed both monitoring the competitive platination of protein and DNA as well as using the platinated proteins as reactive species. Such a conclusion is extremely significant since it supports the relevance of DNA as final target and, thus, further underlines the DNA-adduct characterization as a valuable tool to describe the mechanism of action of these drugs.
Nevertheless, the overall picture of our results clearly further underlines the importance to take into account the reaction profile towards both classes of investigated macromolecular biological targets to design proper SAR for Pt-complexes. Indeed, the relevant protein reactivity of herein tested TPAs in comparison to cisplatin should allow easy prediction of distinct pharmacological pathway(s) in comparison to the reference drug and, moreover, can help in explaining their observed lack of cross-resistance with the clinically used derivatives.
Supplementary Material
Highlights.
Proteins as target, carrier or reservoir systems for platinum-based drugs.
Oligomerization of proteins is promoted by Pt-adducts formation.
Pt-protein adduct are different according to the ligands and the protein involved.
Pt(II) can be transferred from proteins to DNA.
Acknowledgments
The authors acknowledge University of Padova and AIRC (Associazione Italiana per la Ricerca sul Cancro) for financial support. N. P. F. acknowledges NSF and NIH support. They acknowledge Dr. Giuseppe Marson for informatics assistance. Daniel Lee is thanked for synthesis of TPA derivatives.
Abbreviations
- TPAs
trans-platinum amine derivatives
- α-LA
bovine alpha-lactalbumin
- LYS
hen egg lysozyme
- HSA
human serum albumin
- cis-Pt
cis-diamminedichloroplatinum (II), cisplatin
- trans-Pt
trans-diamminedichloroplatinum (II), transplatin
- FAM
fluorescein amidite
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang D, Lippard SJ. Nat Rev Drug Discov. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 2.Reedijk J. Chem Rev. 1999;99:2499–2510. doi: 10.1021/cr980422f. [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Guo Z. Anti-Cancer Agents Med Chem. 2007;7:19–34. doi: 10.2174/187152007779314062. [DOI] [PubMed] [Google Scholar]
- 4.Ruimin X, Willie J, Lorna R, Murugesan G, Gene SH, Buckley B. Anal Bioanal Chem. 2007;387:2815–2822. doi: 10.1007/s00216-007-1147-9. [DOI] [PubMed] [Google Scholar]
- 5.Ivanov AI, Christodoulou J, Parkinson JA, Barnham KJ, Tucker A, Woodrow J, Sadler PJ. J Biol Chem. 1998;273:14721–14730. doi: 10.1074/jbc.273.24.14721. [DOI] [PubMed] [Google Scholar]
- 6.Roy S, Westmaas JA, Hagen KD, van Wezel GP, Reedijk J. J Inorg Biochem. 2009;103:1288–1297. doi: 10.1016/j.jinorgbio.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Khalaila I, Allardyce CS, Verma CS, Dyson PJ. Chem Bio Chem. 2005;6:1788–1795. doi: 10.1002/cbic.200500067. [DOI] [PubMed] [Google Scholar]
- 8.Casini A, Mastrobuoni G, Temperini C, Gabbiani C, Francese S, Moneti G, Supuran CT, Scozzafava A, Messori L. Chem Commun. 2007:156–158. doi: 10.1039/b611122j. [DOI] [PubMed] [Google Scholar]
- 9.Mandal R, Kalke R, Li X-F. Rapid Commun Mass Spectrom. 2003;17:2748–2754. doi: 10.1002/rcm.1259. [DOI] [PubMed] [Google Scholar]
- 10.Casini A, Gabbiani C, Mastrobuoni G, Messori L, Moneti G, Pieraccini G. Chem Med Chem. 2006;1:413–417. doi: 10.1002/cmdc.200500079. [DOI] [PubMed] [Google Scholar]
- 11.Zhao T, King F. J Biol Inorg Chem. 2011;16:633–639. doi: 10.1007/s00775-011-0767-x. [DOI] [PubMed] [Google Scholar]
- 12.Karotki AV, Vašák M. Biochem. 2008;47:10961–10969. doi: 10.1021/bi801253x. [DOI] [PubMed] [Google Scholar]
- 13.Calderone V, Casini A, Mangani S, Messori L, Orioli PL. Angew Chem Int Ed. 2006;45:1267–1269. doi: 10.1002/anie.200502599. [DOI] [PubMed] [Google Scholar]
- 14.Barnham KJ, Djuran MI, del Socorro Murdoch P, Sadler PJ. Chem Commun. 1994:721–722. [Google Scholar]
- 15.Teuben JM, van Boom GESS, Reedijk J. Dalton Trans. 1997:3979–3980. [Google Scholar]
- 16.van Boom SSGE, Reedijk J. Chem Commun. 1993:1397–1398. [Google Scholar]
- 17.Deubel DV. J Am Chem Soc. 2004;126:5999–6004. doi: 10.1021/ja0499602. [DOI] [PubMed] [Google Scholar]
- 18.Aris SM, Farrell NP. Eur J Org Chem. 2009;19:1293–1302. doi: 10.1002/ejic.200801118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benedetti BT, Quintal S, Farrell NP. Dalton Trans. 2011;40:10983–10988. doi: 10.1039/c1dt10964b. [DOI] [PubMed] [Google Scholar]
- 20.Musetti C, Nazarov AA, Farrell NP, Sissi C. Chem Med Chem. 2011;6:1283–1290. doi: 10.1002/cmdc.201100032. [DOI] [PubMed] [Google Scholar]
- 21.Benedetti BT, Peterson EJ, Kabolizadeh P, Martnez A, Kipping R, Farrell NP. Mol Pharm. 2011;8:940–948. doi: 10.1021/mp2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bulluss GH, Knott KM, Ma ESF, Aris SM, Alvarado E, Farrell N. Inorg Chem. 2006;45:5733–5735. doi: 10.1021/ic060741m. [DOI] [PubMed] [Google Scholar]
- 23.Ma ESF, Bates WD, Edmunds A, Kelland LR, Fojo T, Farrell N. J Med Chem. 2005;8:5651–5654. doi: 10.1021/jm050539d. [DOI] [PubMed] [Google Scholar]
- 24.Quiroga AG, Pérez JM, Alonso C, Navarro-Ranninger C, Farrell N. J Med Chem. 2005;49:224–231. doi: 10.1021/jm050804v. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Gonias SL, Pizzo SV. J Biol Chem. 1981;256:12478–12484. [PubMed] [Google Scholar]
- 27.Peerey LM, Kostić NM. Inorg Chem. 1987;26:2079–2083. [Google Scholar]
- 28.Ohta N, Chen D, Ukai M, Matsuo T, Masuda M, Yotsuyanagi T. Int J Pharm. 1995;124:165–172. [Google Scholar]
- 29.Ikeda K, Hamaguchi K, Miwa S, Nishina T. J Biochem. 1972;71:371–378. [PubMed] [Google Scholar]
- 30.Kelly SM, Jess TJ, Price NC. BBA Proteins Proteom. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Lala AK, Kaul P. J Biol Chem. 1992;267:19914–19918. [PubMed] [Google Scholar]
- 32.Longworth JW. Ann N Y Acad Sci. 1981;366:237–245. doi: 10.1111/j.1749-6632.1981.tb20757.x. [DOI] [PubMed] [Google Scholar]
- 33.Cubo L, Groessl M, Dyson PJ, Quiroga AG, Navarro-Ranninger C, Casini A. Chem Med Chem. 2010;5:1335–1343. doi: 10.1002/cmdc.201000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.