Abstract
Aldh1l1, also known as 10-formyltetrahydrofolate dehydrogenase (FDH), contains the carboxy-terminal domain (Ct-FDH), which is a structural and functional homolog of aldehyde dehydrogenases (ALDHs). This domain is capable of catalyzing the NADP+-dependent oxidation of short chain aldehydes to their corresponding acids, and similar to most ALDHs it has two conserved catalytic residues, Cys707 and Glu673. Previously, we demonstrated that in the Ct-FDH mechanism these residues define the conformation of the bound coenzyme and the affinity of its interaction with the protein. Specifically, the replacement of Cys707 with an alanine resulted in the enzyme lacking the ability to differentiate between the oxidized and reduced coenzyme. We suggested that this was due to the loss of a covalent bond between the cysteine and the C4N atom of nicotinamide ring of NADP+ formed during Ct-FDH catalysis. To obtain further insight into the functional significance of the covalent bond between Cys707 and the coenzyme, and the overall role of the two catalytic residues in the coenzyme binding and positioning, we have now solved crystal structures of Ct-FDH in the complex with thio-NADP+ and the complexes of the C707S mutant with NADP+ and NADPH. This study has allowed us to trap the coenzyme in the contracted conformation, which provided a snapshot of the conformational processing of the coenzyme during the transition from oxidized to reduced form. Overall, the results of this study further support the previously proposed mechanism by which Cys707 helps to differentiate between the oxidized and reduced coenzyme during ALDH catalysis.
Keywords: Aldehyde dehydrogenase, 10-Formyltetrahydrofolate dehydrogenase, NADP+/NADPH, catalytic residues, coenzyme conformation, crystal structure
1. Introduction
Aldehyde dehydrogenases are a family of NAD(P)+-utilizing enzymes that catalyze conversion of a variety of aldehydes to their corresponding carboxylic acids. In humans, this family is represented by 19 distinct genes, which encode protein products that form homodimers or homotetramers [1]. ALDHs have a very similar structure, consisting of catalytic, nucleotide binding and oligomerization domains [2-8]. While structural boundaries between these domains can be clearly seen, the functional separation between them is somewhat arbitrary. For example, the nicotinamide ring of bound NAD(P)+ protrudes into the catalytic domain to come in close proximity to catalytic residues. Of note, the nicotinamide moiety forms only a few contacts with an ALDH molecule and most of the interactions involve two ribose rings and the adenine moiety forming contacts with the protein within a classical Rossmann fold conserved throughout the family [3, 5, 6, 8, 9].
In addition to the structural conservation, there is also a functional conservation with regard to ALDH mechanism. Thus, ALDHs have two highly conserved catalytic residues, a cysteine and a glutamate [1]. Numerous structural, site-directed mutagenesis and enzymatic studies of ALDHs have established the role of the cysteine as the active site nucleophile (reviewed in [1]). The proposed mechanism for ALDH catalysis includes two steps, (i) acylation and (ii) deacylation [3, 5, 10-13]. The cysteine forms a thiohemiacetal intermediate with the substrate during the acylation step, which requires deprotonation of its sulfhydryl group (Reviewed in [1]). The glutamate has been proposed to facilitate the cysteine deprotonation, either directly or indirectly, and has also been suggested as a residue involved in the deacylation step of the reaction by activating a water molecule in the active site [3, 5, 8, 10, 14, 15]. The dual role of the glutamate residue in combination with the conserved geometry of the active site created the requirement for coenzyme isomerization during catalysis [9]. In the first stage of the reaction the nicotinamide ring of the cofactor must be sufficiently close to the catalytic cysteine to receive the hydride. The corresponding stretched conformation of NAD(P)+ was named “extended”. Once the hydride transfer is accomplished, the nicotinamide ring vacates the catalytic pocket to allow access of a water molecule that hydrolyzes the thioester intermediate, releasing the reaction product. This conformation of NAD(P)H was named “contracted” because the isomerization brings the nicotinamide moiety closer to the adenine part. In some crystal structures of ALDHs the nicotinamide ring of the reduced coenzyme is not seen due to disorder [8, 16]. This conformation of NADP(H) is a functional analogue of the contracted conformation, as it also allows space for the deacylation stage to proceed.
A member of ALDH family, Aldh1l1 (FDH, 10-formyltetrahydrofolate dehydrogenase) is a folate-metabolizing enzyme, which is a product of a natural fusion of three unrelated genes [17]. One of these genes, an ancient aldehyde dehydrogenase, encodes for the carboxyl-terminal portion (residues 400-902, identified thereafter as Ct-FDH) of Aldh1l1. This domain has high sequence similarity (up to 50%) with ALDH1 and ALDH2, and can catalyze both NADP+-dependent ALDH or NADP+-independent esterase reactions using short-chain aldehydes as substrates [17-19]. Like many other ALDHs, Ct-FDH possesses two key catalytic residues, a cysteine (Cys707) and a glutamate (Glu673); replacement of either resulted in the catalytically inactive enzyme [8, 20]. The ALDH reaction catalyzed by this domain is an integral part of the entire enzyme mechanism, which is the NADP+-dependent conversion of 10-formyltetrahydrofolate to tetrahydrofolate and CO2 [17]. Of note, higher animals have the second gene encoding for a similar enzyme, Aldh1L2, which is a mitochondrial protein (in contrast to Aldh1l1, which is a cytosolic enzyme) [21, 22].
Crystal structures of apo Ct-FDH and the holo enzyme in complexes with either NADP+ or NADPH have demonstrated a high similarity of the protein molecule to other ALDHs [8]. Ct-FDH (as well as full-length Aldh1l1 [20]) forms a homotetramer, with each monomer displaying catalytic, coenzyme binding and oligomerization domains. Previous studies have demonstrated an unusual feature of the complex of Ct-FDH with the oxidized coenzyme: a transient covalent bond between the sulfur atom of the catalytic cysteine and the C4N atom of the nicotinamide (Fig. 1) [8, 23]. Formation of such a bond has been also predicted for mitochondrial ALDH2 using in silico simulations [24] and was experimentally shown for other ALDHs [25]. It has been further demonstrated that the ability to form such a covalent bond is important for the discrimination between oxidized and reduced coenzyme bound to Ct-FDH [23]. In particular, replacement of the catalytic cysteine of Ct-FDH with an alanine resulted in an enzyme that bound both NADP+ and NADPH in the extended conformation. This study also suggested that the conserved catalytic glutamate controls the binding and discharging of the coenzyme, presumably through long-range communications with helix G that interacts with its adenine moiety. In the present work we aimed to clarify the role of the cysteine and the functional significance of its covalent bond with NADP+ in proper positioning of the coenzyme. We further wanted to elucidate if the nicotinamide ring of the coenzyme, which is known to only loosely bind to ALDHs, plays a significant part in defining the overall conformation of NADP(H). To this end, we solved crystal structures of the C707S mutant of Ct-FDH in complex with NADP+ or NADPH and a structure of wild-type Ct-FDH in complex with thio-NADP+, an NADP+ analogue with an altered nicotinamide group. Here we report the structural analysis of these proteins with regard to coenzyme binding and effects of Cys707 and Glu673 on the conformation of bound dinucleotide, and we expand the model for the conformational processing of the coenzyme during the transition from oxidized to reduced form.
Fig, 1.

The covalent bond between Cys707 of Ct-FDH and C4N of the nicotinamide ring of NADP+. Two positions of the sulfur, one forming the covalent bond (1.6 Å distance to C4N) and another oriented away from the nicotinamide ring (4 Å distance to C4N) are shown (green).
2. Materials and methods
2.1. Protein preparation
Site-directed mutagenesis was carried out using a QuickChange site-directed mutagenesis kit (Agilent Technologies) and confirmed by DNA sequencing of the mutant constructs. Wild type and mutant Ct-FDH were expressed in E. coli as constructs with 5xHis tag at the amino-terminus and purified on Ni-NTA resin (GE-Healthcare) as we previously described [8].
2.2. Crystallization and data collection
Crystals were grown by the vapor diffusion method in hanging drops over wells containing 1.5-1.6 M ammonium sulfate and 0.1 M MES-NaOH, pH 6.4 or 0.1 M HEPES-NaOH, pH 7.2, as described [8]. Crystals of binary complexes with NADP+, NADPH or thio-NADP were produced by overnight soaking of protein crystals in the presence of 5 mM of the corresponding coenzyme. Prior to mounting, crystals were passed through the mother liquor solution supplemented with the corresponding coenzyme and containing 27.5% glycerol and flash-cooled in situ at 100 K using an X-Stream Cryostream (Rigaku MSC).
Data sets were collected on an RAXIS IV++ image plate detector mounted on a RU-H3R rotating anode X-ray generator operating at 50 kV and 100 mA (Medical University of South Carolina). All the crystals belonged to space group C2 with cell dimensions a=258.6, b=194.8, c=97.4, and β=108.8. The data were processed using HKL2000 [26]. The statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics.
| C707S NADP+ |
C707S NADPH |
WT Thio-NADP+ |
|
|---|---|---|---|
| Resolution (Å) | 2.3 | 3.4 | 2.3 |
| Completeness % | 95.9(94.3) | 98.3(97.9) | 100.0(100.0) |
| Mean I/σI | 7.7(1.7) | 6.4(2.1) | 11.7(1.9) |
| Redundancy | 2.0(2.0) | 2.5(2.4) | 2.9(2.8) |
| Rmerge (%) | 9.6(39.9) | 19.9(55.3) | 9.6(56.4) |
| Rcryst (%) | 18.1 | 19.8 | 17.7 |
| Rfree (%) | 21.0 | 25.2 | 19.6 |
| Rmsd bond lengths (Å) | 0.012 | 0.018 | 0.012 |
| Rmsd bond angles (°) | 1.37 | 1.92 | 1.38 |
| Ramachandran plot: | |||
| % favored | 97.1 | 91.0 | 97.7% |
| % allowed | 2.7 | 7.9 | 2.1% |
| % outliers | 0.2 | 1.1 | 0.2% |
The figures in parentheses refer to the highest resolution shell of the data.
2.3. Model building and refinement
Structures were obtained by molecular replacement using Molrep [27] with the structure of apo Ct-FDH (PDB code 2O2P) [8] as the search model. Coenzyme molecules were identified by examining the ∣FO∣-∣FC∣ and 2∣FO∣-∣FC∣ electron density maps. Models were refined by alternating rounds of manual building using O [28] and Coot [29] and restrained refinement in REFMAC5 [30]. Water molecules were introduced with Coot [29] and verified manually. During refinement, 5% of the data were reserved to calculate the free R-factor. Table 1 shows the refinement statistics. Stereochemistry of the models was verified by PROCHECK [31]. The coordinates and structure factors have been deposited in the Protein Data Bank with codes 4GNZ, 4GO0 and 4GO2 for the C707S in complex with NADP+, the C707S in complex with NADPH and wild type Ct-FDH in complex with thio-NADP+, respectively. Figures were prepared using PyMol (www.pymol.org).
2.4. Assay of ALDH activity
The ALDH activity of Ct-FDH and the C707S mutant was measured by following the increase in absorbance at 340 nm as a result of formation of NADPH. The standard assay contained 22 μg/ml the enzyme, 120 μM NADP+, and 5 mM propanal in 50 mM HEPES-NaOH buffer, pH 9.0. The reaction was initiated by the addition of propanal. All measurements were done at 25 °C using a Shimadzu 2401PC double beam spectrophotometer. The activity of wild type Ct-FDH with thio-NADP+ (Sigma) was measured by monitoring the increase in absorbance at 398 nm as a result of formation of thio-NADPH.
2.5. Coenzyme binding study
Wild type Ct-FDH or the mutated proteins were incubated with increasing concentrations of NADP+ (up to 4.2×10−5 M), NADPH (up to 5×10−5 M) or thio-NADP (up to 4.2×10−5 M) for 1 hour at room temperature in 50 mM Tris-HCl buffer, pH 7.5 containing 100 mM NaCl and 0.5 mM DTT. Emission fluorescence spectra of proteins were recorded on a Hitachi F-2500 fluorescence spectrophotometer by scanning from 300 to 400 nm with excitation at 295 nm. Changes in NADPH fluorescence at 450 nm were recorded upon excitation at 330 nm. Kd values were determined by non-linear fitting of experimental data to equations described in our previous publication [23].
3. Results
3.1. Overall structure
All crystals obtained in this study belong to the C2 space group with unit cell dimensions identical to those of the wild type Ct-FDH crystals [8]. Protein subunits share the same fold and show no appreciable structural rearrangements compared to these previously published models.
3.2. Structures of the C707S mutant of Ct-FDH in complex with NADP+ and NADPH
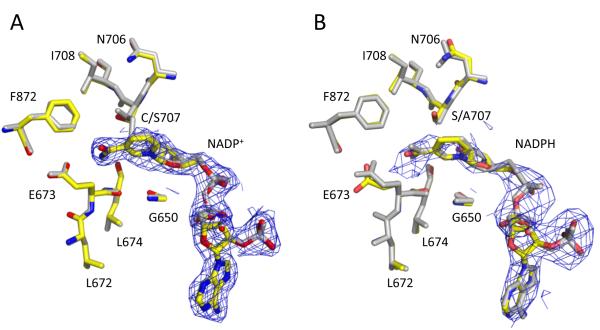
We determined two structures of the C707S mutant of Ct-FDH, the complex with NADP+ at 2.3 Å and the complex with NADPH at 3.4 Å resolution. These structures show no significant conformational deviations from wild type Ct-FDH, and their active sites are virtually identical (Fig. 2). Similarly to the structure of the wild-type enzyme with bound NADP+ [8] and to the structure of the corresponding mutant of human mitochondrial ALDH [9], in the C707S-NADP+ complex the coenzyme is bound in the extended conformation, with the nicotinamide ring displacing the side chain of the catalytic Glu673 to attain close proximity to Ser707 (Fig. 2A). However, the serine residue in place of the catalytic cysteine does not form a covalent bond with the C4N atom of the nicotinamide ring.
Fig. 2.
NADP+ and NADPH molecules in the structures with C707S Ct-FDH. A superposition of the active sites of the C707S enzyme (colored) with bound NADP+ (A) or NADPH (B) and wt Ct-FDH-NADP+ complex or C707A enzyme-NADPH complex (grey), respectively. The 2∣FO∣-∣FC∣ electron density map of the coenzyme contoured at 1 σ is shown in blue.
A remarkable feature of the C707S-NADPH complex is the extended conformation of the reduced coenzyme (Fig. 2B). In contrast, in the wild type Ct-FDH the nicotinamide ring of the reduced coenzyme is disordered, presumably due to the lack of contacts with the protein [8]. We have previously demonstrated the identical arrangement of the reduced coenzyme (extended conformation) bound to another Ct-FDH, the C707A enzyme [23]. In that enzyme, however, the extended conformation of the reduced coenzyme could be possible because the smaller side-chain of alanine allows additional space in the catalytic pocket for the nicotinamide ring. Here our data demonstrated that despite a higher structural and chemical similarity between a serine and a cysteine versus an alanine and a cysteine, NADPH still is in the extended conformation when bound to the C707S enzyme. Of note, the average distance between the hydroxyl oxygen of Ser707 and the C4N atom of NADPH in this structure is 2.9±0.2 Å, which provides enough space to accommodate the nicotinamide ring. Thus, the sulfur atom of Cys707 (likely due to the ability to form the transient covalent bond with the C4N atom of nicotinamide) seems to be crucial for discriminating between the oxidative states of the coenzyme. This sensory function either allows the oxidized nicotinamide ring to enter the catalytic pocket or repels the reduced ring.
3.3. Structure of wild type Ct-FDH in complex with thio-NADP+
We used a coenzyme analogue, thio-NADP+ (Fig. 3), to further probe the role of the two catalytic residues, Cys707 and Glu673, in the coenzyme binding. The structure of wild-type Ct-FDH in complex with thio-NADP+, determined at 2.3 Å resolution, maintains the same protein fold, but the coenzyme is bound in two different conformations with approximately equal occupancies. While one of these conformers is mostly disordered, the other forms a “crooked” structure with the nicotinamide part showing minimal contacts with the protein (Fig. 4A). In this new conformation the nicotinamide ring is shifted 4.6 Å outward from the catalytic center, and it closely resembles the canonical contracted conformation of NADH in the complex with ALDH2 [9] (Fig. 4B). Furthermore, as was previously shown in the structure of the Ct-FDH/NADP+ complex, binding of the nicotinamide ring in the extended conformation of the coenzyme is associated with a movement of the side chain of Glu673 out of the active site (Fig. 4C) [8]. In this new position, Glu673 forms tight contacts with several other residues, water molecules and the carboxamide group of the coenzyme (Fig. 4C). In contrast, in the structure of the Ct-FDH/thio-NADP+ complex the side chain of Glu673 remains in contact with Cys707 as was shown previously for the ligand-free structure [8], blocking the entrance to the catalytic center (Fig. 4A). It is likely that the bulky sulfur atom of thio-NADP+ substituting for the carboxamide oxygen of NADP+ cannot be accommodated in the extended conformation, and its repulsion from the carboxyl group of the glutamate results in the nicotinamide moiety being expelled from the active site. Of note, thio-NADP+ appears in the extended conformation in the structures of DHFR [32] and transhydrogenase [33], the fact suggesting that this conformation is possible. Thus, we suggest that thio-NADP+ bound to Ct-FDH represents the conformation of the natural coenzyme on its way out after the completion of the hydride transfer.
Fig. 3.
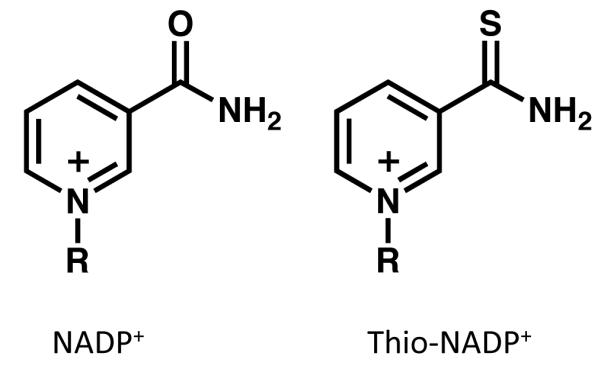
Comparison of structures of NADP+ (left) and thio-NADP+(right).
Fig. 4.
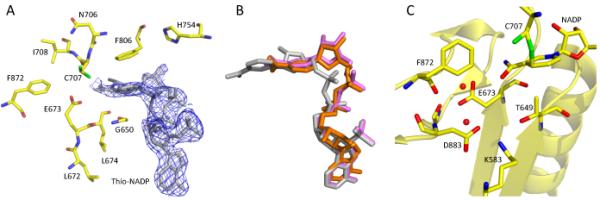
Binding of thio-NADP+ to Ct-FDH. (A) The active site of wild-type Ct-FDH with bound thio-NADP+ (the 2∣FO∣-∣FC∣ electron density map of thio-NADP+ contoured at 1 σ is shown in blue). (B) Overlay of coenzymes bound to ALDHs: grey, NADP+ (extended conformation) bound to wt Ct-FDH; orange, NADPH (contracted conformation) bound to ALDH2 [9]; violet, thio-NADP+ (contracted conformation) bound to wt Ct-FDH. (C) Environment of Glu673 in the complex of Ct-FDH with NADP+ (PDB 2O2Q).
3.4. Enzymatic activity and coenzyme binding
We have previously demonstrated that C707A Ct-FDH is catalytically inactive [20]. A similar effect was observed when the catalytic cysteine of other aldehyde dehydrogenases was replaced with alanine [34]. However, the replacement of the cysteine with a serine in mitochondrial rat liver aldehyde dehydrogenases (Aldh2) resulted in an enzyme that retained residual catalytic activity [34]. In the present study, analysis of the C707S Ct-FDH revealed that it was catalytically inactive. We have also determined Kds for binding both oxidized and reduced coenzymes by the mutant enzyme. The K for NADP+ d was determined based on quenching of tryptophan fluorescence of the C707S enzyme upon binding of the coenzyme as we previously described [20]. The decrease in fluorescence was likely due to quenching of fluorescence of two tryptophans near the coenzyme binding site, Trp573 located about 4 Å from the pyrophosphate group of the coenzyme and Trp582 located about 8 Å from the nicotinamide ring. The replacement of Cys707 with a serine resulted in a significant decrease in the affinity for coenzyme with Kd of 11.0 ± 1.5 μM (Kd for the wild-type enzyme is 0.9 ± 0.1 μM). The above method cannot be used to measure the Kd for NADPH binding due to the fluorescence resonance energy transfer from the tryptophan residues to NADPH [23]. Therefore, changes in NADPH fluorescence at 450 nm were monitored after excitation at 330 nm (binding to Ct-FDH leads to enhanced NADPH fluorescence) [35]. These experiments demonstrated that the wild-type and C707S enzymes have similar Kd values for NADPH (data not shown). This is in contrast to C707A enzyme, which binds NADPH more tightly than the wild-type enzyme does [23]. We suggest the difference is due to the larger side chain of the serine as compared to the alanine residue.
We have also evaluated the aldehyde dehydrogenase activity of wild-type Ct-FDH using thio-NADP+ as the coenzyme. The enzyme did not exhibit any catalytic activity with this derivative, a finding that is in agreement with our structural data demonstrating that the nicotinamide moiety is positioned too far from the catalytic cysteine to allow the hydride transfer; the closest distance between the C4N atom of the nicotinamide of thio-NADP+ and the sulfur of the catalytic cysteine is 4.5 Å compared to 1.6 Å in the case of NADP+ (Fig. 1) [8]. The Kd value (17.0 ± 0.8 μM) for the binding of thio-NADP+ evaluated by quenching of tryptophan fluorescence was significantly increased compared to the Kd value for wild-type enzyme.
4. Discussion
Nicotinamide dinucleotide molecules, NAD+/NADH and NADP+/NADPH, transfer a hydride ion in oxido-reductive reactions and are among the most common coenzymes found in the cell, being utilized by several hundreds of different enzymes [36, 37]. While the precise mode of the accommodation of these coenzymes in their respective binding pockets varies among enzymes and is defined by structural arrangements of catalytic and nucleotide binding centers, the enzyme mechanism, and by specific residues involved in the coenzyme binding and catalysis [38], there are several common features of dinucleotide binding. For example, the most common structural motif involved in the binding of NAD(P)+ molecules is the Rossmann fold (reviewed in [36, 39]), which is found in many different classes of dehydrogenases including the ALDH family [2, 3]. Furthermore, while the nicotinamide adenine dinucleotide is a flexible molecule that can adopt different conformations depending on its environment [40], the coenzyme is bound to most proteins preferentially in the extended conformation [38]. An interesting feature of the coenzyme binding by ALDHs, though, is two distinct conformations of the dinucleotide molecule, extended and contracted [2, 3, 5, 8, 9, 41]. It is generally accepted that the former represents the hydride transfer position of the oxidized coenzyme in the acylation step of the catalysis while the latter corresponds to the reduced coenzyme in the second, deacylation, step when the nicotinamide ring has already left the catalytic pocket.
Crystal structures of the holo Ct-FDH in complex with NADP+ or NADPH demonstrated two conformations of the dinucleotide molecule as well, an extended one for the oxidized coenzyme and one which was interpreted as the contracted conformation for the reduced coenzyme [8]. Of note, in that presumably contracted conformation, there was no electron density seen for the nicotinamide ring of NADPH, suggesting the flexibility of positioning of the reduced ring in this structure. Apparently, the nicotinamide ring in the oxidized coenzyme forms additional contacts with the enzyme that fix it in the extended conformation, whereas the lack of such contacts for the ring in the reduced coenzyme allows it to move into various positions. In the present study, however, the contracted conformation of the coenzyme, with a well-ordered nicotinamide ring, has been observed in the complex of Ct-FDH with thio-NADP+.
Our recent studies of Ct-FDH shed light on the mechanism by which ALDHs could retain the oxidized coenzyme in the extended conformation and also discriminate against the binding of the reduced coenzyme [23]. This novel mechanism of the interaction between ALDHs and NAD(P)+ involves a transient covalent bond formed between the catalytic nucleophile (the sulfhydryl group of the cysteine residue) and the C4N atom of the nicotinamide ring [8]. Since this finding was reported, covalent bonds between the catalytic cysteine and the nicotinamide ring have been observed in crystal structures of several other aldehyde dehydrogenases [24, 25], the fact implicating this phenomenon as an integral part of the catalytic mechanism. We initially suggested that such a covalent bond facilitates stronger binding of the coenzyme and the proper orientation of the nicotinamide ring within the catalytic center [8]. The follow up study from our laboratory has further demonstrated that the covalent bond between the cysteine and nicotinamide ring plays an additional role as the sensor recognizing the oxidative state of the coenzyme [23]. This hypothesis has received further support in the present study.
Based on our data, we propose the following mechanism for discharging the coenzyme from its binding site (schematically depicted in Fig. 5). This mechanism presents two catalytic residues, Cys707 and Glu673, as the key determinants defining the mode of interaction between the enzyme and the coenzyme, and can be described as a two-phase process: (i) the displacement of the reduced nicotinamide ring from the catalytic center, and (ii) dissociation of the coenzyme from the enzyme. In the first phase, upon binding the oxidized coenzyme, the sulfhydryl group of the cysteine forms a transient covalent bond with the C4N atom. This bond, which was also demonstrated for the hydrolytic ALDH from Pseudomonas aeruginosa, can easily dissociate via an elimination reaction [25]. The fact that we observed two positions of the sulfur atom of the cysteine with the 50% probability might suggest that the formation and dissociation of this bond is a constant process. We hypothesize that the ability to form such a temporary bond serves as a sensor of the oxidized state of the coenzyme (it cannot be formed with the reduced form, NADPH). This step is schematically depicted in Fig. 5 (panels A and B) as the balance between two stages, with and without the covalent bond. In the second step of this phase, following the hydride transfer and the reduction of the coenzyme, the sulfhydryl group comes into steric hindrance with the nicotinamide ring (Fig. 5, panel C). In the third step, this hindrance results in the displacement of the ring from the catalytic center thus changing the coenzyme conformation from extended to contracted (Fig. 5, panel D).
Fig. 5.
Proposed changes in the active site of Ct-FDH with the oxidized or reduced coenzyme bound at different stages of catalysis. A and B, bound NADP+ without and with formation of the covalent bond between Cys707 and the nicotinamide ring, respectively. C and D, NADPH bound in the extended (modeled from the C707A enzyme structure, PDB 3RHR [23]) or contracted (modeled from wt Ct-FDH/thio-NADP+ complex) conformation, correspondingly.
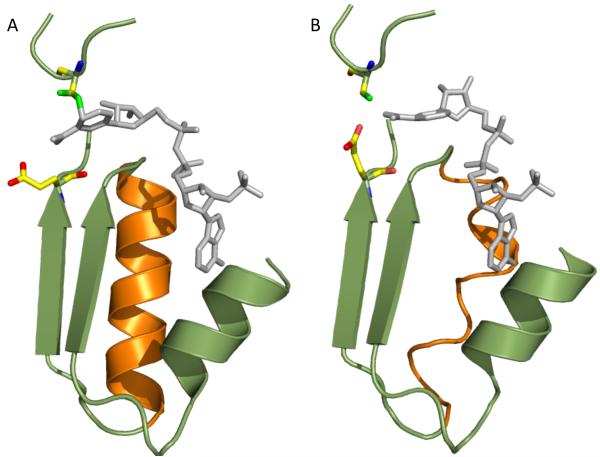
In our crystal structures, the latter stage is characterized by the lack of well-defined electron density for the reduced nicotinamide ring, but well-defined density for the adenosine diphophosphoribose. We interpreted this conformation of NADPH as a disordered nicotinamide ring and the rest of the coenzyme rigidly bound to helix G, which itself is well-ordered at this stage (Fig. 6, panel A). Based on our previous studies [23], we hypothesize that such an arrangement (an ordered helix G) is caused by the distant effect of the catalytic glutamate, which at this point is displaced from the catalytic pocket by the nicotinamide ring (Fig. 6, panel A). The second phase of the coenzyme dissociation involves the rotation of the glutamate back into the nicotinamide-binding pocket within the catalytic center, which may initiate remote conformational alterations in helix G (as shown previously for the E673Q enzyme [23]) thus causing its disorder (Fig. 6, panel B). The disordered helix is losing contact with the adenine moiety, which allows coenzyme dissociation from the enzyme (there would be no strong contacts between the enzyme and the coenzyme with such an arrangement). It should be noted that Glu673 is also likely to participate in the first stage of coenzyme discharging, through energetically unfavorable interactions with carboxamide of nicotinamide, thus cooperating with Cys707 in dissociation of the nicotinamide from the catalytic center. In this regard, the glutamate could discriminate between different orientations of the planes of the carboxamide group relative to the nicotinamide ring in the oxidized and reduced coenzyme, a mechanism described for other enzymes [32, 33].
Fig. 6.
Model depicting interactions of the coenzyme with ordered (A) or distorted (B) helix G. The model proposes that more ordered helix G is formed upon the coenzyme binding and the movement of catalytic glutamate outside of the nicotinamide binding part of the catalytic pocket. The helix presumably becomes distorted upon the return of catalytic glutamate back into the pocket that allows the dissociation of the coenzyme from the protein. Panel B was modeled based on the structure of the apo E673Q enzyme (PDB 3RHM [23]) and the structure of wt Ct-FDH in complex with thio-NADP+
It is not clear at present whether the proposed mechanism of discrimination between the reduce and oxidize states of the coenzyme is unique for Ct-FDH, limited to certain members of ALDH family, or whether it is a more common phenomenon for this class of enzymes. In this regard, while the catalytic cysteine is conserved in all active aldehyde dehydrogenases, the glutamate is less conserved [42]. It is also not clear whether the mechanism is extended to other classes of oxido-reductases. Of note, the ALDH reaction is essentially unidirectional [43], whereas for example alcohol dehydrogenases catalyze a reversible reaction [44]. Thus, the specific feature of the ALDH catalysis could be the necessity to discharge the reduced coenzyme without the requirement to bind and utilize it for the reverse reaction. Toward this end, our study underscores the role of the cysteine and glutamate in discriminating between the reduced and oxidized coenzyme, a feature that perhaps would not be required for enzymes catalyzing the reaction in both directions. Curiously, it appears that one of the ALDHs, ALDH18A1, catalyzes the reversal of ALDH reaction. ALDH18A1, an NADPH-dependent γ-glutamyl phosphate reductase, is a part of the bifunctional pyrroline-5-carboxylate synthase, the key enzyme in the biosynthesis of proline and ornithine [45-48]. It is attractive to suggest that the reductase does not have the catalytic glutamate analogous to Glu673 of Ct-FDH and binds NADPH in the extended conformation. However, while the crystal structure of the apo enzyme from Thermotoga maritima has been solved [49], the structure of its complex with the coenzyme awaits future studies.
Highlights.
Structures of the C707S Ct-FDH mutant were solved in complex with NADP+ and NADPH.
The structure of wild type Ct-FDH in complex with thio-NADP+ has been solved.
A contracted conformation of the coenzyme bound to Ct-FDH has been observed.
We propose a mechanism for binding oxidized and discharging reduced coenzyme.
Acknowledgments
The authors would like to thank Dr. John Hempel, University of Pittsburg for helpful discussions. This work was supported by The National Institutes of Health grant DK54388. Kyle C. Strickland was supported by a Ruth L. Kirschstein National Research Service Award for Individual Predoctoral MD/PhD Fellows F30DK083215. The X-ray crystallography facility used for this work is supported by the Medical University of South Carolina’s Research Resource Facilities program.
Abbreviations
- ALDH
aldehyde dehydrogenase
- Aldh1l1
10-formyltetrahydrofolate dehydrogenase
- Ct-FDH
the carboxy-terminal ALDH domain of 10-formyltetrahydrofolate dehydrogenase
Footnotes
Conflict of interest statement None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Koppaka V, Thompson DC, Chen Y, Ellermann M, Nicolaou KC, Juvonen RO, Petersen D, Deitrich RA, Hurley TD, Vasiliou V. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol. Rev. 2012;64:520–539. doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, Kuo I, Perozich J, Lindahl R, Hempel J, Wang BC. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat. Struct. Biol. 1997;4:317–326. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- [3].Steinmetz CG, Xie P, Weiner H, Hurley TD. Structure of mitochondrial aldehyde dehydrogenase: the genetic component of ethanol aversion. Structure. 1997;5:701–711. doi: 10.1016/s0969-2126(97)00224-4. [DOI] [PubMed] [Google Scholar]
- [4].Lamb AL, Newcomer ME. The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity. Biochemistry. 1999;38:6003–6011. doi: 10.1021/bi9900471. [DOI] [PubMed] [Google Scholar]
- [5].Moore SA, Baker HM, Blythe TJ, Kitson KE, Kitson TM, Baker EN. Sheep liver cytosolic aldehyde dehydrogenase: the structure reveals the basis for the retinal specificity of class 1 aldehyde dehydrogenases. Structure. 1998;6:1541–1551. doi: 10.1016/s0969-2126(98)00152-x. [DOI] [PubMed] [Google Scholar]
- [6].Johansson K, El-Ahmad M, Ramaswamy S, Hjelmqvist L, Jornvall H, Eklund H. Structure of betaine aldehyde dehydrogenase at 2.1 A resolution. Protein Sci. 1998;7:2106–2117. doi: 10.1002/pro.5560071007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cobessi D, Tête-Favier F, Marchal S, Azza S, Branlant G, Aubry A. Apo and holo crystal structures of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. J. Mol. Biol. 1999;290:161–173. doi: 10.1006/jmbi.1999.2853. [DOI] [PubMed] [Google Scholar]
- [8].Tsybovsky Y, Donato H, Krupenko NI, Davies C, Krupenko SA. Crystal structures of the carboxyl terminal domain of rat 10-formyltetrahydrofolate dehydrogenase: implications for the catalytic mechanism of aldehyde dehydrogenases. Biochemistry. 2007;46:2917–2929. doi: 10.1021/bi0619573. [DOI] [PubMed] [Google Scholar]
- [9].Perez-Miller SJ, Hurley TD. Coenzyme isomerization is integral to catalysis in aldehyde dehydrogenase. Biochemistry. 2003;42:7100–7109. doi: 10.1021/bi034182w. [DOI] [PubMed] [Google Scholar]
- [10].Muñoz-Clares RA, González-Segura L, Díaz-Sánchez AG. Crystallographic evidence for active-site dynamics in the hydrolytic aldehyde dehydrogenases. Implications for the deacylation step of the catalyzed reaction. Chem. Biol. Interact. 2011;191:137–146. doi: 10.1016/j.cbi.2010.12.024. [DOI] [PubMed] [Google Scholar]
- [11].Feldman RI, Weiner H. Horse liver aldehyde dehydrogenase. II. Kinetics and mechanistic implications of the dehydrogenase and esterase activity. J. Biol. Chem. 1972;247:267–272. [PubMed] [Google Scholar]
- [12].Sheikh S, Ni L, Hurley TD, Weiner H. The potential roles of the conserved amino acids in human liver mitochondrial aldehyde dehydrogenase. J. Biol. Chem. 1997;272:18817–18822. doi: 10.1074/jbc.272.30.18817. [DOI] [PubMed] [Google Scholar]
- [13].Marchal S, Cobessi D, Rahuel-Clermont S, Tête-Favier F, Aubry A, Branlant G. Chemical mechanism and substrate binding sites of NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. Chem. Biol. Interact. 2001;130-132:15–28. doi: 10.1016/s0009-2797(00)00218-0. [DOI] [PubMed] [Google Scholar]
- [14].Wang X, Weiner H. Involvement of glutamate 268 in the active site of human liver mitochondrial (class 2) aldehyde dehydrogenase as probed by site-directed mutagenesis. Biochemistry. 1995;34:237–243. doi: 10.1021/bi00001a028. [DOI] [PubMed] [Google Scholar]
- [15].Marchal S, Rahuel-Clermont S, Branlant G. Role of glutamate-268 in the catalytic mechanism of nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans. Biochemistry. 2000;39:3327–3335. doi: 10.1021/bi9914208. [DOI] [PubMed] [Google Scholar]
- [16].Cobessi D, Tête-Favier F, Marchal S, Branlant G, Aubry A. Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans. J. Mol. Biol. 2000;300:141–152. doi: 10.1006/jmbi.2000.3824. [DOI] [PubMed] [Google Scholar]
- [17].Krupenko SA. FDH: an aldehyde dehydrogenase fusion enzyme in folate metabolism. Chem. Biol. Interact. 2009;178:84–93. doi: 10.1016/j.cbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Krupenko SA, Wagner C, Cook RJ. Expression, purification, and properties of the aldehyde dehydrogenase homologous carboxyl-terminal domain of rat 10-formyltetrahydrofolate dehydrogenase. J. Biol. Chem. 1997;272:10266–10272. doi: 10.1074/jbc.272.15.10266. [DOI] [PubMed] [Google Scholar]
- [19].Strickland KC, Krupenko NI, Dubard ME, Hu CJ, Tsybovsky Y, Krupenko SA. Enzymatic properties of ALDH1L2, a mitochondrial 10-formyltetrahydrofolate dehydrogenase. Chem. Biol. Interact. 2011;191:129–136. doi: 10.1016/j.cbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Krupenko SA, Wagner C, Cook RJ. Cysteine 707 is involved in the dehydrogenase activity site of rat 10-formyltetrahydrofolate dehydrogenase. J. Biol. Chem. 1995;270:519–522. doi: 10.1074/jbc.270.2.519. [DOI] [PubMed] [Google Scholar]
- [21].Krupenko NI, Dubard ME, Strickland KC, Moxley KM, Oleinik NV, Krupenko SA. ALDH1L2 is the mitochondrial homolog of 10-formyltetrahydrofolate dehydrogenase. J. Biol. Chem. 2010;285:23056–23063. doi: 10.1074/jbc.M110.128843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Strickland KC, Holmes RS, Oleinik NV, Krupenko NI, Krupenko SA. Phylogeny and evolution of aldehyde dehydrogenase-homologous folate enzymes. Chem. Biol. Interact. 2011;191:122–128. doi: 10.1016/j.cbi.2010.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tsybovsky Y, Krupenko SA. Conserved Catalytic Residues of the ALDH1L1 Aldehyde Dehydrogenase Domain Control Binding and Discharging of the Coenzyme. J. Biol. Chem. 2011;286:23357–23367. doi: 10.1074/jbc.M111.221069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wymore T, Deerfield DW, 2nd, Hempel J. Mechanistic implications of the cysteine-nicotinamide adduct in aldehyde dehydrogenase based on quantum mechanical/molecular mechanical simulations. Biochemistry. 2007;46:9495–9506. doi: 10.1021/bi700555g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Díaz-Sánchez AG, González-Segura L, Rudiño-Piñera E, Lira-Rocha A, Torres-Larios A, Muñoz-Clares RA. Novel NADPH-cysteine covalent adduct found in the active site of an aldehyde dehydrogenase. Biochem. J. 2011;439:443–452. doi: 10.1042/BJ20110376. [DOI] [PubMed] [Google Scholar]
- [26].Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- [27].Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- [28].Jones TA, Zou JY, Cowan SW. Kjeldgaard, Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- [29].Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- [30].Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- [31].Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- [32].McTigue MA, Davies JF, 2nd, Kaufman BT, Kraut J. Crystal structures of chicken liver dihydrofolate reductase: binary thioNADP+ and ternary thioNADP+.biopterin complexes. Biochemistry. 1993;32:6855–6862. doi: 10.1021/bi00078a008. [DOI] [PubMed] [Google Scholar]
- [33].Singh A, Venning JD, Quirk PG, van Boxel GI, Rodrigues DJ, White SA, Jackson JB. Interactions between transhydrogenase and thio-nicotinamide Analogues of NAD(H) and NADP(H) underline the importance of nucleotide conformational changes in coupling to proton translocation. J. Biol. Chem. 2003;278:33208–33216. doi: 10.1074/jbc.M303061200. [DOI] [PubMed] [Google Scholar]
- [34].Farrés J, Wang TT, Cunningham SJ, Weiner H. Investigation of the active site cysteine residue of rat liver mitochondrial aldehyde dehydrogenase by site-directed mutagenesis. Biochemistry. 1995;34:2592–2598. doi: 10.1021/bi00008a025. [DOI] [PubMed] [Google Scholar]
- [35].Takahashi K, Weiner H, Hu JH. Increase in the stoichiometry of the functioning active sites of horse liver aldehyde dehydrogenase in the presence of magnesium ions. Arch Biochem Biophys. 1980;205:571–578. doi: 10.1016/0003-9861(80)90140-x. [DOI] [PubMed] [Google Scholar]
- [36].Bellamacina CR. The nicotinamide dinucleotide binding motif: a comparison of nucleotide binding proteins. FASEB J. 1996;10:1257–1269. doi: 10.1096/fasebj.10.11.8836039. [DOI] [PubMed] [Google Scholar]
- [37].Mu F, Unkefer PJ, Unkefer CJ, Hlavacek WS. Prediction of oxidoreductase-catalyzed reactions based on atomic properties of metabolites. Bioinformatics. 2006;22:3082–3088. doi: 10.1093/bioinformatics/btl535. [DOI] [PubMed] [Google Scholar]
- [38].Kuppuraj G, Sargsyan K, Hua YH, Merrill AR, Lim C. Linking distinct conformations of nicotinamide adenine dinucleotide with protein fold/function. J. Phys. Chem. B. 2011;115:7932–7939. doi: 10.1021/jp1118663. [DOI] [PubMed] [Google Scholar]
- [39].Jörnvall H, Hedlund J, Bergman T, Oppermann U, Persson B. Superfamilies SDR and MDR: from early ancestry to present forms. Emergence of three lines, a Zn-metalloenzyme, and distinct variabilities. Biochem. Biophys. Res. Commun. 2010;396:125–130. doi: 10.1016/j.bbrc.2010.03.094. [DOI] [PubMed] [Google Scholar]
- [40].Smith PE, Tanner JJ. Conformations of nicotinamide adenine dinucleotide (NAD(+)) in various environments. J. Mol. Recognit. 2000;13:27–34. doi: 10.1002/(SICI)1099-1352(200001/02)13:1<27::AID-JMR483>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- [41].González-Segura L, Rudiño-Piñera E, Muñoz-Clares RA, Horjales E. The crystal structure of a ternary complex of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa Provides new insight into the reaction mechanism and shows a novel binding mode of the 2′-phosphate of NADP+ and a novel cation binding site. J. Mol. Biol. 2009;385:542–557. doi: 10.1016/j.jmb.2008.10.082. [DOI] [PubMed] [Google Scholar]
- [42].Hempel J, Kuo I, Perozich J, Wang BC, Lindahl R, Nicholas H. Aldehyde dehydrogenase. Maintaining critical active site geometry at motif 8 in the class 3 enzyme. Eur. J. Biochem. 2001;268:722–726. doi: 10.1046/j.1432-1327.2001.01926.x. [DOI] [PubMed] [Google Scholar]
- [43].Hempel J, Stanley S, Perozich J, Wymore T, Nicholas HB., Jr . Residue conservations in aldehyde dehydrogenase gene products reemphasize functional interpretations. In: Weiner EMH, Lindahl R, Plapp B, editors. Enzymology and Molecular Biology of Carbonyl Metabolism 12. Purdue University Press; West Lafayette IN: 2006. [Google Scholar]
- [44].Duester G, Farres J, Felder MR, Holmes RS, Hoog JO, Pares X, Plapp BV, Yin SJ, Jornvall H. Recommended nomenclature for the vertebrate alcohol dehydrogenase gene family. Biochem. Pharmacol. 1999;58:389–395. doi: 10.1016/s0006-2952(99)00065-9. [DOI] [PubMed] [Google Scholar]
- [45].Pérez-Arellano I, Carmona-Alvarez F, Martínez AI, Rodríguez-Díaz J, Cervera J. Pyrroline-5-carboxylate synthase and proline biosynthesis: from osmotolerance to rare metabolic disease. Protein Sci. 2010;19:372–382. doi: 10.1002/pro.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Hu CA, Khalil S, Zhaorigetu S, Liu Z, Tyler M, Wan G, Valle D. Human Delta1-pyrroline-5-carboxylate synthase: function and regulation. Amino Acids. 2008;35:665–672. doi: 10.1007/s00726-008-0075-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bicknell LS, Pitt J, Aftimos S, Ramadas R, Maw MA, Robertson SP. A missense mutation in ALDH18A1, encoding Delta1-pyrroline-5-carboxylate synthase (P5CS), causes an autosomal recessive neurocutaneous syndrome. Eur. J. Hum. Genet. 2008;16:1176–1186. doi: 10.1038/ejhg.2008.91. [DOI] [PubMed] [Google Scholar]
- [48].Skidmore DL, Chitayat D, Morgan T, Hinek A, Fischer B, Dimopoulou A, Somers G, Halliday W, Blaser S, Diambomba Y, Lemire EG, Kornak U, Robertson SP. Further expansion of the phenotypic spectrum associated with mutations in ALDH18A1, encoding Delta(1)-pyrroline-5-carboxylate synthase (P5CS) Am. J. Med. Genet. A. 2011;155A:1848–1856. doi: 10.1002/ajmg.a.34057. [DOI] [PubMed] [Google Scholar]
- [49].Page R, Nelson MS, von Delft F, Elsliger MA, Canaves JM, Brinen LS, Dai X, Deacon AM, Floyd R, Godzik A, Grittini C, Grzechnik SK, Jaroszewski L, Klock HE, Koesema E, Kovarik JS, Kreusch A, Kuhn P, Lesley SA, McMullan D, McPhillips TM, Miller MD, Morse A, Moy K, Ouyang J, Robb A, Rodrigues K, Schwarzenbacher R, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, Vincent J, Wang X, West B, Wolf G, Hodgson KO, Wooley J, Wilson IA. Crystal structure of gamma-glutamyl phosphate reductase (TM0293) from Thermotoga maritima at 2.0 A resolution. Proteins. 2004;54:157–161. doi: 10.1002/prot.10562. [DOI] [PubMed] [Google Scholar]