Abstract
Human pluripotent stem cells represent an accessible cell source for novel cell-based clinical research and therapies. With the realization of induced pluripotent stem cells (iPSCs), it is possible to produce almost any desired cell type from any patient's cells. Current developments in gene modification methods have opened the possibility for creating genetically corrected human iPSCs for certain genetic diseases that could be used later in autologous transplantation. Promising preclinical studies have demonstrated correction of disease-causing mutations in a number of hematological, neuronal and muscular disorders. This review aims to summarize these recent advances with a focus on iPSC generation techniques, as well as gene modification methods. We will then further discuss some of the main obstacles remaining to be overcome before successful application of human pluripotent stem cell-based therapy arrives in the clinic and what the future of stem cell research may look like.
A Brief History of Pluripotent Stem Cells
Stem cells are defined by both their ability to indefinitely self-renew, while maintaining the capacity to differentiate into one or more differentiated cell types. The potency of stem cells can range from totipotent, which are able to give rise to all of the cells in an organism, including extraembryonic tissues, (e.g. zygote) to unipotent, which are only able to differentiate into one type of cell (e.g. spermatogonia). Pluripotent stem cells are defined by their capacity to differentiate into all three germ layers. Due to their tremendous potential for therapeutic use, research on deriving, expanding and manipulating human pluripotent stem cells, including embryonic stem cells (hESCs) and the related induced pluripotent stem cells (hiPSCs), has grown exponentially.
In 1981 the first pluripotent, embryonic stem cell (ESC) lines were established from mouse blastocysts (mESC) [1, 2]. Culture conditions for long-term maintenance of mESC pluripotency were significantly improved during the late 1980s, when leukemia inhibitory factor (LIF) or other agonists of the gp130-Jak-Stat signaling pathway were shown to promote self-renewal of mESCs in vitro [3–6]. Nearly two decades later, James Thomson’s group accomplished the long sought after goal of isolating and fully characterizing the first hESCs from donated human embryos [7]. Thomson’s isolation and establishment of hESCs enabled translational and clinical research with human pluripotent stem cells. Interestingly, hESCs do not require LIF/gp130 agonists to prevent differentiation. Instead hESCs use bFGF as a key mediator of pluripotency [8].
Another significant breakthrough in human pluripotent stem cells research occurred in 2006, when Takahashi and Yamanaka transformed terminally differentiated murine fibroblasts into iPSCs (miPSCs) [9]. These miPSCs look and function almost identically to mESCs, including the generation of fertile adult mice derived entirely from miPSCs by tetraploid complementation assays, just as is done for mESCs [10]. The following year Yamanaka’s group and Thomson’s group described the derivation of hiPSCs using terminally differentiated human fibroblasts [11, 12]. Yamanaka’s initial studies found that only four transcription factors (Oct3/4, Sox2, Klf4, and c-Myc; OSKM) were necessary and sufficient to transform terminally differentiated fibroblasts into iPSCs. Yamanaka ascribed this remarkable discovery to the convergence of at least three distinct areas of stem cell research [13]. The first area was the knowledge that differentiated cells were competent to undergo reprogramming/de-differentiation when exposed to a previously known, but elusive combination of factors present in oocytes during nuclear transfer [14, 15]. These factors are also present in mESCs, which are able to direct reprogramming of terminally differentiated T-cells when fused together [16]. The second area of research enabling the formulation of iPSCs was the finding that a master regulator factor(s) could define the differentiation state of a given cell [17, 18]. Finally, the third important stream in establishing iPSCs was the cumulative knowledge from 25 years of ESC cultivation conditions. Since the initial description of iPSCs, a variety of transcription factors and different types of cells have been used to generate iPSCs [19]. Defining improved methods to derive iPSCs remains an area of active research, as will be discussed later in this review.
Dr. Yamanaka was awarded a share of the 2012 Nobel Prize in Physiology or Medicine alongside Sir John B. Gurdon for their landmark work demonstrating the potential for terminally differentiated cells to regain pluripotency. In 1962 Gurdon provided the first evidence of the ability of mature, differentiated cells to return to a pluripotent state. He did this by replacing the nucleus of a frog oocyte with the nucleus from a mature intestinal epithelium cell, from which developed a normal tadpole [14]. This breakthrough experiment changed the dogma of the irreversible process of cell differentiation and set up a whole new scientific discipline of cloning, eventually leading to the generation of a cloned mammal [15]. However, cloning via somatic cell nuclear transfer is technically challenging and requires the use of a large number of oocytes, leading several groups to seek the identity of the pluripotency genes that would drive the de-differentiation of mature cells. Finally, more than 40 years later, Takahashi and Yamanaka identified the correct combination of genes sufficient to accomplish this task to generate the first iPSCs [9]. It is important to recognize that the power and potential of hiPSCs would not be possible without the ground-breaking work on ESCs that facilitated the development of hiPSCs.
Although both hESCs and hiPSCs are pluripotent stem cells, there remain concerns that hiPSCs are not equivalent to the gold-standard set by hESCs. Some studies using larger sample sets of hESCs and hiPSCs demonstrate that, like mESC clones, independent hESC and hiPSC clones have inherent variability in their gene expression and DNA methylation states but do not differ substantially between each other [20–23]. These studies attribute this variability to both the initial method of hiPSC derivation and the subsequent lab-specific maintenance of these cells rather than from any actual differences between hESCs and hiPSCs [22, 23]. However, other studies describe differences in gene expression and DNA methylation profiles between hESCs and hiPSCs [24–29]. A separate group has proposed that these gene expression and DNA methylation differences between many hESC and hiPSC lines create a bias in their pluripotent potential. Nichols and Smith have proposed that two phases of pluripotency can be defined: naive and primed [30]. While naïve (or ground) state stem cells are truly pluripotent, primed cells are predestined for lineage specification and commitment. In the case of hiPSCs, primed hiPSCs could still be useful therapeutically, since they would be poised to generate differentiated cells in greater numbers. Additionally, primed cells can regain their naïve state by neutralizing inductive differentiation stimuli or by transfection with Klf4 [31–34].
Methods to Derive iPSCs
There remains a need for more efficient and rapid generation of patient-specific hiPSCs to advance successful clinical application of hiPSC-based therapies. A number of factors play a role in determining which method will satisfy these criteria. These include iPSC-generating transcription factor combinations, transcription factor delivery methods, and optimal iPSC culture conditions. In the first report of miPSC generation, Takahashi and Yamanaka used the OSKM transcription factors to generate the miPSCs from murine fibroblasts [9]. They used a similar approach to generate hiPSCs a year later [11]. However shortly thereafter, the Thomson group generated hiPSCs with similar efficiency using a slightly different combination of transcription factors, substituting Nanog and Lin28 for Klf4 and c-Myc [12]. Although a number of different transcription factors combination have been used, the OSKM transcription factor combination is predominately used to generate iPSCs currently (Reviewed in [19]). Interestingly, subtle and temporary differences in the transcriptional level and stoichiometry of the OSKM reprogramming factors may have a profound impact on their protein expression resulting in qualitative differences between generated iPSCs, even when the same reprogramming factors are used. Jaenisch's group demonstrated that high Oct3/4 and Klf4 combined with lower Sox2 and c-Myc expression increases the efficiency to generate high-quality iPSCs [35]. Another group has shown that increasing the activating efficiency of Oct3/4 by fusing it to the transactivation domain of MyoD creates a protein chimera that also increases the number and quality of the iPSCs when combined with the remaining OSKM factors [36]. These studies show that different OSKM factor stoichiometries can produce higher quality iPSCs.
In generating the first miPSCs and subsequently in hiPSCs, Takahashi and Yamanaka used retroviruses to deliver the OSKM transcription factors, which integrated in approximately 20 sites throughout the genome in each iPSC line [9, 11]. In spite of the high copy number of exogenous OSKM, many of these lines were able to robustly differentiate. The Thomson group opted to use lentiviruses to deliver their transcription factors, which also resulted in multiple genome integrations [12]. In an attempt to minimize multiple viral integrations during iPSC generation and increase overall iPSC generation efficiency, Mostoslavsky's group designed a "stem cell cassette" (STEMCCA) containing all of the OSKM transcription factors on a single lentiviral vector [37]. The STEMMCCA cassette is able to consistently generate hiPSCs with a single genomic integration site [38]. While these methods provide high efficacy and reproducibility of iPSCs, the development of non-integrating vectors to generate iPSCs is rapidly becoming more standard [13].
While retro- and lentiviral vectors have successfully produced iPSCs, these methods invariably require integration into the genome, which comes with an inherent risk of insertional mutagenesis [39]. Therefore for clinical use of hiPSCs in gene therapy, these integration sites either need to be repaired or avoided altogether. In order to minimize the integration footprint, second-generation lentiviral cassettes are flanked by loxP sites (ex. STEMCCA-loxP vectors), which after iPSC establishment can be removed by Cre-mediated excision [40]. Unfortunately, this vector excision scars the host DNA by leaving residual sequence behind, which could still potentially result in tumorigenesis. A modified Sendai virus has been presented as an alternative to integrating retro- and lentiviruses for iPSC generation [41]. Sendai virus is a negative-strand RNA virus that replicates in the cytoplasm of infected cells and does not integrate into the host genome at any point during its lifecycle. Adenovirus is another non-integrating vector that has also been used to generate iPSCs, allowing for transient expression of OSKM without integrating into the host genome [42]. However, adenovirus does not seem to be very efficient in iPSC generation. Another study finds adenovirus can integrate into the genome at low levels, bringing any advantage adenovirus has over retro- and lentiviruses to generate iPSCs into question [43].
A number of non-viral methods have also been used for iPSC reprogramming. iPSC generation using plasmids containing the coding sequence for the OSKM transcription factors was first reported by the Yamanaka group [44]. Thomson’s group has also successfully created hiPSCs using a combination of three episomal-based vectors derived from the Epstein-Barr virus that contained an altered OSKM ratio (2:2:2:1) in combination with Nanog and Lin28 [45]. In addition, iPSCs have been derived by the addition of recombinant OSKM proteins [46, 47] and synthetic mRNAs [48]. Regrettably, these non-viral methods for iPSC generation suffer from low efficiency to date.
Methods for Gene Correction
Although a number of clinically relevant cells can potentially be produced from hiPSCs, diseases that result from somatic genetic mutations would not be cured due to the presence of the underlying mutation in the resulting hiPSCs. Therefore, it is necessary to provide an efficient and precise method to correct those genetic changes in patient hiPSCs. In this scenario, a much larger number of diseases could then be clinically treated by autologous transplantation. Research in homologous recombination and gene therapy has developed several new tools that allow for the sequence-specific excision and repair of almost any mutated sequence. These tools will be described in more detail below.
Since the late 1980s, homologous recombination has been a common technique used to integrate exogenous DNA templates with long regions of homology into the host cell genome by native cellular repair mechanisms [49]. In 1994 it was reported that the induction of double-stranded (ds) DNA breaks stimulates homologous recombination in mammalian cells [50]. This discovery led to the development of zinc finger nucleases (ZFNs) capable of creating targeted dsDNA breaks in a gene of interest, which greatly increases both insertional efficacy and site-specificity [51].
The first nucleases for generating site-specific dsDNA breaks, ZFNs, are commonly used to enhance the efficacy of gene targeting by inducing a dsDNA break at a specific DNA sequence [52]. ZFNs contain the zinc finger motif, which is able to recognize a nucleotide triplet [53]. These zinc finger motifs are linked to the nonspecific FokI nuclease, which creates the dsDNA breaks. DNA cleavage by ZFNs requires protein dimerization, which has allowed for the recognition of DNA cleavage sequences of up to 36bp by ZFNs containing 6 tandem zinc finger motifs. The resulting dsDNA breaks are repaired by the intrinsic cellular repair mechanisms of the non-homologous end joining repair pathway (NHEJ) or the homology directed repair pathway (HR) [54, 55]. Whereas the first mechanism simply ligates the ends of the broken DNA strands together in an error-prone fashion, the latter mechanism uses undamaged sister chromatids or exogenous donor DNA as a template for repair, which facilitates site-specific transgene incorporation of DNA sequences as long as 8 kb into the cell [56]. A common human genomic site for donor DNA insertion is the adeno-associated virus type 2 target region (AAVS1 locus) on chromosome 19, which is also known as "safe harbor". The expression of inserted genes in this locus has been demonstrated to be more stably maintained during hESCs differentiation in comparison to random integration, and no pathologic events in cells have been observed by disrupting this site [57]. Both viral and non-viral systems effectively deliver ZFN template and donor DNA into human pluripotent stem cells [52].
Recently, another novel class of nucleases, the transcription activator-like effector (TALE) nuclease (TALEN), was developed to allow for more flexible targeted genome engineering [58–60]. TALEs are bacterial proteins containing multiple tandem repeats of ~34 amino acids, which when combined together form a DNA binding domain. Within each repeat there exists polymorphism within a repeat variable diresidues (RVD) module that determines the nucleotide binding specificity. Upon fusing TALE repeats with the FokI nuclease used in ZFNs, the Voytas lab created sequence-specific nucleases they called TALENs [58]. Modification of the RVDs in the TALE tandem repeats can be done to generate novel sequence-specific designer TALENs, which allows researchers to have a much larger pool of DNA sequences for gene modification compared to using ZFNs [61]. When compared to ZFNs, TALENs offer higher DNA binding specify and lower toxicity [62]. Use of the "safe harbor" AAVS1 locus as an integration site for TALENs, will avoid functional perturbation of the host-cell transcriptome in human pluripotent stem cells.
Potential problems have been identified with using ZFNs and TALENs, such as off-target cleavage of sequences similar to the target sequence [63] and transgene addition caused by the combination of HDR and NHEJ repair mechanisms [64]. The use of TALENs with strong, distinct sequence specificity and verification of transgene insertion in only the correct locus through molecular biology techniques should be employed well before actual clinical use of modified human pluripotent stem cells to ensure their continued clinical relevance [52].
hiPSCs and Gene Therapy: Hurdles for Clinical Translation
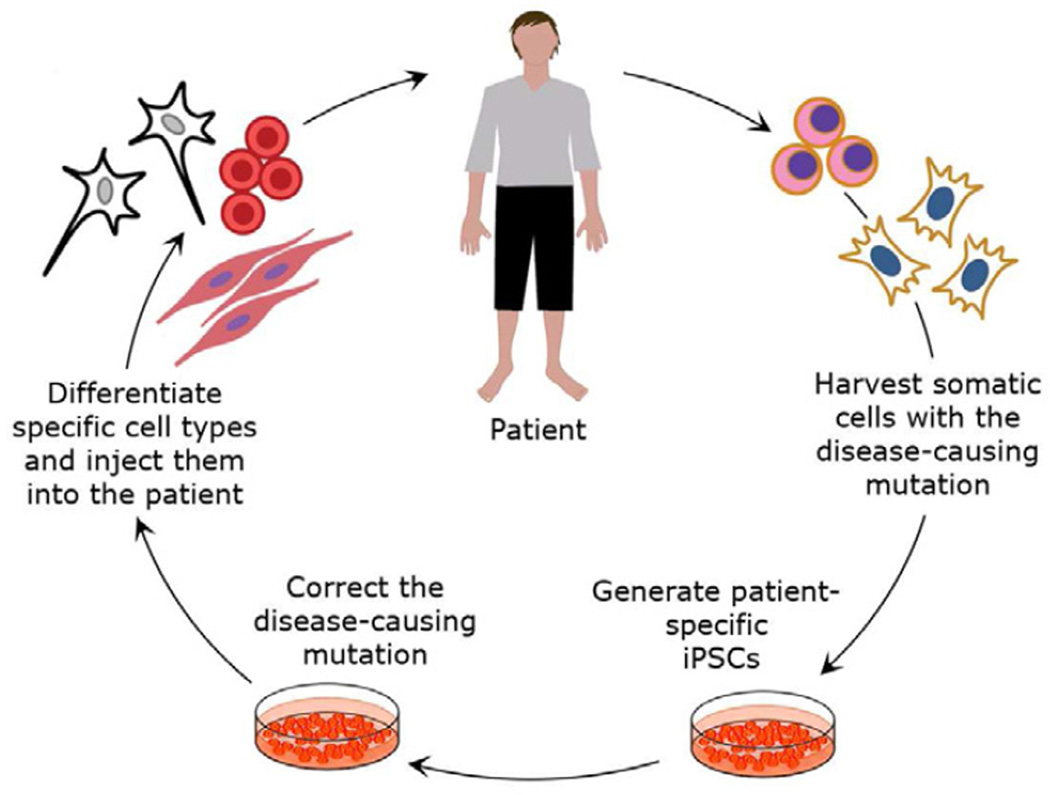
Combining the methods of stem cell technology with gene correction techniques offers an extraordinary potential for transplantation therapy, disease modeling and drug screening. iPSC technology provides the promise for treatment of genetic diseases by autologous transplantation of genetically corrected hiPSCs (Figure 1). In this regard, the main advantages of using hiPSCs are (i) eliminating the need for immunosuppression as is currently done during hematopoietic stem cells (HSCs) transplantation from human leukocyte antigen (HLA)-matched donors, (ii) the unlimited proliferative ability in cell culture to generate an unlimited supply of syngeneic patient-derived transplantable cells, and (iii) the ability for gene editing of disease-causing genetic mutations. The potential to make genetically corrected human pluripotent stem cells from diverse genetic subtypes from any number of diseases also allows for the development of reliable models for studying the development and progression of genetic diseases in vitro. For example, disease-causing gene correction in hiPSCs would make a "perfect control" for pharmaceutical in vitro comparative studies.
Figure 1. Schematic representation of iPSC-based combined gene and cell therapy.
The autologous transplantation of genetically corrected iPSC-derived cells can potentially be used to treat patients with certain genetic diseases, such as sickle cell anemia, Fanconi anemia, β-thalassemia, Parkinson's disease, Huntington's disease, and muscular dystrophies. Patient's somatic cells (e.g. skin fibroblasts or mononuclear blood cells), containing the disease-causing mutation, are in vitro transformed into iPSCs by introduction of transcription factors. The mutation is corrected by gene-specific targeting of iPSCs, using methods such as zinc finger nucleases (ZNFs) or transcription activator-like effector nucleases (TALENs). Corrected iPSCs are differentiated into the required cell type precursors or mature cells, such hematopoietic, neural or muscle cells. Healthy differentiated cells would be injected back into the patient where they would need to integrate and provide improved function.
There are several obstacles to be overcome before stem cell gene therapy can be considered for a clinical application. Safe methods for hiPSC generation and high reprogramming efficacy are of the utmost importance. Viral delivery of transcription factors that integrate into the host DNA is currently the most efficient reprogramming method. However, insertional mutagenesis and incomplete pluripotent gene silencing may occur, which are associated with the risk of tumorigenesis. Once efficient and sound hiPSC generation methods are established, it will also be vital to develop robust and reliable differentiation protocols for human pluripotent stem cells into a variety of different cell types. In regards to transgene correction, screening methods will need to be employed to verify correct DNA insertion and the exclusion of off-target effects.
As the first hESCs trials have shown, the use of hiPSCs in clinic would have to overcome a number of barriers before successfully transitioning there [65–67]. Unlike the majority of small molecules and antibodies, human pluripotent stem cell-derived cells likely need to integrate into the body once they are administrated and do not leave the body after treatment ceases. The irreversible nature of stem cell based therapy therefore requires special precautions to be taken in any clinical trials. Companies such as Geron and Advanced Cell Technology have pioneered clinical application of hESCs aimed at treating such intractable conditions as the loss of limb function following spinal cord injury and progressive vision loss in degenerative retinal diseases [68].
Based on extensive preclinical data of hESC-derived oligodendrocyte progenitors [69, 70], Geron Corporation (California, USA) began using oligodendrocytes derived from hESCs to treat patients with spinal cord injuries. Four patients were enrolled into a phase I clinical trial and none suffered serious adverse side effects after hESC-derived oligodendrocyte injection. In these trials the risk of teratoma formation was the main concern. Despite overcoming the rigorous safety requirements demanded by the FDA and overall encouraging results, the company unexpectedly abandoned their hESCs clinical trials in late 2011 [71]. The underlying reason was reported to be due to financial reasons and not due to problems with the medical procedure.
Advanced Cell Technology (California, USA) is currently carrying out two phase I/II clinical trials for the treatment of dry age-related macular degeneration and Stargardt's macular dystrophy with hESCs [68]. These two diseases are the leading causes of blindness in the developed world. The goal of these prospective studies is to assess the safety profiles and tolerance of subretinal injection of hESC-derived retinal pigment epithelial cells in patients. A four month follow-up of patients did not reveal any signs of hyperproliferation, tumorigenicity, ectopic tissue formation, or apparent rejection, while at the same time some patients reported a degree of vision restoration. These pioneering trials have overcome a number of regulatory hurdles and set high standards for cell characterization and quality control for any following stem cell based therapeutic trials.
Disease Candidates for Combined iPSC and Gene Therapy
Unlike most other stem cell clinical applications, there has been remarkable success using the transplantation of HSCs to cure disease [65]. With this experience, blood disorders with a genetic basis represent an attractive target for further stem cell research and therapeutic gene-corrected autologous stem cell transplantation. Significant progress has especially been made in gene therapy of red blood cell disorders [72]. The proof of principle results for treatment of genetic diseases by autologous transplantation of genetically corrected miPSCs was provided in 2007 by the Jaenisch and Townes labs only one year after the first successful miPSC generation was demonstrated [73]. In these break-through experiments in mice, it was shown that sickle cell anemia can be treated by the correction of the sickle hemoglobin allele by gene-specific targeting in miPSCs. A humanized knock-in mouse model of sickle cell anemia was used in this experiment to derive miPSCs from tip-tail fibroblasts. One copy of the βS (sickle) globin gene was corrected in these miPSCs using homologous recombination and differentiated into hematopoietic progenitors, which were then injected back into irradiated sickle mice. These miPSC-derived hematopoietic progenitors were able to completely reconstitute normal hematopoiesis. In order to increase the efficiency of gene targeting at specific genomic loci, ZFNs were recently used to correct the sickle cell anemia mutation in hiPSCs derived from human patients [74].
Mutations in any of the 13 genes in the Fanconi anemia DNA repair pathway makes Fanconi anemia an attractive target for gene therapy [75]. A collaboration between groups in Spain, Italy and the US published a proof-of-principle study for the treatment of Fanconi anemia using hiPSCs [76]. Through reprogramming and gene correction of somatic cells from Fanconi anemia patients, the groups managed to generate disease-free hematopoietic progenitors of the myeloid and erythroid lineages. Correction of mutations in globin genes causing β-thalassemia in hiPSCs have also been successfully demonstrated by Wang et al. [77]. CD34+ cells differentiated from corrected hiPSCs were able to engraft into irradiated SCID mice and in vivo differentiated erythrocytes showed improved hemoglobin production over the uncorrected hiPSCs counterparts. However, transplanted CD34+ cells bear limited erythropoietic potential and long-term maintenance of erythroid differentiation has yet to be established. The disease-specific phenotype of a neutrophil defect, X-linked chronic granulomatous disease, was recapitulated in vitro using hiPSCs [78]. A ZFN strategy was then employed to correct the oxidase function with a lentivirus vector encoding the gp91phox minigene, resulting in abrogation of the disease phenotype. An advantage to using gene correction in neutrophils is the reduction of the oncogenic potential to extremely low levels as hiPSCs-derived mature neutrophils have no potential to proliferate or permanent engraftment.
Although blood diseases are one of the most prominent fields in stem cell research, gene correction in hiPSCs derived from various other monogenic diseases have also been accomplished. Soldner et al. corrected in hiPSCs the point mutation in the α-synuclein gene, which can cause some types of Parkinson's disease [79]. ZFN technology was used to modify fibroblast-derived hiPSCs from the patient carrying the A53T (G209A) α-synuclein mutation with no observed evidence of off-target genome disruption. Additionally, they were also able to generate Parkinson's disease-related cell lines by inducing either the A53T (G209A) or E46K (G188A) mutation in the genome of wild-type hESCs. These experiments hint at the great potential of human pluripotent stem cells to generate an unlimited supply of patient-derived, disease-relevant cells. The Ellerby lab derived hiPSCs from a Huntington's disease patient and replaced the disease-causing expanded CAG trinucleotide repeat with a normal sized repeat allowing for the derivation of normal healthy neurons [80]. In vitro testing of these repeat corrected hiPSC-derived neural stem cells rescued defective signaling pathways and reversed the disease phenotype, while retaining pluripotent characteristics. Subsequent in vivo experiments in mice suggested that the corrected hiPSC-derived neural stem cells are able to populate the striatum of murine Huntington's disease model brains and undergo differentiation to neurons and glial cells.
Muscular dystrophies therapy using hiPSC gene-correction is another auspicious area of stem cell research. Promising results were achieved in using genetically corrected hiPSCs for treatment of muscular dystrophies [81]. The mouse model of limb-girdle muscular dystrophy was employed for transplantation of hiPSCs-derived muscle progenitor cells, mesoangioblasts. hiPSCs were generated from fibroblasts and myoblasts of limb-girdle muscular dystrophy patients and genetically corrected in vitro with a lentiviral vector carrying the gene encoding human α-sarcoglycan, the gene responsible for the development of the disease. The hiPSC-derived corrected mesoangioblasts were proved to generate muscle fibers in α-sarcoglycan–null immunodeficient mice. These results suggest that gene and cell therapy employing hiPSCs might be applicable for treating various types of recessive muscular dystrophies. The point mutation in the gene encoding ornithine-δ-aminotransferase causes the retinal degenerative disease gyrate atrophy. hiPSCs generated from patients were used to assess the mutational load caused by homologous recombination during gene correction [82]. Encouragingly, their data suggests that hiPSC expansion and gene correction do not increase genomic instability.
One key issue that will need to be solved for many of these new hiPSC-based therapies is not only the efficient derivation of the mature cell population of interest, but also the ability to transplant these cells into a host in a manner that will allow effective disease correction. These possible hurdles include deriving enough therapeutic cells and also transplanting them in a manner where they will effectively integrate with the host. This is potentially a significant problem with neuronal diseases such as Parkinson’s disease. Indeed, previous trials using neuronal transplants for Parkinson’s illustrate this issue. While dopaminergic cells could be transplanted, they often did not function properly with either too much or too little dopamine being released [83]. Similarly therapeutic trials for muscular dystrophy using varying muscle cell populations have not achieved a significant therapeutic result [84–86]. Indeed, since muscular dystrophy affects essentially all the muscles in the body over time, how systemic delivery of these cells will be achieved remains a key question. Notably for hematopoietic therapies these barriers for integration may be less as has been proven by decades of experience with hematopoietic cell transplantation. From this experience we know that transplanted hematopoietic cells can home to the bone marrow and tissues and function essentially normally for many years or decades following the therapy. Complicating the issue of therapeutic cell engraftment is the methods which have been employed to successfully cause engraftment so far. In the case of the use of the hiPSC-derived corrected mesoangioblasts to correct limb-girdle muscular dystrophy described above, the authors overexpressed the myogenic regulator, MyoD, to derived these cells [81]. In the case of hematopoietic therapies, the overexpression of HoxB4 in vitro has been necessary to facilitate sufficient engraftment of hematopoietic cells in vivo [73, 87]. Taking these realities into account, there is still much to do to ensure that engraftable and functional cells of sufficient quantity and quality can be efficiently derived from iPSCs for the desired therapies.
Problems with immune rejection also need to be better understood for therapies using hiPSCs and hESCs. Indeed this was a key issue with the two hESC-based therapies described above. While patients may likely require at least low dose immunosuppression for allogeneic transplantation, how and when immunosuppressant should be dosed will only really be able to be solved through well designed clinical trials. While hiPSCs can potentially be derived on a patient specific basis to create autologous cells, the newly inserted gene could be seen as foreign to the host. Indeed, this has been seen in treatment of some diseases such as hemophilia. Currently, these are important issues that will be addressed with future preclinical and clinical studies.
Conclusion, Future Perspectives
hiPSCs have extraordinary potential for clinical applications in regenerative medicine. In this current perspective, we briefly discussed how decades of ESC research paved the way for the derivation of iPSCs. To this end hiPSCs may be considered as one evolutionary step higher than hESCs since they are much more appropriate for clinical applications. hiPSCs offer many advantages over hESCs in terms of the cell source availability and patient specificity. However it remains to be seen whether the use of hiPSCs advances into routine clinical practice in the next few decades, or whether they become a stepping stone between the isolation of hESCs and other innovative technologies. Direct cellular reprogramming represents one of the latest and very promising streams in regenerative medicine [88–92]. This approach circumvents the pluripotent cell stage and aims to directly transform one mature cell type into another, shortening the time needed to generate the clinically relevant cells. Moreover, direct reprogramming may also allow for in vivo reprogramming of cells [92, 93]. Together, these advances highlight the exciting potential for stem cell research combined with gene therapy approaches to produce new therapies in the future.
Acknowledgements
Research in the Kaufman lab is supported by NIH R01-HL77923, R01-DE022556, R01-HL067828, the Leukemia Research Fund of the University of Minnesota Cancer Center, the William L. and Blanche Hughes Foundation, the European Social Fund within the project Young Talent Incubator III (reg. no. CZ.1.07/2.3.00/20.0239), the Ministry of Education of the Czech Republic (MSM- 0021622430), and by the European Regional Development Fund - Project FNUSA-ICRC (No. CZ.1.05/1.1.00/02.0123). We thank Carol Taubert for production of the figure.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have read the journal’s policy on conflicts of interest and have no conflicts of interest to declare.
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, et al. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- 4.Smith AG, Hooper ML. Buffalo rat liver cells produce a diffusible activity which inhibits the differentiation of murine embryonal carcinoma and embryonic stem cells. Dev Biol. 1987;121:1–9. doi: 10.1016/0012-1606(87)90132-1. [DOI] [PubMed] [Google Scholar]
- 5.Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, et al. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 6.Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 7.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 8.Xu C, Rosler E, Jiang J, Lebkowski JS, Gold JD, O'Sullivan C, et al. Basic fibroblast growth factor supports undifferentiated human embryonic stem cell growth without conditioned medium. Stem Cells. 2005;23:315–323. doi: 10.1634/stemcells.2004-0211. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Boland MJ, Hazen JL, Nazor KL, Rodriguez AR, Gifford W, Martin G, et al. Adult mice generated from induced pluripotent stem cells. Nature. 2009;461:91–94. doi: 10.1038/nature08310. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 13.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- 15.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 16.Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Current biology : CB. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 17.Schneuwly S, Klemenz R, Gehring WJ. Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia. Nature. 1987;325:816–818. doi: 10.1038/325816a0. [DOI] [PubMed] [Google Scholar]
- 18.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 19.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward CM, Barrow KM, Stern PL. Significant variations in differentiation properties between independent mouse ES cell lines cultured under defined conditions. Exp Cell Res. 2004;293:229–238. doi: 10.1016/j.yexcr.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 21.Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman AM, Cooper JB. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell. 2010;7:258–262. doi: 10.1016/j.stem.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature genetics. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nishino K, Toyoda M, Yamazaki-Inoue M, Fukawatase Y, Chikazawa E, Sakaguchi H, et al. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002085. e1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 31.Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H, Lee G, Ganat Y, Papapetrou EP, Lipchina I, Socci ND, et al. miR-371-3 expression predicts neural differentiation propensity in human pluripotent stem cells. Cell Stem Cell. 2011;8:695–706. doi: 10.1016/j.stem.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 35.Carey BW, Markoulaki S, Hanna JH, Faddah DA, Buganim Y, Kim J, et al. Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell. 2011;9:588–598. doi: 10.1016/j.stem.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Hirai H, Tani T, Katoku-Kikyo N, Kellner S, Karian P, Firpo M, et al. Radical acceleration of nuclear reprogramming by chromatin remodeling with the transactivation domain of MyoD. Stem Cells. 2011;29:1349–1361. doi: 10.1002/stem.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sommer CA, Stadtfeld M, Murphy GJ, Hochedlinger K, Kotton DN, Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somers A, Jean JC, Sommer CA, Omari A, Ford CC, Mills JA, et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells. 2010;28:1728–1740. doi: 10.1002/stem.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 40.Sommer CA, Sommer AG, Longmire TA, Christodoulou C, Thomas DD, Gostissa M, et al. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28:64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harui A, Suzuki S, Kochanek S, Mitani K. Frequency and stability of chromosomal integration of adenovirus vectors. J Virol. 1999;73:6141–6146. doi: 10.1128/jvi.73.7.6141-6146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 45.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, et al. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capecchi MR. Altering the genome by homologous recombination. Science. 1989;244:1288–1292. doi: 10.1126/science.2660260. [DOI] [PubMed] [Google Scholar]
- 50.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 52.Collin J, Lako M. Concise review: putting a finger on stem cell biology: zinc finger nuclease-driven targeted genetic editing in human pluripotent stem cells. Stem Cells. 2011;29:1021–1033. doi: 10.1002/stem.658. [DOI] [PubMed] [Google Scholar]
- 53.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Dekelver RC, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci U S A. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith JR, Maguire S, Davis LA, Alexander M, Yang F, Chandran S, et al. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells. 2008;26:496–504. doi: 10.1634/stemcells.2007-0039. [DOI] [PubMed] [Google Scholar]
- 58.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mussolino C, Cathomen T. TALE nucleases: tailored genome engineering made easy. Curr Opin Biotechnol. 2012 doi: 10.1016/j.copbio.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 61.Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 2011;39:5790–5799. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mussolino C, Morbitzer R, Lütge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- 65.Daley GQ. The promise and perils of stem cell therapeutics. Cell Stem Cell. 2012;10:740–749. doi: 10.1016/j.stem.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strauss S. Geron trial resumes, but standards for stem cell trials remain elusive. Nat Biotechnol. 2010;28:989–990. doi: 10.1038/nbt1010-989. [DOI] [PubMed] [Google Scholar]
- 67.Ben-David U, Kopper O, Benvenisty N. Expanding the boundaries of embryonic stem cells. Cell Stem Cell. 2012;10:666–677. doi: 10.1016/j.stem.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 68.Schwartz SD, Hubschman JP, Heilwell G, Franco-Cardenas V, Pan CK, Ostrick RM, et al. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379:713–720. doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 69.Keirstead HS, Nistor G, Bernal G, Totoiu M, Cloutier F, Sharp K, et al. Human embryonic stem cell-derived oligodendrocyte progenitor cell transplants remyelinate and restore locomotion after spinal cord injury. J Neurosci. 2005;25:4694–4705. doi: 10.1523/JNEUROSCI.0311-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McDonald JW, Howard MJ. Repairing the damaged spinal cord: a summary of our early success with embryonic stem cell transplantation and remyelination. Prog Brain Res. 2002;137:299–309. doi: 10.1016/s0079-6123(02)37023-7. [DOI] [PubMed] [Google Scholar]
- 71.Baker M. Stem-cell pioneer bows out. Nature. 2011;479:459. doi: 10.1038/479459a. [DOI] [PubMed] [Google Scholar]
- 72.Xu X, Qu J, Suzuki K, Li M, Zhang W, Liu GH, et al. Reprogramming based gene therapy for inherited red blood cell disorders. Cell Res. 2012;22:941–944. doi: 10.1038/cr.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 74.Sebastiano V, Maeder ML, Angstman JF, Haddad B, Khayter C, Yeo DT, et al. In situ genetic correction of the sickle cell anemia mutation in human induced pluripotent stem cells using engineered zinc finger nucleases. Stem Cells. 2011;29:1717–1726. doi: 10.1002/stem.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 76.Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53–59. doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y, Zheng CG, Jiang Y, Zhang J, Chen J, Yao C, et al. Genetic correction of β-thalassemia patient-specific iPS cells and its use in improving hemoglobin production in irradiated SCID mice. Cell Res. 2012;22:637–648. doi: 10.1038/cr.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zou J, Sweeney CL, Chou BK, Choi U, Pan J, Wang H, et al. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells: functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood. 2011;117:5561–5572. doi: 10.1182/blood-2010-12-328161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soldner F, Laganière J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.An MC, Zhang N, Scott G, Montoro D, Wittkop T, Mooney S, et al. Genetic Correction of Huntington's Disease Phenotypes in Induced Pluripotent Stem Cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tedesco FS, Gerli MF, Perani L, Benedetti S, Ungaro F, Cassano M, et al. Transplantation of Genetically Corrected Human iPSC-Derived Progenitors in Mice with Limb-Girdle Muscular Dystrophy. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003541. 140ra89. [DOI] [PubMed] [Google Scholar]
- 82.Howden SE, Gore A, Li Z, Fung HL, Nisler BS, Nie J, et al. Genetic correction and analysis of induced pluripotent stem cells from a patient with gyrate atrophy. Proc Natl Acad Sci U S A. 2011;108:6537–6542. doi: 10.1073/pnas.1103388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Freed CR, Greene PE, Breeze RE, Tsai WY, DuMouchel W, Kao R, et al. Transplantation of embryonic dopamine neurons for severe Parkinson's disease. N Engl J Med. 2001;344:710–719. doi: 10.1056/NEJM200103083441002. [DOI] [PubMed] [Google Scholar]
- 84.Mendell JR, Kissel JT, Amato AA, King W, Signore L, Prior TW, et al. Myoblast transfer in the treatment of Duchenne's muscular dystrophy. N Engl J Med. 1995;333:832–838. doi: 10.1056/NEJM199509283331303. [DOI] [PubMed] [Google Scholar]
- 85.Gussoni E, Blau HM, Kunkel LM. The fate of individual myoblasts after transplantation into muscles of DMD patients. Nat Med. 1997;3:970–977. doi: 10.1038/nm0997-970. [DOI] [PubMed] [Google Scholar]
- 86.Blau HM. Cell therapies for muscular dystrophy. N Engl J Med. 2008;359:1403–1405. doi: 10.1056/NEJMcibr0805708. [DOI] [PubMed] [Google Scholar]
- 87.Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 88.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 90.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Szabo E, Rampalli S, Risueño RM, Schnerch A, Mitchell R, Fiebig-Comyn A, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 92.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]