Abstract
Tuberculosis (TB) remains one of the most devastating infectious diseases and its eradication is still unattainable given the limitations of current technologies for diagnosis, treatment and prevention. The World Health Organization’s goal to eliminate TB globally by 2050 remains an ongoing challenge as delayed diagnosis and misdiagnosis of TB continues to fuel the worldwide epidemic. Despite considerable improvements in diagnostics for the last few decades, a simple and effective point-of-care TB diagnostic test is yet not available. Here, we review the current assays used for TB diagnosis, and highlight the recent advances in nanotechnology and microfluidics that potentially enable new approaches for TB diagnosis in resource-constrained settings.
1. Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (MTB), is a major global health problem with an estimated 8.8 million new active TB cases, and approximately 1.1 million deaths in 2011 (WHO, 2011b). Among these new TB cases, the majority occurred in the South-East Asia region, which accounted for 40% of the global incidence. Further, only 65% (5.7 million) of the estimated TB cases in 2010 were reported, indicating the need for improved diagnosis (Van Rie et al., 2010). The WHO estimates that there will be more than 2 million new cases of MDR-TB between 2011 and 2015 (WHO/Global Fund, 2012). These numbers, combined with increasing spread of infection with multi drug-resistant (MDR) and extensively drug-resistant (XDR) strains, demonstrate an urgent need for new approaches for early diagnosis, therapy monitoring, and disease supervision.
Diagnosis of TB infection (active or latent) is essential not only for treatment of the infected individual, but also for controlling its spread among various populations. In TB endemic areas, TB control programs aim to treat active TB patients rather than individuals with latent TB infection (LTBI), which is only evidenced by the immunological responses to MTB proteins (Barry et al., 2009). Although LTBI individuals are not infectious, their identification is equally important since 10% of these individuals, particularly the immunosuppressed, can subsequently develop active tuberculosis. The routine diagnosis of active TB infection employs various approaches including smear microscopy, culture of MTB bacilli, detection of MTB nucleic acids (NAATs, nucleic acid amplification tests), and clinical symptoms. Conversely, LTBI is identified via tuberculin skin test (TST) and interferon gamma (IFNγ) release assays (IGRAs). However, currently TB diagnosis is limited by three primary limitations: (i) low specificity of clinical diagnosis, (ii) unavailability of high performing diagnostic methods in developing world laboratories, and (iii) incapability to monitor patient compliance to the 6-9 month long therapy.
Although the present pipeline of tuberculosis diagnostics is promising (e.g., a POC manual NAAT kit using loop-mediated isothermal amplification from Eiken/FIND (Foundation for Innovation New Diagnostics) and a handheld NAAT device from Epistem / Xcelris (Pai and Pai, 2012, WHO, 2011b), absence of an instrument-free test and high cost continue to be a bottleneck. Thus, there is an unmet need to develop a simple, inexpensive, sensitive and portable assay for the detection of active MTB infection, and for the differentiation of active TB from LTBI at the point-of-care (POC), where sustainable financial support, laboratory infrastructure, and well-trained operators are limited (Wang et al., 2010).
In this review, we (i) focus on the present TB diagnostic technologies in terms of their potential for detecting active TB infection at the POC in the near future, (ii) highlight the gap between present assays and clinical need to manage TB patients, and (iii) evaluate the potential of nanotechnology and microfluidics to develop POC diagnostics for TB.
2. Present assays for TB diagnosis
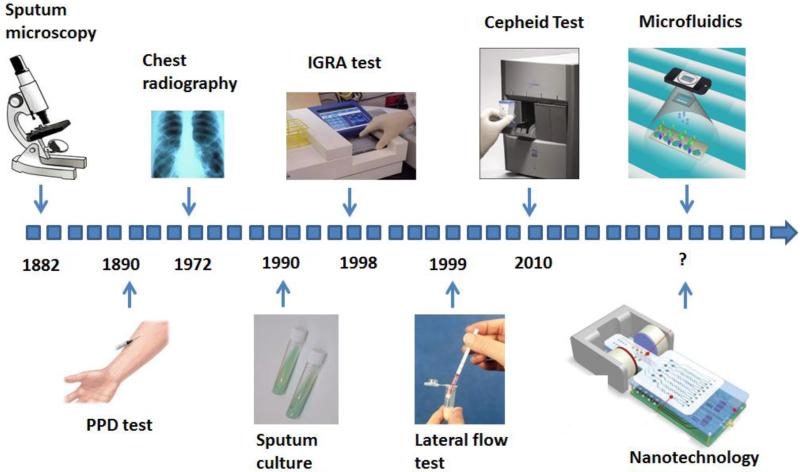
Over time, diagnostic methods for TB have evolved from sputum microscopy to the latest WHO-endorsed GeneXpert based test (Figure 1). These technologies are based on the detection of (i) whole MTB bacillus, (ii) MTB nucleic acid, or (iii) MTB-specific immune responses in TB patients. Here, we categorized these TB diagnostics according to their potential (low, moderate and high) for POC testing with consideration of sensitivity, specificity and accuracy, as well as cost and rapidity. Although urine based antigen assay and sputum smear are simple and widely used in resource-constrained settings, they have limited utility in detecting active TB infection in high-risk populations at the POC. Hence, we categorized these two assays in the group of “assays with moderate potential for POC testing” with the current formats. On the other hand, we categorized nucleic acid amplification tests as “assays with high potential for POC testing”, because of high sensitivity and specificity in the current formats. In addition, these assays (eg., GeneXpert) can be made inexpensive, rapid and automated for POC testing in decentralized settings.
Figure 1.
Timeline of the evolution of MTB diagnostic technologies. The advent of each representative technology for MTB diagnosis is indicated by an arrow. Among them, sputum culture remains to be the gold standard. Miniaturized devices based on microfluidics/nanotechnology are emerging.
2. 1. Assays with low potential for POC testing
Tuberculin skin test (TST)
TST, also known as the purified protein derivative (PPD) test, was developed over 100 years ago. It is based on the use of the PPD tuberculin, a precipitate of non-species-specific molecules extracted from sterilized and concentrated cultures of MTB, to detect immune reactivity in subjects (Andersen et al., 2000). This test is simple and widely used in the developed world to detect MTB exposure. If the subject is exposed to MTB, intradermal injection of PPD tuberculin leads to induration of the skin after 48 to 72 hours, due to an immune response. Although the procedure is relatively simple, the assay requires well-trained operators to perform and analyse results. False-positive and false-negative results can arise from the presence of nontuberculous mycobacterial infection or Bacillus Calmette-Guérin (BCG) vaccination (Farhat et al., 2006). As reported, BCG vaccination in infancy causes a TST false positive rate of 8.5% and 1% when tested before or after 10 years of age, respectively. These findings show that the results of TST in high-prevalence countries such as India and China should be interpreted with caution, since BCG vaccination is given to infants after birth in these countries (Zwerling et al., 2011). It should also be noted that the sensitivity of TST is low in HIV-infected patients due to compromised immune responses (Cobelens et al., 2006).
Interferon-Gamma (IFN-γ) Release Assays (IGRA)
IFN-γ release assays have been developed and implemented since 2001 with varying degrees of success (Mazurek et al., 2005). These tests measure T cell release of IFN-γ (host cellular immune response) upon stimulation of whole blood with MTB-specific antigens, including early-secreted antigenic target 6 (ESAT6) and culture filtrate protein 10 (CFP10) and TB7.7 (an additional antigen used in modified IGRA kits). IGRA assays are available in two formats, ELISA (e.g., QuantiFERON®-TB, GFT) and enzyme-linked immunospot assay (ELISPOT, e.g., T-SPOT® TB test). QuantiFERON®-TB (including QFT-Gold and QFT-Gold In-Tube) quantifies the concentration of IFN-γ, while T-SPOT® TB test counts the number of IFN-γ-producing antigen-specific T lymphocytes. Although these assays are not affected by prior BCG vaccination as the antigens are absent in all BCG strains, they cannot distinguish individuals having LTBI from those with active disease (Dheda et al., 2009). In addition, systematic review and meta-analyses have shown that neither of IGRA assays has better sensitivity than TST in detecting active or latent TB infection in HIV-infected individuals (Cattamanchi et al., 2011, Metcalfe et al., 2011). Thus IGRA assays cannot be used to rule-in or rule-out active TB cases, especially in HIV-infected individuals in high-burden settings (Chen et al., 2011, Ling et al., 2011). This limitation has led a WHO expert group to discourage the use of IGRA assays for active pulmonary TB diagnosis in low- and middle-income countries (Pinto et al., 2012). Furthermore, the assay takes 24 hr to produce results and requires significant instrumentation and well-trained personnel.
2. 2. Assays with moderate potential for POC testing
Sputum smear microscopy
Conventional smear microscopy, in combination with Ziehl-Neelsen stain, has been widely used in resource-constrained settings owing its inexpensiveness and rapidity. However, this method has a low sensitivity (34-80%) (Davies and Pai, 2008), as it can detect only a high bacillary burden (5,000-10,000 CFU/mL) in the sputum (Hobby et al., 1973, Tostmann et al., 2008). Thus, the active pulmonary TB patients having low bacillary burden at the early stage of the disease often go undetectable by this test. Although adding fluorescence staining has increased the sensitivity, it requires a fluorescence microscope. In addition, at least two sputum smears from two separate clinical visits are required for microscopy-based assays, rending great challenges for rapid clinical decision-making at the POC. Specificity of this method also needs to be confirmed by other methods, as the morphology detection cannot differentiate MTB from other mycobacterial strains (such as other strains of MTB complex; M. kansasii; M. marinum; and M. avium complex) and Nocardia sp. Despite its disadvantages, this method can be potentially used for POC testing provided improvements on simplicity, portability and cost (section 4.2).
Urine based antigen detection assay
Detection of lipoarabinomannan (LAM, lipopolysaccharide component of MTB cell wall) antigen in urine has been used to develop a simple lateral flow assay known as Determine TB-LAM (Alere, Waltham, MA, USA) (Lawn et al., 2012). Analysis of urine has many advantages over sputum samples, as urine is simpler to collect and safer to handle at resource-constrained settings. This kit only need a single clinic visit, is inexpensive (~US $ 3.5/test), and yields results in half an hour. Further, the assay requires minimal training without the use of complex instruments or electric supply. Thus, it has potential to be used as a POC assay (Lawn et al., 2012, Peter et al., 2012). On the other hand, this test has a low sensitivity of 28.2% in patients with culture-confirmed TB (Lawn et al., 2012). In contrast, the sensitivity improved to 66.7%, when CD4 cell count was below 50 cells/μL. In another study, the LAM lateral flow test also had improved sensitivity in AIDS patients with CD4 cell count below 200 cells/μL. These studies indicate that the LAM lateral flow test may add incremental value of diagnosing TB in AIDS patients with advanced diseases (Lawn et al., 2012, Peter et al., 2012). Further, both studies demonstrate that LAM lateral flow test in combination with smear microscopy can increase the sensitivity comparable to GeneXpert MTB/RIF in this particular AIDS population (Lawn et al., 2012, Peter et al., 2012). Thus, the LAM lateral flow test may aid management of AIDS patients in Sub-Saharan countries where TB is the leading cause of death. Nevertheless, rigorous field evaluation is needed to assess the diagnostic utility of this test as a rule-in assay for detection of active TB cases in various AIDS populations (e.g., women and children) in resource-constrained settings (Denkinger and Pai, 2012, Lawn et al., 2012, Peter et al., 2012).
2. 3. Assays with high potential for POC testing
Antibody detection assay
Owing to low-cost, simplicity and rapidity, lateral-flow immunochromatographic commercial tests for detection of antibodies in blood specific for mycobacterial antigens have been widely used in the developing world having high burden of TB. The antibody-antigen interaction triggers the aggregation of detection particles, which generate a visible signal without complicated equipment. These assays can be operated with minimal training, are disposable and have short testing time, making them suitable for POC testing. However, these commercial serological tests still face challenges such as accuracy and/or cost-effectiveness for the diagnosis of TB (Dowdy et al., 2011, Steingart et al., 2011b). These challenges have prompted the WHO to advise against their use for TB diagnosis in 2011 (WHO, 2011a). Despite a poor sensitivity of 53% (95% confidence interval (CI), 42%-64%) in detecting active pulmonary TB, a meta-analysis showed that immunochromatographic tests had a comparable pooled specificity of 98% (95% CI, 96%-99%) compared to ELISA (Steingart et al., 2011a). As such, future research efforts should be committed to addressing current sensitivity limitations and increasing the specificity in the presence of co-infection with multiple mycobacterial strains. With regards to immunochromatographic tests, limiting factors also include multiplex capability and waste disposal in resource-constrained settings (McNerney and Daley, 2011).
Nucleic acid amplification test (NAAT)
Nucleic acid amplification test (NAAT) has become a routine analytical tool for diagnosis, detection of drug resistance, and treatment monitoring. Among various NAAT technologies, reverse-transcription polymerase chain reaction (RT-PCR) and line probe assays are the most commonly used for rapid detection of TB and drug resistance. The two line-probe assays for TB, namely the Inno-LiPA Mycobacteria assay (Innogenetics NV, Ghent, Belgium) and the GenoType Mycobacterium assay (Hain Lifescience GmbH, Nehren, Germany), are based on nucleic acid hybridization of amplified products on a nitrocellulose membrane (Padilla et al., 2004, Sarkola et al., 2004). For rapid diagnosis, these assays detect 16S-23S spacer and 23S gene, respectively. Line-probe assays have also been adapted to detect mutations causing rifampicin, isoniazid, fluoroquinolone, and ethambutol drug resistance with satisfactory sensitivity and specificity. Compared to traditional MTB NAAT assays such as the Amplicor MTB (Roche Diagnostics) and the Amplified Mycobacterium tuberculosis direct test (Gen-Probe Inc.), the two line-probe assays are simpler and more affordable. In addition, these line-probe assays are designed to detect multiple MTB strains simultaneously. Thus, they have an advantage over culture-based drug-resistance testing to differentiate pathogenic and non-pathogenic strains (Parsons et al., 2011). Despite the advantages, line-probe assays require temperature control and multiple testing steps for stringent nucleic acid hybridization conditions. Thus, they are restricted to use in centralized laboratories. Furthermore, they are also not recommended for testing smear-negative samples due to low sensitivity and risk of bacterial contamination.
Recently, an automated cartridge-based NAAT (GeneXpert MTB/RIF, Cepheid Inc.) has been endorsed by the WHO for simultaneous detection of MTB and rifampicin resistance from sputum. This assay significantly improves the accessibility of molecular testing in the developing world, where the majority of patients cannot be tested due to the need for expensive equipment, laboratory infrastructure, and skilled operators. This assay is essentially molecular beacon based RT-PCR, which utilizes five overlapping probes spanning the entire core region of rpoB (81bp) (Piatek et al., 1998). Each probe is a fragment of DNA oligonucleotides designed to have a stem-loop structure, with a fluorophore and a quencher at either end. During the amplification, these 5 probes go under conformational change and thus lead to increase in fluorescence, depending on the presence of their complementary sequences. In addition to high sensitivity in detecting drug-resistance mutations, the assay is further integrated in a disposable cartridge preloaded with reagents for sample processing, DNA extraction and PCR amplification. A multi-sited clinical study showed that this assay had a specificity of 99.2% for non-TB patients, and a sensitivity of 98.2% and 72.5% for smear-positive and smear-negative TB, respectively (Boehme et al., 2010). Another clinical study further demonstrated the deployment of almost fully automated GeneXpert MTB/RIF assay in resource-constrained settings (Boehme et al., 2011). However, the cost associated with implementing GeneXpert MTB/RIF assay (< $10/cartridge), requirement of an electrical supply, and maintenance of instruments makes this assay less ideal for POC testing in the developing world (Niemz et al., 2011). This test is noteworthy because it has revived the interest in rapid molecular tests for detecting active TB and drug resistance.
Another advancement in the development of POC NAAT assays for TB is the loop-mediated isothermal amplification (LAMP) assay (Iwamoto et al., 2003, Notomi et al., 2000). In contrast to PCR, this assay utilizes 4 primers to specifically initiate synthesis of a large amount of DNA by Bst DNA polymerase without thermal cycling, which eliminates the need for an expensive thermal cycler to perform NAAT at the POC. In addition, this assay is compatible with turbidity or fluorescence-based detection (Tomita et al., 2008). In the former format, pyrophosphate is produced during DNA amplification and it leads to a white precipitate of magnesium pyrophosphate. The turbidity-based approach can be detected qualitatively by the naked eye. This assay can be modified for fluorescence detection and quantification by adding SYBR Green. A field evaluation in resource-constrained settings showed that this assay had a sensitivity of 97.7% in smear- and culture-positive sputum specimens, and a specificity of 99% in culture-negative samples (Boehme et al., 2007). Another study performed in Nepal showed that MTB-LAMP had a sensitivity of 100 % in culture-positive samples and a specificity of 94.2 % in culture-negative samples. These studies demonstrated the feasibility of performing TB LAMP assay in resource-constrained settings (Pandey et al., 2008). However, this DNA-based detection assay cannot distinguish between live and dead bacteria, and further optimization needs to be carried out on initial denaturation and assay time for improved sensitivity (Aryan et al., 2010, Geojith et al., 2011).
3. Diagnostic gaps between present technologies and unmet clinical need
Although MTB was identified as the pathogen of TB more than a century ago, the detection of TB in the developing world remains a significant healthcare issue owing to a number of challenges. First, MTB is a slow-growing bacterium, and therefore culture, despite high sensitivity, cannot provide guidance for on-site patient care. For example, MTB takes 4-8 weeks to grow on traditional solid culture and 10-14 days even with rapid liquid culture. Second, pulmonary TB causes relatively low clinical symptoms early in the course of disease, which leads to delays in seeking patient care. Third, active pulmonary TB may present low bacillary burden at the early stage, which often leads to low sensitivity for sputum smear microscopy and other POC tests commonly used in the developing world. Fourth, the use of sputum samples for diagnosis of TB with existing methodology is more complex compared to the use of blood and urine samples. Particularly, standardized sputum collection, transportation and storage procedures are required to ensure consistent diagnostic results. Lastly, there is lack of financial incentive to develop a diagnostic assay for TB that is predominant in resource-constrained settings.
In addition to the challenges posed by the slow-growing nature of TB, the lack of reliable and validated biological markers (for detection of active TB and identification of LTBI), either derived from the host or pathogen, hampers advances in TB diagnostic assays. The unavailability of reliable biomarkers is mainly due to insufficient understanding of the complex interaction between MTB and its host, pathogenesis, and protective immune responses during infection. Furthermore, heterogeneous immune responses from individuals with different disease statuses such as latent infection/re-infection or with different immunization records may confound the interpretation of immunoassay results. An ideal POC diagnostic test for MTB would (i) detect early infection/disease with high sensitivity and specificity, (ii) quickly yield results at low cost, (iii) require a single visit, (iv) cause little or no patient discomfort, (v) use specimens other than sputum, (vi) detect smear negative cases and assess drug susceptibility, (vii) detect multiple biomarkers to increase sensitivity and specificity, and (viii) be available to all persons with symptoms suggestive of TB in remote communities with poor access to a reference laboratory (Baltz et al., 2011). Furthermore, proactive diagnosis of patients at an early stage, while still smear negative, would offer additional advantages since these patients are less contagious (Behr et al., 1999) and may yield better treatment outcomes, thus reducing overall morbidity and mortality (Siddiqi et al., 2003).
Despite existing technologies and advances over the last few decades, development of a simple POC test in the near future is still challenging in the current pipeline of tuberculosis diagnostics (Pai and Pai, 2012, WHO, 2011b). In addition, novel diagnostic tools and reliable biomarkers are required to provide better performance in diagnosing TB in smear-negative patient groups (such as those with HIV and extrapulmonary infections, as well as children from whom sputum samples are difficult to collect). Although the molecular assay like GeneXpert allows the same-day clinical decision, it is limited by cost for sustainable routine diagnosis. An alternative strategy to develop inexpensive TB diagnostic tests for resource-constrained settings is to miniaturize TB diagnosis by using on-chip microfluidic technologies or by integrating novel nanotechnologies (Figure 1).
4. What nanotechnology/microfluidics holds for TB diagnostics?
With vast advances in nanotechnology and microfluidics, a variety of biosensors have been developed for detection of MTB (Table 1), foreshadowing the inevitable era of nanotechnology and microfluidics in TB diagnosis. In principle, a biosensor platform consists of an analytic device coupled with a biological sensor, which responds to physicochemical changes on the sensing area. Depending on the employed signal-producing mechanism, TB biosensors can be placed into one of the following categories: mass/piezoelectric, biochemical, electrical, and optical sensors as discussed below. These sensing platforms are based on detecting antibody-antigen interactions, whole mycobacteria or nucleic acid hybridization.
Table 1. Comparison of different biosensors for MTB detection.
| Technology | Biomarker | Limit of detection | Reference |
|---|---|---|---|
| QCM | Whole MTB bacilli | 105 CFUmL | (He and Zhang, 2002) |
| MSPQC | NH3 & CO2 absorption |
10 CFU/mL | (Ren et al., 2008a) |
| RBS Breathalyzer Fluorometry |
Ag85B antigen | 50-75 CFU/mL | (McNerney et al., 2010) |
| Interferometric Biosensor | 38-kDa antigen | NR | (Nagel et al., 2008) |
| SPR | CFP-10 antigen | 100 ng/mL | (Hong et al., 2011) |
| DMR | NR | 20 CFU/mL | (Chun, 2009, Lee et al., 2008) |
| SPCE | Ag360 & Ag231 antigens |
1 ng/ml | (Diaz-Gonzalez et al., 2005) |
| Enzymatic Immonosensor |
Antibody to mycolic acid |
NR | (Thanyani et al., 2008) |
| Electroosmosis mediated Microtip |
Whole MTB bacilli | 8 × 103 CFU/mL | (Yeo et al., 2009) |
| Acoustic Sensor | Whole MTB bacilli | 2 × 103 CFU/mL | (Stocher et al., 2003) |
Abbreviations: QCM, quartz crystal microbalance; MSPQC, multi-channel series piezoelectric quartz crystal; RBS, Rapid Biosensor Systems; SPR, surface plasmon resonance; DMR, diagnostic magnetic resonance; SPCE, screen-printed carbon electrode; MTB, Mycobacterium tuberculosis; CFU, colony forming unit; NR, Not reported.
4.1 Biosensors based on detecting antigen/antibody/whole mycobacteria
4.1.1 Mass/piezoelectric detection technologies
Mass/piezoelectric sensors utilize quartz crystals that have high sensitivity to changes in mass and surface characteristics. These sensors can detect molecular interaction between target and ligand, and monitor biochemical reactions occurring on the surface of a sensing platform. Two main quartz crystal-based biosensing platforms have been utilized to detect MTB, including (i) quartz crystal microbalance (QCM) technology, and (ii) series piezoelectric crystal based sensors.
In the QCM technology, changes in gravitational load on the sensor and viscoelastic properties of the sample cause a frequency shift of a quartz crystal resonator (Höök et al., 2001, Peh et al., 2007). In an immuno-piezosensor which has been used to detect MTB, a QCM sensing surface was first coated by a styrene-butadiene-styrene copolymer as a membrane-mimicking layer to immobilize anti-MTB antibodies on the sensor (He and Zhang, 2002). During the incubation with MTB on the platform, the capture of MTB cells was monitored in real-time by observing the frequency shift as a result of the change in mass loading on the sensor. The limit-of-detection of this system was observed to be 105 CFU/mL (He and Zhang, 2002). Although this technology is rapid, simple, and label-free, the accuracy is affected by a number of factors such as density, viscosity, dielectric constant, and electrical conductivity of the sample (Ren et al., 2008b).
Another MTB detection platform based on the piezoelectric technology was constructed by using a multi-channel series piezoelectric quartz crystal (MSPQC) sensor system (Ren et al., 2008a). The MSPQS system consisted of a multiple-sample detection platform, a microprocessor system, and a data output system. The silver-coated oscillator had a key role in detection, and it was sensitive to changes in the frequency. The system was designed to detect volatile metabolic products (i.e., NH3 and CO2), which are produced as a result of MTB growth. These metabolites were then absorbed by the presence of KOH in the media and caused a frequency shift, which was recorded by a frequency counter. With a cutoff of 100 Hz change in the frequency shift, this assay had a wide linearity ranging from 102 to 107 CFU/mL and a detection limit of 10 CFU/mL. Compared to the conventional assays such as BACTEC™ MGIT™ 960 and Lowenstein–Jensen (L–J) slants, MSPQC assay is more inexpensive (less than $1,000 for the setup and $4.2 per assay) and sensitive (Ren et al., 2008a). However, this assay still requires 2-4 days to culture MTB, and needs sample pretreatment to eliminate potential contamination with other bacteria, which may be less suitable for POC testing.
4.1.2 Optical detection technologies
Optical biosensors based on Raman spectroscopy have been used to detect cultured MTB and have proven to be rapid and highly specific in differentiating MTB from other Mycobacterium strains (Buijtels et al., 2008). In this sensor, a Raman spectra module was coupled to a custom-built inverted microscope with an automated XY-stage. To excite the samples, a laser light was used, and the wavelength range was measured from 750 nm to 1000 nm. A beam of monochromatic light passed through the sample and generated spectrographic fingerprints of multiple mycobacterial strains. The fingerprints represented the molecular composition of the viable microorganism at both the species and strain level. The sensitivity of the described method for various Mycobacterium strains in culture media was observed to be 95.2%, compared to 16S rRNA identification (sequencing), which has drawbacks such as high cost, complexity, and unambiguous interpretations (Buijtels et al., 2008). Further, the spectra of heat-inactivated samples showed minimal difference from that of viable mycobacteria samples, which can allow this platform to be used outside Biosafety Level 3 laboratories with heat-inactivated samples. However, this approach is limited by several challenges for POC testing, including the cost (use of laser and a fluorescence microscope), lengthy procedure (approximately 3 hours) and requirement to establish optical fingerprints of nontuberculous mycobacteria and other bacterial pathogens for differentiation. In addition, the sensitivity for detection of MTB in sputum needs to be further evaluated.
Another optical detection technology for MTB diagnosis is RBS breathalyzer fluorometry (Rapid Biosensor Systems, Cambridge, UK) (McNerney et al., 2010). This system consists of a portable device including a disposable plastic collection tube (3.5 × 10 cm) into which the patient coughed. The collection tube was designed to collect aerosols and particles coughed out by the patient, and it was inserted into a small battery-operated instrument containing a diode laser and a photo-multiplier tube for optical sensing. In the collection tube, the cough sample is pressed down by a plunger and distributed onto the surface of a prism at the bottom. Since the prism was coated with fluorescence labeled analogue, the presence of native MTB antigen (e.g., Ag85B) displaced the analogue and led to a fluorescence signal change. The digital readout of fluorescence change was obtained within 10 minutes. In the field trial, this RBS breathalyzer fluorometry detected 23 samples collected from 31 tuberculosis patients, with a sensitivity of 74% (95% CI 55-87). Of 29 negative samples, 6 were false positive, resulting in a specificity of 79% (95% CI 60-91). Although this assay cannot replace the use of sputum microscopy in resource-constrained settings, it can certainly assist the diagnosis of patients (e.g., children) who cannot provide sputum samples. The drawback of this assay is that the specificity and sensitivity of this assay still needs to be improved. Further, the use of fluorescence-labeled antibody inherently increases the cost, which might be an issue for wide use in remote settings.
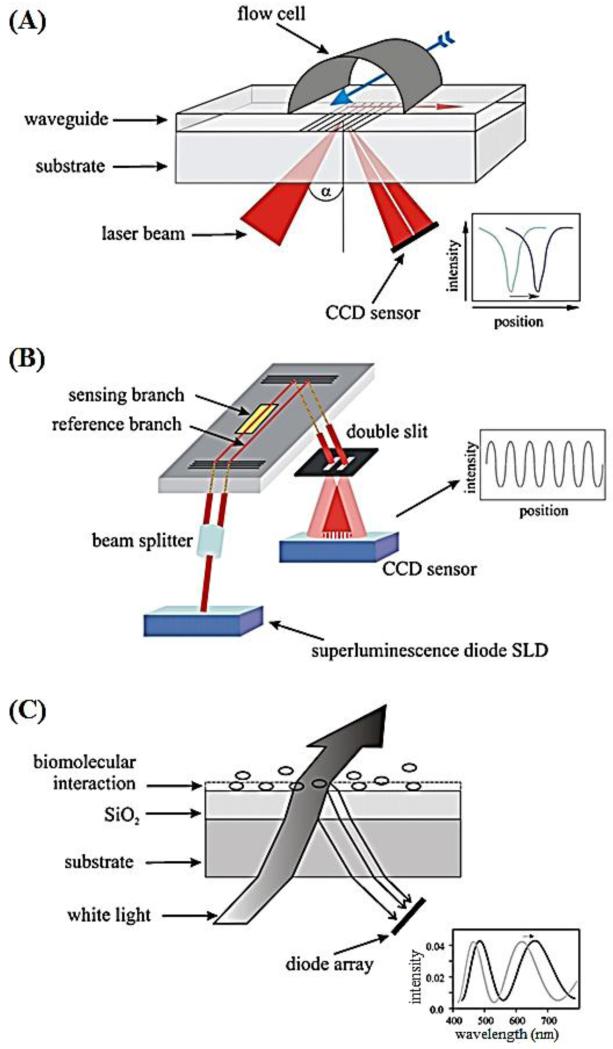
Nagel et al. developed three different label-free optical immunosensors for rapid detection of tuberculosis-specific antibodies from serum (Nagel et al., 2008). These optical immunosensors included a grating coupler, an interferometric biosensor, and a reflectometric interference spectroscopy (RIfS) (Figure 2). The grating coupler and the interferometric biosensor, both working in a refractometric mode, monitored the changes in the effective refractive index at the sensor surface (Ta2O5 and SiO2). The grating coupler biosensor determined the shift of the coupling angle, and the interferometric biosensor detected phase change of two waves travelling through a sensing branch and a reference branch within a waveguide. The RIfS based on a reflectometric approach determined the optical thickness of a thin sensor layer by detecting light reflection at interfaces. Although these three sensors differed in detection mechanisms, they utilized the same surface chemistry, in which 1,4-phenylenediisothiocyanate, a crosslinker, was used to immobilize a 38-kDa MTB lipoprotein antigen. These three different technologies were used to detect MTB-specific antibodies that might be present in serum. As reported, the grating coupler had a specificity of 100% and a sensitivity of 75% compared to ELISA. The other two sensors showed similar sensorgrams. Since these three optical sensors were label-free technologies, they required stringent specific conditions. In this study, the use of 1,4-phenylenediisothiocyanate achieved the effect of complete surface coverage to remove non-specific binding. Despite little non-specific binding, high chip-to-chip variation was observed. Adding to this challenge, the requirement of expensive couplers, lasers, and a pump system limit the utility of this system at the POC.
Figure 2.
Schematic of three optical label-free devices for MTB detection (Nagel et al., 2008).
(A) Grating coupler biosensor. A laser beam is shed onto the grating through a waveguide and the reflected light is detected by a charge-coupled device (CCD). The binding of analyte on the sensor surface changes the refractive index, leading to a shift in the coupling angle. (B) Interferometric biosensor. The light (wavelength at 675 nm) is introduced into a waveguide, passed through two branches (one sensing branch and one reference branch) and guided through a double slit. Binding of analyte on the sensor surface causes a change in the interference pattern, which is detected by a CCD sensor. (C) The reflectometric interference spectroscopy (RIfS) biosensor. This sensor is adaptable for detection of white-light interference. When bound to the interface, the analyte causes a shift in the interference pattern in the optical path, which is detected by a diode array.
Surface plasmon resonance (SPR) is an optical method based on the real-time monitoring of changes in surface refractive index caused by association or dissociation of molecules onto/from the sensor (Homola, 2008). This optical technology had also been developed to detect MTB specific antigens (e.g., CFP-10 antigen) in tissue fluid (Hong et al., 2011). In this immuno-based detection method, monoclonal anti-CFP10 antibodies were first immobilized on a commercial immunosensor chip. The chip was integrated with an SPR-based optical immunosensor system (BiaCore 3000, Sweden) and utilized the CFP10 antigen as a sensitive TB marker. The results indicated that the change in SPR angle increased linearly with CFP10 concentrations, i.e., in the range from 0.1 to 1 μg/mL (Hong et al., 2011). SPR-based immunosensors can also be adapted to sense other antigens and pathogens by altering the detection molecules (e.g., antibodies), thus offering highly versatile platforms. Although this SPR immunosensor offers the advantages of simplicity, small sample consumption, label-free, high sensitivity, specificity, and reusability, it requires a well-equipped laboratory infrastructure. Thus, portable and inexpensive SPR-based MTB biodetection systems are needed to minimize laboratory requirement for POC testing.
Alternatively, a microtip-based system has been shown to concentrate and capture MTB (Yeo et al., 2009). The system was composed of microelectrodes, a microtip, and a coil. MTB cells were concentrated to the end of a microtip due to electro-osmosis, resulting from the application of an alternating current (AC) field. The concentrated cells were aspirated into a microtip by a capillary force. Addition of fluorophore-labeled, MTB-specific, polyclonal antibodies facilitated fluorescence detection. Via this approach, MTB cells were detected within 10 minutes at a concentration as low as 8,000 CFU/mL, which is comparable to sputum smear microscopy. Although the electro-osmotic concentration approach is promising to detect culture-free TB, its application to concentrate MTB in sputum samples needs to be further evaluated. The requirement of fluorescence-labeled antibodies also restricts this system from being used in for remote settings.
4.1.3 Chip–Nuclear Magnetic Resonance (NMR) biosensor
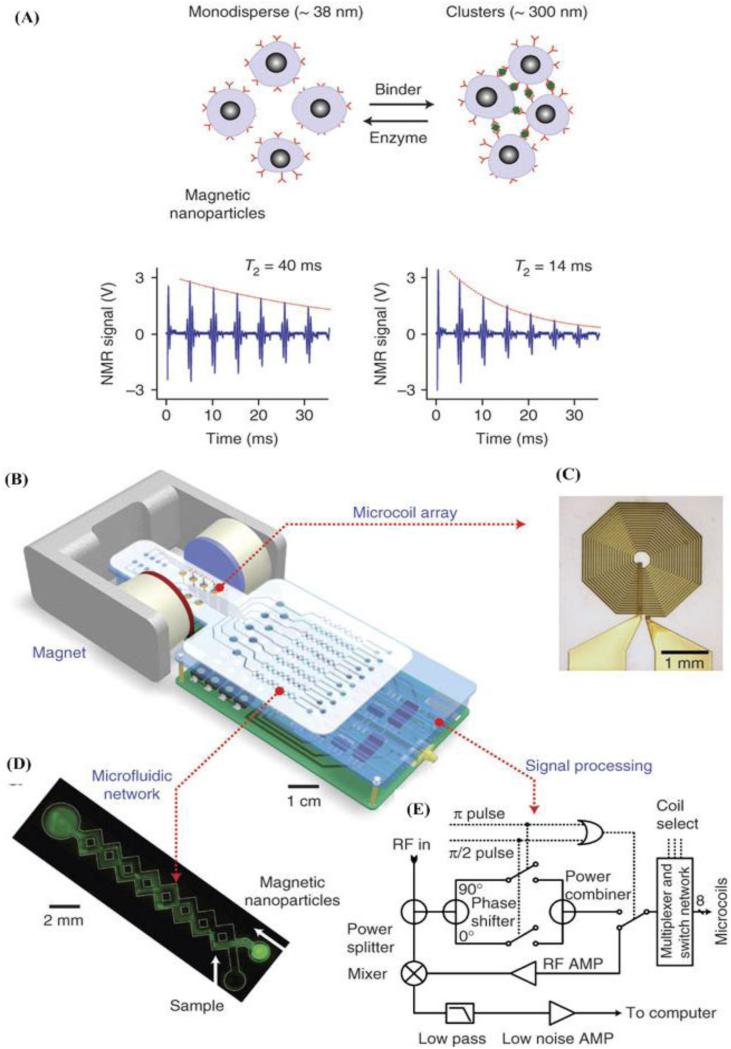
Recently, the development of a new, miniaturized diagnostic magnetic resonance (DMR) system (Figure 3), which can detect MTB as few as 20 CFU/ml in unprocessed sputum sample within 30 minutes, has been reported (Chun, 2009, Lee et al., 2008). The DMR system was essentially a proximity assay that can detect the bulk change in spin-spin relaxation time of surrounding water molecules, when magnetic nanoparticles aggregated due to the presence of target biomarkers. The system consisted of 3 modules, including a microcoil array, microfluidic networks and on-board NMR electronics. Magnetic particles were coated with antibodies specific for target biomarkers, which can be mammalian cells, bacteria, or protein. Because the assay works on the principle of NMR, turbid samples such as blood, sputum or urine can be used. Compared to the benchtop relaxometer, this system exhibited 80-fold increase in mass sensitivity in detecting avidin (Lee et al., 2008). The superior sensitivity and decreased assay time make this system suitable for POC testing, that is, this device has potential to be translated into a MTB POC assay (Chun, 2009). Other notable features include handling small sample volumes (5-10 μL) and short turnaround time, versatility and multiplexing capability. Also, the NMR component costs less than $200 and the disposable microchip costs less than $1, making the technology potentially suitable for the developing world.
Figure 3.
Design of the diagnostic magnetic resonance (DMR) system (Lee et al., 2008).
(A) Principle of the magnetic particle based proximity assay. Due to the presence of targets, functionalized magnetic nanoparticles aggregate and result in a decrease in spin-spin relaxation time. (B) Schematic of the DMR system. This system includes microcoils for NMR measurements, microfluidic networks for sample preparation, NMR electronics and a magnetic field generated by a permanent magnet. (C) Picture of the microcoil. (D) Image of the microfluidic network. (E) Schematic of the nuclear magnetic resonance circuit.
4.1.4 Enzymatic immunosensor
The enzymatic immunosensors represent the most common approach for TB diagnostics to date. Diaz-Gonzalez et al. used a sandwich ELISA device to detect the immuno-complex, which was captured by a streptavidin modified screen-printed carbon electrode (SPCE) (Diaz-Gonzalez et al., 2005). In this study, two antigens (Ag360 and Ag231) of MTB were used in combination with their monoclonal antibodies to form a respective immuno-complex. The immuno-complex was captured by biotinylated anti-MTB antibodies immobilized on the sensor surface via streptavidin-biotin interaction. Then, the immuno-complex was detected using a generic detection antibody that was conjugated to an alkaline phosphatase (AP). The enzyme digested an electrochemical substrate 3-indoxyl phosphate (3-IP), to achieve voltammetric detection. The limit of detection of this immunosensor was shown to be 1.0 ng/mL for the pair of Ag231 and its monoclonal antibody. This approach leverages well-established screen-printing microfabrication, which allows for mass production of inexpensive electrodes (Diaz-Gonzalez et al., 2005). In addition, the miniaturized detection is integrated into a disposable portable device, which is promising for detection of TB at the POC. However, the assay time is more than 4 hours. Thus, this assay could benefit from shorting the turnaround time for rapid results at the POC.
Recently, a new enzymatic TB sensor that utilized a natural BlaC enzyme from tubercular bacilli was developed (Xie et al., 2012). BlaC, an enzyme from class A β-lactamase family, hydrolyzes all classes of β-lactam substrates, including cephalosporins. Owing to the unique flexibility of BlaC, chemically modified BlaC-specific fluorogenic substrates (i.e., cephalosporins) were used as fluorescence probes, which differentiated MTB from other bacteria such as Pseudomonas, Staphylococcus and environmental Mycobacterium like M. smegmatis. In addition, the use of these fluorescence probes also improved the sensitivity of detecting BlaC from MTB other than homologue TEM-1 BlaC and β-lactamases from other bacteria. As reported, the modified probes enhanced the fluorescence intensity by 100-200 folds, and improved the selectivity by 1,000 folds, compared to TEM-1 β-lactamase. As low as 100 bacilli spiked in unprocessed patient sputum were detected in less than ten minutes. To eliminate the need for fluorescence detection, an LED-based, inexpensive, and portable imaging setup was constructed. Clearly, this system offers the opportunity for rapid MTB detection in resource-constrained setting. However, this system needs to be further evaluated in the field using unprocessed sputum samples compared to culture, smear microscopy or PCR.
4.1.5 Resonant mirror immunosensor
An affinity based immunosensor detecting antibodies against mycolic acids (MA) of MTB in TB patient samples has been described (Thanyani et al., 2008). This resonant mirror biosensor, IAsys (Affinity Sensors, Cambridge, UK), measured the binding and disassociation of antibody-antigen on the sensor. Briefly, the surface of a twin-celled biosensor cuvette was activated with cetyl-pyridinium chloride (CPC). Liposomes containing MA were then immobilized on the surface followed by blocking with saponin. A highly diluted (1:1000) serum sample was used to calibrate the binding signal of the two cuvettes prior to the addition of patient serum at lower dilution (1:500). The patient serum was pre-incubated with MA-containing liposomes or liposomes only before addition to the cuvette. Thus, anti-MA antibodies in TB patient serum were inhibited from binding to the biosensor surface, which resulted in a decreased signal. Via the inhibition of antibody binding, this biosensor showed a sensitivity of 86.7% and a specificity of 48.4% in patients co-infected with TB and HIV, i.e., TB+HIV+ patients. Upon exclusion of HIV+ individuals, the specificity increased to 76.9%. Owing to the capability to detect real-time antibody binding and disassociation including low- and high-affinity antibodies, this biosensor may improve the detection limit compared to traditional ELISA, in which low affinity binding antibody may be washed away. Although the assay appears to be technologically simple and quick for POC testing, this biosensor could benefit from reduced cost and improved consistency between cuvettes.
4.1.6 Acoustic sensor
An acoustic wave-based impedance biosensor was developed to rapidly detect the growth of MTB in culture (He et al., 2003). MTB takes 1-3 weeks to reach confluency depending on the type of media used. However, by using this bulk acoustic wave-based impedance sensor, the detection time was shortened to a day. The device worked on the principle of conductivity changes in the culture media resulting from MTB growth. The growth of MTB yielded production of protein, fatty acids and nucleic acids, as well as metabolic by-products. This caused the reference signal’s strength to decrease and frequency to increase, which was detected by the bulk acoustic wave impedance sensor. Further, the sensor quantified the initial concentration of MTB, with initial calibration, by observing the time at which the detected frequency changed. As demonstrated, the sensor can inexpensively detect and quantify MTB concentrations ranging from 2×103 to 3×107 cells/mL in significantly reduced assay time. In addition, it has been shown that the typical response curve associated with MTB is different from that of other bacteria, such as Escherichia coli, Staphylococcus aureus and Proteus mirabilis. The disadvantage is that this sensor still requires a laborious culture procedure, which is not suitable for POC testing. The utility of this assay needs to be evaluated through additional clinical testing with patient samples.
4.2 Biosensors based on nucleic acid hybridization
In addition to the detection of immune response generated against MTB, nanotechnology have also been utilized to facilitate detection of MTB-specific nucleic acid. For instance, gold nanoparticle-based probe assays have been developed to detect MTB PCR amplification products by analyzing patterns of color change as a result of nanoparticle aggregation (Costa et al., 2010, Soo et al., 2009). Although these methods simplified the detection of MTB PCR products, they still required amplification of MTB nucleic acid by PCR, which remains challenging at the POC. To overcome this challenge, a PCR-free electrochemical biosensor was developed for detection of MTB genomic DNA based on the dual labeling of gold nanoparticles with alkaline phosphatase and specific DNA oligonucleotides (Thiruppathiraja et al., 2011). Briefly, MTB genomic DNA was first extracted and broken into small fragments using ultrasound sonication. The generated MTB DNA fragments were hybridized onto electrodes having immobilized with specific capture probes. Addition of dual-labeled gold nanoparticles allows generation of an electroactive species of para-nitrophenol, which serves as a substrate in the following electrical sensing. As demonstrated, this method detected MTB DNA down to 1.25 ng/ml. Also, this method showed comparable sensitivity and specificity to PCR in detecting MTB from clinical sputum samples. However, the requirements for stringent hybridization conditions (including multiple wash and temperature controls) and long incubation time make it less ideal for POC testing.
SPR has also been used to detect MTB genomic DNA via hybridization with cysteine modified NH2-end peptide nucleic acid (PNA, 24-mer) probe and 5′-thiol end labelled DNA probes (Prabhakar et al., 2008). For nucleic acid hybridization, the DNA probes were designed to detect the sequence of MTB either with or without mutations. In this study, the change in SPR angle was monitored during the hybridization of DNA samples with PNA and DNA immobilized on gold (Au) electrodes. The results showed that there was no non-specific binding of non-complementary sequences to the DNA/Au and PNA/Au electrodes. Further, the PNA/Au electrodes were more efficient for detection of sequences with single-base mismatches, possessing a lower limit of detection (1 ng/mL) than DNA-Au electrode (3 ng/mL) (Prabhakar et al., 2008). Although the SPR technology shows sensitive detection of MTB sequences, further efforts can contribute to miniaturize the detection system to a portable device, which can be implemented at the POC.
4.3 Cell phone based fluorescence microscopy
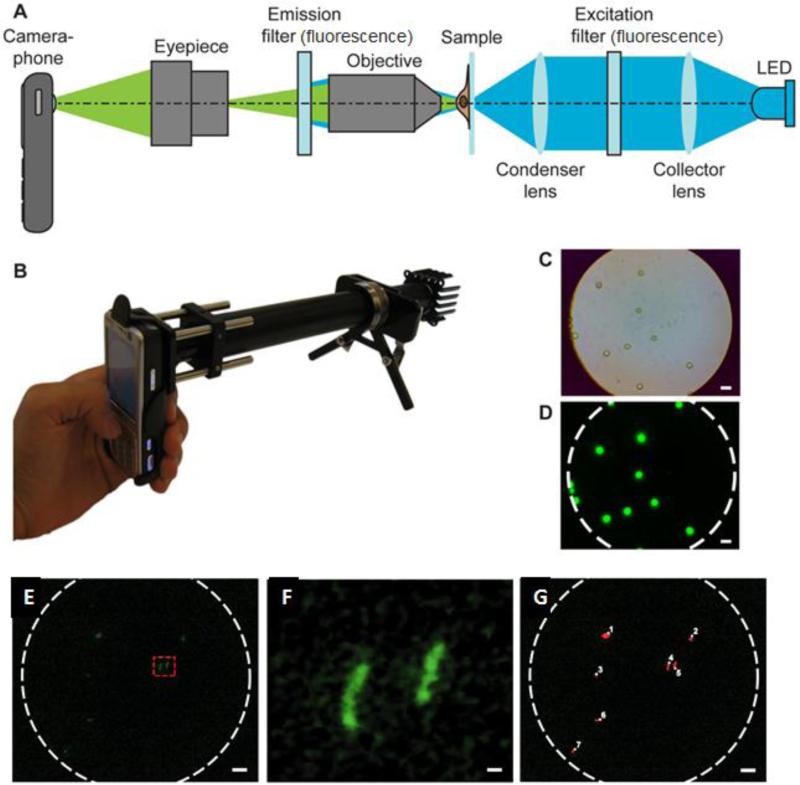
Owing to the challenges associated with smear microscopy (e.g., the need for a bulky light microscopy and microbiology skills for pathogen detection), fluorescence detection of MTB has been developed to demonstrate the feasibility of an integrated and portable mobile phone microscopy system (Breslauer et al., 2009). The platform had a dual microscope mode, which can monitor samples under brightfield and fluorescence. To take an image, a cell phone having 3.2 Megapixels camera was used. The phone was mounted on an optical rail platform (Figure 4). For fluorescence imaging, the system was equipped with an inexpensive LED excitation source, which emits within the excitation range of fluorescent Auramine O-stain commonly used for detection of TB bacilli in sputum smears. To image the sample, the light generated from the LED source first passed through the collector lens, and then the excitation filter with a wavelength range to view TB bacilli. The images are then transferred to a computer for further analysis using the Image J program (http://rsb.info.nih.gov/ij/). Compared to the non-fluorescent Ziehl-Neelsen stain, this method uses a lower power (20X) objective with a larger field-of-view, thus reducing the number of field images to cover the entire screening area. Further, the use of a 20X 0.4 NA objective allows sufficient light-gathering for fluorescence detection without using a traditional fluorescence microscope. However, this system requires expensive filters and lenses, and the sample images need to be transported to a computer for further analysis.
Figure 4.
Mobile phone-integrated microscopy (Breslauer et al., 2009).
(A) Schematic of mobile phone-integrated microscopy for fluorescence and brightfield imaging. (B) The presented prototype with filters and LEDs for fluorescence imaging. (C) Brightfield image of 6 mm diameter fluorescent beads. (D) Fluorescent images of beads presented in the C. (E) Fluorescence image of Auramine O-stained TB sputum sample. (F) Magnified view of two TB bacilli from the selected area in E. (G) Automated counting of Auramine O-stained TB bacilli in E. Scale bars in (C), (D), (E), and (G) are 10 mm, scale bar in (F) is 1 mm.
4.4 Other microfluidic approaches for the POC
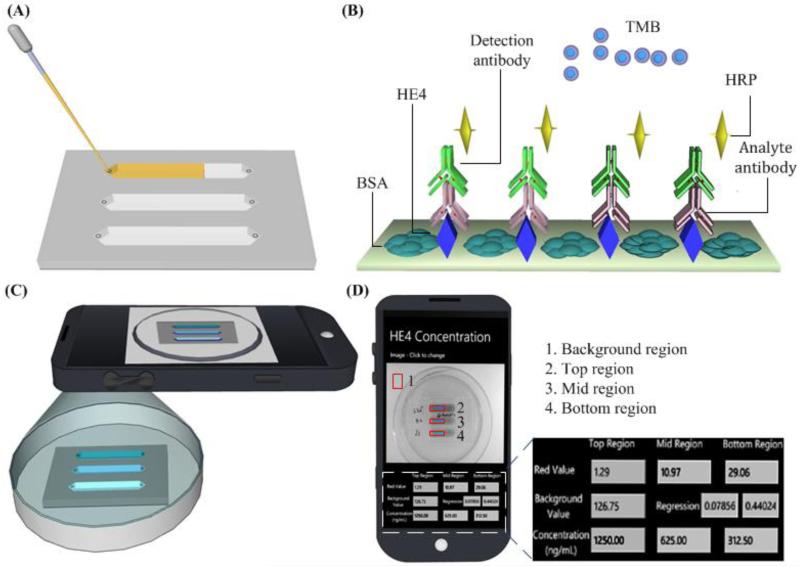
Recently, microfluidic approaches have been used to develop POC diagnostics and monitoring technologies, (Alyassin et al., 2009, Chin et al., 2011, Christodoulides et al., 2005, Dimov et al., 2011, Gurkan et al., 2011, Kim et al., 2009, Moon et al., 2011, Moon et al., 2009, Wang et al., 2012a, Wang et al., 2012b, Wang et al., 2010, Wang et al., 2011). These technology platforms can potentially be employed for identification and tracking of TB in the field. One example is a self-powered integrated microfluidic blood analysis system (SIMBAS), which can achieve sample-to-answer within 10 minutes (Dimov et al., 2011). This device incorporated a filter trench to separate plasma from whole blood, eliminating the need for any external equipment for sample processing. Further, detection of biotin–streptavidin in 5 parallel channels has been demonstrated, which allows the system to simultaneously detect 5 different biomarkers, enabling a broad range of applications. Another example is on-chip ELISA coupled with cell phone detection (Wang et al., 2011). In this study, a generic ELISA approach was developed on a microchip for the detection of HE4, an ovarian cancer biomarker from clinical urine samples (Figure 5). On the microchannel surface, the HE4 analyte was first absorbed. Then, primary antibody and detection antibody were sequentially added. The presence of HE4 was finally indicated by blue colour as a result of the hydrolysis of 3,3′,5,5′-Tetramethylbenzidine (TMB) by horseradish peroxidase (HRP) conjugated detection antibody. Similar portable, microchip-based approaches can be potentially developed to detect biomarkers specific to MTB. In addition, development of cell phone based colorimetric detection with an integrated mobile application eliminates the need for specialized expensive detection equipment for obtaining and displaying results. Given the widespread use of cell phones across the developing world, this detection platform holds promise to enable POC testing for global health applications.
Figure 5.
Schematic of microchip-ELISA couple with cell phone detection (Wang et al., 2011).
(A) Loading clinical urine samples onto a microchip. (B) On-chip sandwich ELISA is performed by flowing samples, reagents and buffers. (C) The result is imaged using a smart cell phone. (D) The concentration of analyte is calculated and reported on the screen using an integrated mobile application.
5. Future perspective
It has been reported that 40-50% of TB cases remain undetected due to the absence of TB bacilli in sputum (Dheda et al., 2009). As such, new tests based on the detection of whole TB bacilli in the sputum will face the significant challenge. This line of clinical evidence strongly suggests that the detection of either bacterial byproducts or changes in host immune response might be an effective diagnostic approach at POC. In addition, diagnostic tests based on monitoring MTB-derived byproducts and MTB- specific host immune response has the capacity to follow disease progression, thus enhancing clinical utility. Several attempts have been made on these lines to improve current diagnostic tools, but none of these attempts have demonstrated clear clinical utility. Additionally, new biomarkers are needed for specific and sensitive detection of MTB (McNerney and Daley, 2011).
Nanotechnology is a fast-advancing field, which attracts multidisciplinary teams to target various medical challenges in infectious diseases, cancer, and cardiovascular diseases. In miniaturizing devices, nanotechnology holds the promise to change the landscape of TB diagnosis and treatment, specifically to reach the sensitivity and specificity required clinically, to lower the cost, and to enable portable microfluidic platforms suitable for resource-constrained settings. If successfully adapted, nanotechnology holds the key to the development of a long-awaited POC test for TB. Moreover, the devices based on nanotechnology holds the promise to develop a platform that can detect simultaneously TB as well as HIV viral loads within the same sample in clinical settings. This could have great implication due to the increase in TB and HIV co-infection across the globe. On the other hand, it should be noted that nanotechnology is still at a stage of translating the proof-of-concept to clinical usage. Future successes in developing reliable biosensors for POC TB diagnosis could benefit from close collaborations between biomarker developers and sensing technologists, and also from the active involvement of policy makers and funding agencies. This has been recently highlighted in the revised Global Plan to Stop TB 2011-2015, which calls for an investment of ~ $1.7 billion for improving TB diagnostics (Stop TB Partnership, 2010). With these joint efforts and fast advancement in nanotechnology/microfluidics, diagnosis and treatment of TB at the POC remains promising for the future. As optimistically predicted, TB POC tests, which can be performed at the peripheral level, might be available in 2-3 years (Pai and Pai, 2012).
Acknowledgements
We would like to acknowledge the W.H. Coulter Foundation Young Investigator Award, RO1A1081534, RO1AI093282, U54EB15408 and R21AI087107. This work was supported by (i) the Center for Integration of Medicine and Innovative Technology (CIMIT) under U.S. Army Medical Research Acquisition Activity Cooperative Agreements DAMD17-02-2-0006, W81XWH-07-2-0011, and W81XWH-09-2-0001, (ii) A*STAR, Science and Engineering Institutes (SCEI), BMRC– SERC diagnostic grant No. 1121480006, (iii) A*STAR-CIMIT grant No. 1187. Further the work was made possible by a research grant that was awarded and administered by the U.S. Army Medical Research & Materiel Command (USAMRMC) and the Telemedicine & Advanced Technology Research Center (TATRC), at Fort Detrick, MD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alyassin MA, Moon S, Keles HO, Manzur F, Lin RL, Haeggstrom E, et al. Rapid automated cell quantification on HIV microfluidic devices. Lab Chip. 2009;9:3364–9. doi: 10.1039/b911882a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen P, Munk ME, Pollock JM, Doherty TM. Specific immune-based diagnosis of tuberculosis. Lancet. 2000;356:1099–104. doi: 10.1016/s0140-6736(00)02742-2. [DOI] [PubMed] [Google Scholar]
- Aryan E, Makvandi M, Farajzadeh A, Huygen K, Bifani P, Mousavi SL, et al. A novel and more sensitive loop-mediated isothermal amplification assay targeting IS6110 for detection of Mycobacterium tuberculosis complex. Microbiological Research. 2010;165:211–20. doi: 10.1016/j.micres.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Barry CE, 3rd, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nature reviews Microbiology. 2009;7:845–55. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, et al. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–3. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- Boehme CC, Nabeta P, Henostroza G, Raqib R, Rahim Z, Gerhardt M, et al. Operational feasibility of using loop-mediated isothermal amplification for diagnosis of pulmonary tuberculosis in microscopy centers of developing countries. Journal of Clinical Microbiology. 2007;45:1936–40. doi: 10.1128/JCM.02352-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme CC, Nabeta P, Hillemann D, Nicol MP, Shenai S, Krapp F, et al. Rapid molecular detection of tuberculosis and rifampin resistance. The New England journal of medicine. 2010;363:1005–15. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme CC, Nicol MP, Nabeta P, Michael JS, Gotuzzo E, Tahirli R, et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet. 2011;377:1495–505. doi: 10.1016/S0140-6736(11)60438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslauer DN, Maamari RN, Switz NA, Lam WA, Fletcher DA. Mobile Phone Based Clinical Microscopy for Global Health Applications. PLoS ONE. 2009:4. doi: 10.1371/journal.pone.0006320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijtels PC, Willemse-Erix HF, Petit PL, Endtz HP, Puppels GJ, Verbrugh HA, et al. Rapid identification of mycobacteria by Raman spectroscopy. J Clin Microbiol. 2008;46:961–5. doi: 10.1128/JCM.01763-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattamanchi A, Smith R, Steingart KR, Metcalfe JZ, Date A, Coleman C, et al. Interferon-gamma release assays for the diagnosis of latent tuberculosis infection in HIV-infected individuals: a systematic review and meta-analysis. J Acquir Immune Defic Syndr. 2011;56:230–8. doi: 10.1097/QAI.0b013e31820b07ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang R, Wang J, Liu L, Zheng Y, Shen Y, et al. Interferon-gamma release assays for the diagnosis of active tuberculosis in HIV-infected patients: a systematic review and meta-analysis. Plos One. 2011;6:e26827. doi: 10.1371/journal.pone.0026827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CD, Laksanasopin T, Cheung YK, Steinmiller D, Linder V, Parsa H, et al. Microfluidics-based diagnostics of infectious diseases in the developing world. Nat Med. 2011;17:1015–9. doi: 10.1038/nm.2408. [DOI] [PubMed] [Google Scholar]
- Christodoulides N, Mohanty S, Miller CS, Langub MC, Floriano PN, Dharshan P, et al. Application of microchip assay system for the measurement of C-reactive protein in human saliva. Lab Chip. 2005;5:261–9. doi: 10.1039/b414194f. [DOI] [PubMed] [Google Scholar]
- Chun AL. Nanoparticles offer hope for TB detection. Nature nanotechnology. 2009;4:698–9. doi: 10.1038/nnano.2009.322. [DOI] [PubMed] [Google Scholar]
- Cobelens FG, Egwaga SM, van Ginkel T, Muwinge H, Matee MI, Borgdorff MW. Tuberculin skin testing in patients with HIV infection: Limited benefit of reduced cutoff values. Clin Infect Dis. 2006;43:634–9. doi: 10.1086/506432. [DOI] [PubMed] [Google Scholar]
- Costa P, Amaro A, Botelho A, Inacio J, Baptista PV. Gold nanoprobe assay for the identification of mycobacteria of the Mycobacterium tuberculosis complex. Clin Microbiol Infect. 2010;16:1464–9. doi: 10.1111/j.1469-0691.2009.03120.x. [DOI] [PubMed] [Google Scholar]
- Davies PD, Pai M. The diagnosis and misdiagnosis of tuberculosis. Int J Tuberc Lung Dis. 2008;12:1226–34. [PubMed] [Google Scholar]
- Denkinger CM, Pai M. Point-of-care tuberculosis diagnosis: are we there yet? The Lancet infectious diseases. 2012;12:169–70. doi: 10.1016/S1473-3099(11)70257-2. [DOI] [PubMed] [Google Scholar]
- Dheda K, van Zyl Smit R, Badri M, Pai M. T-cell interferon-gamma release assays for the rapid immunodiagnosis of tuberculosis: clinical utility in high-burden vs. low-burden settings. Curr Opin Pulm Med. 2009;15:188–200. doi: 10.1097/MCP.0b013e32832a0adc. [DOI] [PubMed] [Google Scholar]
- Diaz-Gonzalez M, Gonzalez-Garcia MB, Costa-Garcia A. Immunosensor for Mycobacterium tuberculosis on screen-printed carbon electrodes. Biosensors & Bioelectronics. 2005;20:2035–43. doi: 10.1016/j.bios.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Dimov IK, Basabe-Desmonts L, Garcia-Cordero JL, Ross BM, Park Y, Ricco AJ, et al. Stand-alone self-powered integrated microfluidic blood analysis system (SIMBAS) Lab Chip. 2011;11:845–50. doi: 10.1039/c0lc00403k. [DOI] [PubMed] [Google Scholar]
- Dowdy DW, Steingart KR, Pai M. Serological testing versus other strategies for diagnosis of active tuberculosis in India: a cost-effectiveness analysis. PLoS Med. 2011;8:e1001074. doi: 10.1371/journal.pmed.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhat M, Greenaway C, Pai M, Menzies D. False-positive tuberculin skin tests: what is the absolute effect of BCG and non-tuberculous mycobacteria? The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 2006;10:1192–204. [PubMed] [Google Scholar]
- Geojith G, Dhanasekaran S, Chandran SP, Kenneth J. Efficacy of loop mediated isothermal amplification (LAMP) assay for the laboratory identification of Mycobacterium tuberculosis isolates in a resource limited setting. Journal of Microbiological Methods. 2011;84:71–3. doi: 10.1016/j.mimet.2010.10.015. [DOI] [PubMed] [Google Scholar]
- Gurkan UA, Moon S, Geckil H, Xu F, Wang S, Lu TJ, et al. Miniaturized lensless imaging systems for cell and microorganism visualization in point-of-care testing. Biotechnol J. 2011;6:138–49. doi: 10.1002/biot.201000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Zhang L. Rapid diagnosis of M. tuberculosis using a piezoelectric immunosensor. Anal Sci. 2002;18:397–401. doi: 10.2116/analsci.18.397. [DOI] [PubMed] [Google Scholar]
- He F, Zhao J, Zhang L, Su X. A rapid method for determining Mycobacterium tuberculosis based on a bulk acoustic wave impedance biosensor. Talanta. 2003;59:935–41. doi: 10.1016/S0039-9140(02)00643-4. [DOI] [PubMed] [Google Scholar]
- Hobby GL, Holman AP, Iseman MD, Jones JM. Enumeration of tubercle bacilli in sputum of patients with pulmonary tuberculosis. Antimicrobial agents and chemotherapy. 1973;4:94–104. doi: 10.1128/aac.4.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homola J. Surface plasmon resonance sensors for detection of chemical and biological species. Chemical Reviews. 2008;108:462–93. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- Hong SC, Chen HX, Lee J, Park HK, Kim YS, Shin HC, et al. Ultrasensitive immunosensing of tuberculosis CFP-10 based on SPR spectroscopy. Sens Actuator B-Chem. 2011;156:271–5. [Google Scholar]
- Höök F, Kasemo B, Nylander T, Fant C, Sott K, Elwing H. Variations in Coupled Water, Viscoelastic Properties, and Film Thickness of a Mefp-1 Protein Film during Adsorption and Cross-Linking: A Quartz Crystal Microbalance with Dissipation Monitoring, Ellipsometry, and Surface Plasmon Resonance Study. Analytical Chemistry. 2001;73:5796–804. doi: 10.1021/ac0106501. [DOI] [PubMed] [Google Scholar]
- Iwamoto T, Sonobe T, Hayashi K. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol. 2003;41:2616–22. doi: 10.1128/JCM.41.6.2616-2622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Moon S, Kuritzkes DR, Demirci U. Quantum dot-based HIV capture and imaging in a microfluidic channel. Biosens Bioelectron. 2009;25:253–8. doi: 10.1016/j.bios.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawn SD, Kerkhoff AD, Vogt M, Wood R. Diagnostic accuracy of a low-cost, urine antigen, point-of-care screening assay for HIV-associated pulmonary tuberculosis before antiretroviral therapy: a descriptive study. The Lancet infectious diseases. 2012;12:201–9. doi: 10.1016/S1473-3099(11)70251-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14:869–74. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling DI, Pai M, Davids V, Brunet L, Lenders L, Meldau R, et al. Are interferon-gamma release assays useful for diagnosing active tuberculosis in a high-burden setting? Eur Respir J. 2011;38:649–56. doi: 10.1183/09031936.00181610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek GH, Jereb J, Lobue P, Iademarco MF, Metchock B, Vernon A. Guidelines for using the QuantiFERON-TB Gold test for detecting Mycobacterium tuberculosis infection, United States. MMWR Recomm Rep. 2005;54:49–55. [PubMed] [Google Scholar]
- McNerney R, Daley P. Towards a point-of-care test for active tuberculosis: obstacles and opportunities. Nat Rev Microbiol. 2011;9:204–13. doi: 10.1038/nrmicro2521. [DOI] [PubMed] [Google Scholar]
- McNerney R, Wondafrash BA, Amena K, Tesfaye A, McCash EM, Murray NJ. Field test of a novel detection device for Mycobacterium tuberculosis antigen in cough. BMC Infect Dis. 2010;10:161. doi: 10.1186/1471-2334-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe JZ, Everett CK, Steingart KR, Cattamanchi A, Huang L, Hopewell PC, et al. Interferon-gamma release assays for active pulmonary tuberculosis diagnosis in adults in low- and middle-income countries: systematic review and meta-analysis. The Journal of infectious diseases. 2011;204(Suppl 4):S1120–9. doi: 10.1093/infdis/jir410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S, Gurkan UA, Blander J, Fawzi WW, Aboud S, Mugusi F, et al. Enumeration of CD4+ T-cells using a portable microchip count platform in Tanzanian HIV-infected patients. PLoS One. 2011;6:e21409. doi: 10.1371/journal.pone.0021409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S, Keles HO, Ozcan A, Khademhosseini A, Haeggstrom E, Kuritzkes D, et al. Integrating microfluidics and lensless imaging for point-of-care testing. Biosens Bioelectron. 2009;24:3208–14. doi: 10.1016/j.bios.2009.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel T, Ehrentreich-Forster E, Singh M, Schmitt K, Brandenburg A, Berka A, et al. Direct detection of tuberculosis infection in blood serum using three optical label-free approaches. Sens Actuator B-Chem. 2008;129:934–40. [Google Scholar]
- Niemz A, Ferguson TM, Boyle DS. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011;29:240–50. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research. 2000:28. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla E, Gonzalez V, Manterola JM, Perez A, Quesada MD, Gordillo S, et al. Comparative evaluation of the new version of the INNO-LiPA Mycobacteria and genotype Mycobacterium assays for identification of Mycobacterium species from MB/BacT liquid cultures artificially inoculated with Mycobacterial strains. J Clin Microbiol. 2004;42:3083–8. doi: 10.1128/JCM.42.7.3083-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai NP, Pai M. Point-of-care diagnostics for HIV and tuberculosis: landscape, pipeline, and unmet needs. Discovery medicine. 2012;13:35–45. [PubMed] [Google Scholar]
- Pandey BD, Poudel A, Yoda T, Tamaru A, Oda N, Fukushima Y, et al. Development of an in-house loop-mediated isothermal amplification (LAMP) assay for detection of Mycobacterium tuberculosis and evaluation in sputum samples of Nepalese patients. Journal of Medical Microbiology. 2008;57:439–43. doi: 10.1099/jmm.0.47499-0. [DOI] [PubMed] [Google Scholar]
- Parsons LM, Somoskovi A, Gutierrez C, Lee E, Paramasivan CN, Abimiku A, et al. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin Microbiol Rev. 2011;24:314–50. doi: 10.1128/CMR.00059-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peh WYX, Reimhult E, Teh HF, Thomsen JS, Su X. Understanding Ligand Binding Effects on the Conformation of Estrogen Receptor α-DNA Complexes: A Combinational Quartz Crystal Microbalance with Dissipation and Surface Plasmon Resonance Study. Biophysical Journal. 2007;92:4415–23. doi: 10.1529/biophysj.106.099382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter JG, Theron G, van Zyl-Smit R, Haripersad A, Mottay L, Kraus S, et al. Diagnostic accuracy of a urine lipoarabinomannan strip-test for TB detection in HIV-infected hospitalised patients. The European respiratory journal : official journal of the European Society for Clinical Respiratory Physiology. 2012;40:1211–20. doi: 10.1183/09031936.00201711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek AS, Tyagi S, Pol AC, Telenti A, Miller LP, Kramer FR, et al. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nature biotechnology. 1998;16:359–63. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- Pinto LM, Grenier J, Schumacher SG, Denkinger CM, Steingart KR, Pai M. Immunodiagnosis of tuberculosis: state of the art. Med Princ Pract. 2012;21:4–13. doi: 10.1159/000331583. [DOI] [PubMed] [Google Scholar]
- Prabhakar N, Arora K, Arya SK, Solanki PR, Iwamoto M, Singh H, et al. Nucleic acid sensor for M. tuberculosis detection based on surface plasmon resonance. The Analyst. 2008;133:1587–92. doi: 10.1039/b808225a. [DOI] [PubMed] [Google Scholar]
- Ren J, He F, Yi S, Cui X. A new MSPQC for rapid growth and detection of Mycobacterium tuberculosis. Biosens Bioelectron. 2008a;24:403–9. doi: 10.1016/j.bios.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Ren J, He F, Yi S, Cui X. A new MSPQC for rapid growth and detection of Mycobacterium tuberculosis. Biosensors & Bioelectronics. 2008b;24:403–9. doi: 10.1016/j.bios.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Sarkola A, Makinen J, Marjamaki M, Marttila HJ, Viljanen MK, Soini H. Prospective evaluation of the GenoType assay for routine identification of mycobacteria. European journal of clinical microbiology & infectious diseases : official publication of the European Society of Clinical Microbiology. 2004;23:642–5. doi: 10.1007/s10096-004-1168-7. [DOI] [PubMed] [Google Scholar]
- Siddiqi K, Lambert ML, Walley J. Clinical diagnosis of smear-negative pulmonary tuberculosis in low-income countries: the current evidence. Lancet Infect Dis. 2003;3:288–96. doi: 10.1016/s1473-3099(03)00609-1. [DOI] [PubMed] [Google Scholar]
- Soo PC, Horng YT, Chang KC, Wang JY, Hsueh PR, Chuang CY, et al. A simple gold nanoparticle probes assay for identification of Mycobacterium tuberculosis and Mycobacterium tuberculosis complex from clinical specimens. Mol Cell Probes. 2009;23:240–6. doi: 10.1016/j.mcp.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Steingart KR, Flores LL, Dendukuri N, Schiller I, Laal S, Ramsay A, et al. Commercial Serological Tests for the Diagnosis of Active Pulmonary and Extrapulmonary Tuberculosis: An Updated Systematic Review and Meta-Analysis. Plos Medicine. 2011a:8. doi: 10.1371/journal.pmed.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingart KR, Flores LL, Dendukuri N, Schiller I, Laal S, Ramsay A, et al. Commercial serological tests for the diagnosis of active pulmonary and extrapulmonary tuberculosis: an updated systematic review and meta-analysis. PLoS Med. 2011b;8:e1001062. doi: 10.1371/journal.pmed.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocher M, Leb V, Berg J. A convenient approach to the generation of multiple internal control DNA for a panel of real-time PCR assays. J Virol Methods. 2003;108:1–8. doi: 10.1016/s0166-0934(02)00266-5. [DOI] [PubMed] [Google Scholar]
- Stop TB Partnership WHO [accessed on October 23rd, 2012];The Global Plan to Stop TB 2011-2015: Transforming the fight towards elimination of tuberculosis. 2010. p. http://www.stoptb.org/global/plan/ [Google Scholar]
- Thanyani ST, Roberts V, Siko DGR, Vrey P, Verschoor JA. A novel application of affinity biosensor technology to detect antibodies to mycolic acid in tuberculosis patients. Journal of Immunological Methods. 2008;332:61–72. doi: 10.1016/j.jim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Thiruppathiraja C, Kamatchiammal S, Adaikkappan P, Santhosh DJ, Alagar M. Specific detection of Mycobacterium sp genomic DNA using dual labeled gold nanoparticle based electrochemical biosensor. Analytical Biochemistry. 2011;417:73–9. doi: 10.1016/j.ab.2011.05.034. [DOI] [PubMed] [Google Scholar]
- Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nature protocols. 2008;3:877–82. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Tostmann A, Kik SV, Kalisvaart NA, Sebek MM, Verver S, Boeree MJ, et al. Tuberculosis transmission by patients with smear-negative pulmonary tuberculosis in a large cohort in the Netherlands. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2008;47:1135–42. doi: 10.1086/591974. [DOI] [PubMed] [Google Scholar]
- Van Rie A, Page-Shipp L, Scott L, Sanne I, Stevens W. Xpert((R)) MTB/RIF for point-of-care diagnosis of TB in high-HIV burden, resource-limited countries: hype or hope? Expert Rev Mol Diagn. 2010;10:937–46. doi: 10.1586/erm.10.67. [DOI] [PubMed] [Google Scholar]
- Wang S, Esfahani M, Gurkan UA, Inci F, Kuritzkes D, Demirci U. Efficient on-chip isolation of HIV subtypes Lab Chip. 2012a doi: 10.1039/c2lc20706k. DOI: 10.1039/c2lc20706k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Inci F, Chaunzwa T, Ramanujam A, Vasudevan A, Subramanian S, et al. Portable Microfluidic chip for Detection of Escherichia coli in Produce and Blood Int J Nanomedicine. 2012b doi: 10.2147/IJN.S29629. http://dx.doi.org/10.2147/IJN.S29629. [DOI] [PMC free article] [PubMed]
- Wang S, Xu F, Demirci U. Advances in developing HIV-1 viral load assays for resource-limited settings. Biotechnology advances. 2010;28:770–81. doi: 10.1016/j.biotechadv.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhao X, Khimji I, Akbas R, Qiu W, Edwards D, et al. Integration of cell phone imaging with microchip ELISA to detect ovarian cancer HE4 biomarker in urine at the point-of-care. Lab Chip. 2011;11:3411–8. doi: 10.1039/c1lc20479c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO [Accessed on January 26th, 2012];Commercial serodiagnostic tests for diagnosis of tuberculosis. 2011a http://whqlibdoc.who.int/publications/2011/9789241502054_eng.pdf.
- WHO [accessed on January 26th, 2012];Global tuberculosis control 2011. 2011b http://www.who.int/tb/publications/global_report/en/
- WHO/Global Fund . Partners call for increased commitment to tackle MDR-TB. 2012. [Google Scholar]
- Xie H, Mire J, Kong Y, Chang M, Hassounah HA, Thornton CN, et al. Rapid point-of-care detection of the tuberculosis pathogen using a BlaC-specific fluorogenic probe. Nature chemistry. 2012;4:802–9. doi: 10.1038/nchem.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo WH, Liu S, Chung JH, Liu YL, Lee KH. Rapid detection of Mycobacterium tuberculosis cells by using microtip-based immunoassay. Analytical and Bioanalytical Chemistry. 2009;393:1593–600. doi: 10.1007/s00216-008-2591-x. [DOI] [PubMed] [Google Scholar]
- Zwerling A, Behr MA, Verma A, Brewer TF, Menzies D, Pai M. The BCG World Atlas: a database of global BCG vaccination policies and practices. PLoS Med. 2011;8:e1001012. doi: 10.1371/journal.pmed.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]