Abstract
In the 45 years since its development, the pyrimidine analog 5-fluorouracil (5-FU) has become an integral component of many cancer treatments, most notably for the management of colorectal cancer. An appreciable fraction of patients who receive 5-FU suffer severe adverse toxicities, which in extreme cases may result in death. Dihydropyrimidine dehydrogenase (DPD, encoded by DPYD) rapidly degrades 85% of administered 5-FU, and as such, limits the amount of drug available for conversion into active metabolites. Clinical studies have suggested that genetic variations in DPYD increase the risk for 5-FU toxicity, however there is not a clear consensus as to which variations are relevant predictors. In the present study, DPYD variants were expressed in mammalian cells, and the enzymatic activity of expressed protein was determined relative to wild type. Relative sensitivity to 5-FU for cells expressing DPYD variations was also measured. The DPYD*2A variant (exon 14 deletion caused by IVS14+1G>A) was confirmed to be catalytically inactive. Compared to wild type, two variants, S534N and C29R, showed significantly higher enzymatic activity. Cells expressing S534N were more resistant to 5-FU mediated toxicity compared to cells expressing wild type DPYD. These findings support the hypothesis that selected DPYD alleles are protective against severe 5-FU toxicity, and, as a consequence, may decrease the effectiveness of 5-FU an anti tumor drug in carriers. Additionally, this study demonstrates a method that may be useful for phenotyping other genetic variations in pharmacologically relevant pathways.
Keywords: DPYD, phenotypic assay, 5-fluorouracil, real-time cellular analysis, individualized medicine
Introduction
The pyrimidine analog 5-florouracil (5-FU) and its prodrug capecitabine are frequently prescribed for the treatment of aggressive cancers, particularly those of the colon and breast. Like many chemotherapy drugs, 5-FU has a narrow therapeutic index, with a very small difference between effective and toxic doses (1). 5-FU is converted to active and inactive metabolites by the uracil anabolic and catabolic pathways, respectively (2, 3). Anabolism of 5-FU generates cytotoxic metabolites that inhibit production of thymidine and uridine and are incorporated during DNA and RNA synthesis (reviewed in 4). Only 1-3% of administered 5-FU enters the anabolic pathway; the vast majority (approximately 85%) is catabolized in the liver (3). Dihydropyrimidine dehydrogenase (DPD, encoded by the DPYD gene) is the initial and rate-limiting enzyme of the uracil catabolic pathway, and as such has a pivotal role in determining the clearance rate and circulating half-life of 5-FU (5, 6).
Early studies showed that DPD deficiency prolonged the half-life of 5-FU by approximately 10-fold (5, 7, 8) and correlated with adverse responses in those treated with the standard dose of the drug (8-10). Complete deficiency of DPD manifests during childhood as a rare neurological disorder (11-13). Partial deficiency is more common and presents as a toxic response (≥ grade 3) to the standard dose of 5-FU (8, 9). These adverse responses range in severity and include diarrhea, hand-foot syndrome, stomatitis, mucositis, myelosuppression, neurotoxicity, and, in extreme cases, death (14).
Genetic variations in DPYD have been identified as a major contributor to DPD deficiency. The most studied DPYD variation, *2A (rs3918290, a G>A base change at the splice acceptor sequence for exon 14), significantly reduces the enzyme activity of the translated protein and has been shown to prolong the clearance of 5-FU leading to increased occurrence of severe toxicity and occasionally death (15, 16). In addition to *2A, 96 nonsynomymous and 2 frameshift variations have been reported for DPYD (17). For many DPYD variations, clinical studies have yielded unclear or contradicting results pertaining to their contribution to severe 5-FU toxicity (18).
The objective of the present study was to develop and validate a cellular model system that would allow rapid phenotypic assessment of DPYD variations for sensitivity to 5-FU. In this study, DPYD alleles were expressed in a human cell line, and enzyme activity of the expressed protein was directly quantified. Sensitivity to 5-FU was measured using real-time cellular analysis (RTCA) of cells expressing DPYD variations. Using the developed system we have functionally classified several commonly studied DPYD alleles. Of the variants studied, two showed significantly higher levels of enzyme activity compared to wild type. This finding suggests that individuals carrying these alleles may be protected against adverse toxicity to 5-FU at the expense of reduced drug efficacy at the standard dose.
Materials and Methods
Cells
A protein expression screen of commonly used laboratory cell lines showed that DPD was not detectible in HEK293T clone 17 (HEK293T/c17) and HCT116 cells (data not shown). Low passage HEK293T/c17 cells were obtained from ATCC (culture CRL-11268) and grown using standard cell culture conditions of 37° C and 5% CO2 in a humidified incubator using Dulbecco’s modified Eagle’s medium (Mediatech) supplemented with 10% fetal bovine serum (Denville Scientific), 100 IU ml−1 penicillin, and 100 μg ml−1 streptomycin (Mediatech). HCT116 cells were obtained from ATCC (culture CCL-247) and cultured in McCoys 5A medium supplemented with 10% fetal bovine serum, 100 IU ml−1 penicillin, and 100 μg ml−1 streptomycin.
ATCC routinely authenticates cell lines in their repository using short tandem repeat (STR) profiling, cell morphology monitoring, karyotyping, and cytochrome C oxidase I (COI) testing. Upon receipt from ATCC, cell lines were observed microscopically to confirm morphology, and population-doubling times were determined by daily counting of viable cells using the trypan blue dye exclusion method. Low passage stocks of both cells lines were prepared and stored in liquid nitrogen within two weeks of receipt from ATCC. For experimental use, cell stocks were thawed and maintained in culture for no longer than two months or a total of 10 passages. Cell lines were periodically monitored for mycoplasma by Hoechst staining (Sigma-Aldrich). Culture identity and health were monitored by microscopy and by comparing the population doubling times to baseline values determined for the original cell stock received. Culture health was additionally monitored for real-time cellular analysis (RTCA) experiments as described by Irelan et al (19).
Vector Construction
Human DPYD was cloned into the pIRES-neo3 expression vector (Clontech) using the Rapid DNA Ligation kit (Roche). The *2A, C29R, S534N, I543V, I560S, and V732I variant constructs were generated using the Phusion Site-Directed Mutagenesis Kit (Finnzymes) using primers listed in Supplementary Table S1. Plasmid sequences were confirmed at the Mayo Clinic Advanced Genomics Technology Center (Rochester, MN) using primers listed in Supplementary Table S2. DPYD variants were subcloned back into parental pIRES-neo3 and reconfirmed to eliminate potential spurious mutations introduced into the vector backbone during PCR. Endotoxin-free plasmid DNA was prepared for transfections using the NucleoBond Extra Midi EF kit (Macherey-Nagel). The pIRES-neo3 vector was used as an “empty vector” control where indicated.
DPD Enzyme Activity Assay
Low passage HEK293T/c17 cells were seeded at a density of 3×106 per 10 cm plates and transfected after 16 hours with 5 μg plasmid DNA using FuGene HD (Roche). Total protein lysates were prepared 48 hours following transfection. Cells were trypsinized (0.05% Trypsin and 0.53 mM EDTA, Mediatech), washed with PBS, and resuspended in buffer consisting of 35 mM potassium phosphate buffer at pH 7.4 supplemented with 2.5 mM MgCl2, 0.035% 2-mercaptoethanol, and Complete EDTA-free protease inhibitor cocktail at the concentration recommended by manufacturer (Roche). Cells were disrupted by sonication on ice and cleared of debris by centrifugation. Total protein concentration was determined using the BioRad Protein Assay. Lysates were diluted to a standard concentration and stored at −80° C.
Enzyme activity was determined using a modification of a method described earlier by our lab (20). Lysates were incubated with 20 μM NADPH (Sigma) and 825 nM [6-C14]-5-Fluorouracil ([6-C14]-5-FU) (Moravek Biochemicals) for 30 minutes at 37° C with constant agitation. Reactions were terminated by the addition of an equivalent volume of ice-cold 100% ethanol, and samples were then frozen at −80 °C. Precipitated material was removed by centrifugation and filtering through a 0.2 μm PVDF Mini-UniPrep syringeless filter (Whatman). Conversion of [6-C14]-FU to [6-C14]-5-Dihydrofluorouracil ([6-C14]-5-DHFU) was determined using two reverse-phase C18 HPLC columns (Grace) connected in serial to a Perkin Elmer Radiomatic 625TR flow scintillation analyzer. DPD activity was calculated by measuring the percent region of interest as the area under the curve for ([6-C14]-5-DHFU) / ([6-C14]-5-FU + [6-C14]-5-DHFU) using ProFSA software. Biological replicates were normalized by z-scores and sample groups compared using two-tailed Student’s t-tests.
Western blotting
Protein lysates were separated on a 10% SDS-PAGE gel and transferred to PVDF-FL membrane (Millipore). Membranes were blocked using Odyssey blocking buffer (LI-COR). Blots were probed with primary antibodies against DPD and alpha-tubulin (both AbCam) and subsequent secondary IRDye800 conjugated goat anti-mouse and IRDye 680 conjugated goat anti-rabbit (both LI-COR). Blots were scanned and band intensities quantified using the LI-COR Odyssey Infrared Imaging system.
Cell viability assays
HEK293T/c17 cells were seeded with a target density of 1×104 cells per well of a 96-well plate. After incubation for 16 hours, cultures were transfected with the indicated expression vectors. Media was removed 24 hours after transfection and replaced with media containing serial dilutions of 5-FU ranging from 1 nM to 20 mM. Viable cell counts were estimated using CellTiter-Blue (Promega) 48 hours after treatment as an indicator of cell proliferation. IC50 was determined by fitting data using a logistic 4P nonlinear model (JMP, SAS Institute Inc.).
Real-Time Cellular Analysis (RTCA)
Cell settlement, morphology, and proliferation were monitored in real-time using an xCELLigence RTCA system (Acea Biosciences). This instrument uses an electrical current that is sent through gold electrodes located on the floor of each well at defined time intervals and the electrical impedances recorded. The electrical impedance readouts are expressed in terms of an arbitrary unit, the Cell Index (CI), and are displayed as kinetic curves. At each time point, the CI is calculated by Zx-Zy / Zy, where Zx is the electrical impedance of the electronic sensor in a particular well containing cells, and Zy is the background impedance of medium alone in that particular well.
Prior to seeding cells, 50 μL media was added to each RTCA plate well and the plates scanned on the xCELLigence to generate a background reading. HEK293T/c17 cells were seeded with a target density of 5×103 cells in 150 μL media per well. Cells were incubated on the xCELLigence in standard cell culture conditions, and CI recorded at 15 minute intervals. After 20-24 hours of culture, cells were transfected with the indicated plasmid amounts in a 1:3 ratio using FuGene HD (Roche). Following transfection, CI was recorded every 2 minutes for 8 hours and then every 15 minutes thereafter. Approximately 20-24 hours after transfection, cells were treated with the indicated concentrations of 5-FU (Sigma) or vehicle. CI was recorded every 2 minutes for 8 hours following treatment and every 15 minutes thereafter.
Objective determination of analysis time points from cell index data
CI profiles were smoothed using a spline smoothing algorithm with λ=1000. The λ coefficient was empirically determined to adequately remove noise from data without altering the overall shape of CI profiles. The slope was subsequently calculated between adjacent data points along the smoothed CI profile and the resultant first order derivative approximation profile was plotted. The change in slope (second derivative of the CI profile) was calculated and used to define statistical endpoints for the slope profile. Time points were established following the initial surge in CI that occurred directly following the media change at the time of 5-FU treatment. The “toxicity” time point was defined as the time when the second derivative of the cell index reached or approached zero. In cases where the second derivative did not reach zero, this time point was observed as a peak on the second derivative curve. The “recovery” time point was similarly defined as the next time when the second derivative of the cell index reached or approached zero. All data analyses and transformations were performed using JMP (SAS Institute Inc.).
Endpoint quantitation at the defined time points
The CI slope was determined for a given time point using the first order derivative approximation of the smoothed CI profile. Data were assessed using a three-effect model to compare the relative contributions of replicate, DPYD variation, and 5-FU treatment level (grouped by DPYD variation) to the observed variability within the overall dataset. In cases where replicate was shown to significantly contribute to model error (P<0.05), data were normalized relative to that for the untreated culture expressing the same DPYD variation within the replicate. In all cases where normalization was deemed necessary, following normalization, replicate no longer contributed to the model variability. All data transformations and analyses were performed using JMP (SAS Institute Inc.).
Statistical tests
Specific statistical tests used for a given experiment are described in the relevant methods subsection above. All data analyses and transformations were performed using JMP (SAS Institute Inc.), unless otherwise noted.
Results
Development of an in vitro system to measure DPD enzymatic activity
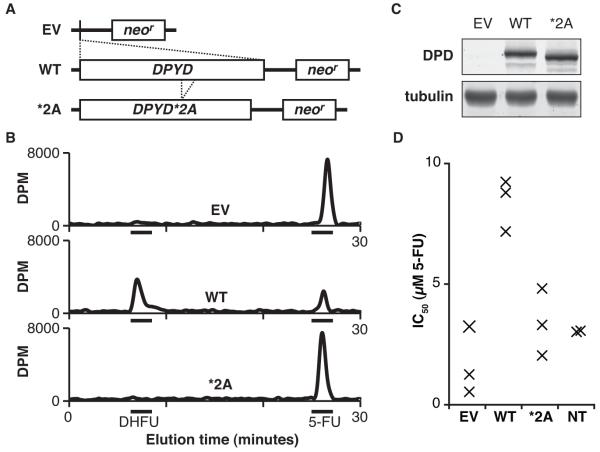
Expression constructs were generated to encode DPYD and a mimic of the *2A splice product lacking the sequence corresponding to exon 14 (Fig. 1A). Constructs were transfected into low passage HEK293T/c17 cells, and crude protein lysates were harvested by sonication after 48 hours of culture. The total protein concentrations of lysates were determined, and samples were diluted to a standard concentration for subsequent analyses. The ability of lysates to catalyze the conversion of radiolabeled 5-FU to DHFU was measured by HPLC as an indicator of DPD enzyme activity (Fig. 1B). Lysates from cells expressing wild type (wt) DPYD were able to efficiently reduce 5-FU to DHFU, whereas lysates from cells transfected with either the empty vector control or the *2A variant did not show any 5-FU conversion to DHFU (Fig. 1B). Expression of wt DPD protein was similar to that of the *2A variant, and no endogenous DPD expression as noted for cell lysates from cells transfected with empty vector (Fig. 1C). Extending the reaction times to 24 hours did not result in any detectible conversion of 5-FU for untransfected cells or for those expressing either the empty vector or *2A (data not shown), further suggesting that HEK293T/c17 cells lack detectible endogenous DPD enzyme activity.
Figure 1. Measurement of DPD enzyme activity in cells transfected with DPYD expression constructs.
A, the human DPYD gene (WT) and the *2A variant lacking sequence corresponding to exon 14 (*2A) were engineered into the pIRES-neo3 expression vector (EV corresponds to the empty parental vector). B, lysates prepared from transfected cells were assayed for DPD-dependent conversion of radiolabeled 5-FU to DHFU as measured by HPLC (DPM, disintegrations per minute). C, DPD expression levels were measured by immunoblot of DPD and alpha-tubulin. D, the concentration of 5-FU that inhibits cell growth by 50% (IC50) was determined by treating transfected and non-transfected (NT) cells with a dilution series of 5-FU and measuring cell viability 48 hours after 5-FU treatment. Each individual data point (represented by an “x”) constitutes the IC50 calculated as the mean of three technical replicates.
To confirm that the transgenically expressed DPD was catalytically active in vivo, the half maximal inhibitory concentration (IC50) for 5-FU was determined (Fig. 1D). Cells transfected with wt DPYD showed significantly higher resistance to 5-FU than untransfected cells (IC50=8.4 μM and 3.0 μM, respectively, P=0.0070). The IC50 for cells transfected with the empty vector and *2A were 1.7 μM and 3.6 μM 5-FU, respectively. These cells were significantly more sensitive to 5-FU than those expressing wt DPYD (P=0.0028 and P=0.0079). IC50 values were not significantly different between empty vector-expressing cells, *2A-expressing cells, and non-transfected cells.
Selection of additional DPYD variants for further functional analyses
We selected five additional DPYD variations for further study. For each of these variants, allele frequencies in the HapMap and 1000 genomes populations are presented in Table 1 along with a summary of references that are relevant to the previously reported contribution of each variation to 5-FU toxicity. For ease of data interpretation, genetic variants are referred to in this manuscript by the amino acid change for which they encode. Of the five selected variants, one (I560S, rs55886062, previously referred to as DPYD*13) has consistently contributed to 5-FU toxicity in clinical reports (21-24). The roles of the additional four selected variants in 5-FU-related toxicities are less clear. The I543V variant (rs1801159, previously referred to as DPYD*5) is widely considered to be a common polymorphism that has not shown association with 5-FU toxicity (25-27), however two reports suggest otherwise (28, 29). The S534N variant (rs1801158, previously referred to as DPYD*4) has an unclear association with 5-FU toxicity. Whereas earlier studies suggested this allele was linked to drug sensitivity (30-33), more recent studies using large clinical cohorts have contradicted this finding (18, 25-27). V732I (rs1801160, previously referred to as DPYD*6) has been shown to contribute to 5-FU toxicity only when the M166V variation (rs2297595) was not present (18). Several other reports have shown no association with toxicity for V732I (25-27). Finally, C29R (rs1801265, previously referred to as DPYD*9A) was identified in a DPD-deficient patient and shown to be catalytically inactive (34, 35); however, clinical studies have failed to establish a link between the variation and 5-FU sensitivity (25-27). Additional studies have suggested that C29R may in fact serve as a protective allele against 5-FU toxicity (18, 32).
Table 1. Allele frequencies of selected DPYD variants.
| Amino acid change |
“Star” allele |
dbSNP identifier |
Minor allele frequency | Previously reported impact on function |
||||
|---|---|---|---|---|---|---|---|---|
| CEU | YRI | HCB | JPT | 1000 genomes |
||||
| Del e14 | *2A | rs3918290 | 0.004 | 0 | 0 | 0 | 0.002 | ↓(18, 21, 26, 45) |
| I560S | *13 | rs55886062 | 0.008 | 0 | 0 | nr | nr | ↓(21-24) |
| I543V | *5 | rs1801159 | 0.165 | 0.133 | 0.293 | 0.244 | 0.204 | ↓(28, 29) = (25-27) |
| S534N | *4 | rs1801158 | 0.058 | 0 | 0 | 0 | 0.012 | ↓(30-33) = (18, 25-27) |
| V732I | *6 | rs1801160 | 0.067 | nr | nr | nr | 0.040 | ↓(18) = (25-27) |
| C29R | *9A | rs1801265 | 0.167 | 0.417 | 0.056 | 0.067 | 0.231 | ↓(34, 35) = (25-27) ↑ (18, 32) |
Relative DPD enzyme activity produced by each of the selected DPYD variants
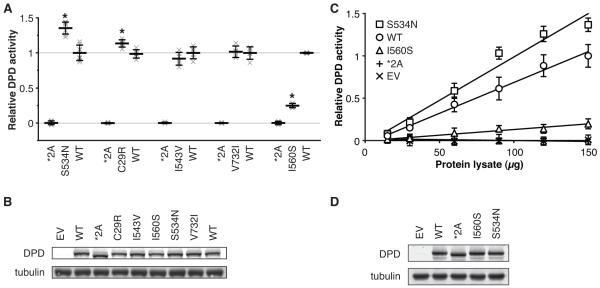
Each selected variant was transiently expressed and assayed for DPD enzyme activity relative to that of wt and the catalytically inactive *2A variation (Fig. 2A). The most striking result was obtained for S534N, which was 36% more active than wt (P=3.4×10−7). C29R was also significantly hyperactive (P=0.0013), exhibiting 13% higher activity than wt. The amino acid substitutions I543V and V732I did not significantly affect enzyme activity. Consistent with previous reports, the I560S substitution impaired DPD enzyme function, showing a 75% reduction in activity relative to wt (P=5.2×10−9). Each variant was expressed at a level similar to that of wt DPD (Fig. 2B).
Figure 2. Screening of five additional single amino acid DPD variants for enzyme activity.
A, cell lysates from cells transfected with vectors encoding the DPD amino acid variants S534N, C29R, I543V, V732I, and I560S were assayed for DPD enzyme activity in parallel with negative (*2A) and positive (WT) controls. Horizontal bars represent the mean of replicate experiments +/− the standard deviation of normalized results. To show the overall variance of data, individual replicate data points are represented by an “x.” Variants that show a significant increase or decrease in DPD enzyme function compared to WT are denoted by an “*”. B, equivalent expression of DPD variants was confirmed by immunoblot against DPD and alpha-tubulin. A representative blot is depicted. C, empty vector (EV), WT, *2A, I560S, and S534N were transfected in parallel and DPD enzyme activity determined using varying quantities of protein lysates. Data was normalized and scaled to the results for the mean of 150 μg *2A and 150 μg WT. Error bars represent the standard deviation of 5 independent biological replicates. D, representative immunoblot depicting equivalent expression of DPD variants used for panel c.
To confirm that the observed results were not due to saturation of the enzymatic reaction, varying amounts of cellular lysate from cells transfected with empty vector, wt, *2A, I560S, and S534N were assayed for DPD activity (Fig. 2C). For all concentrations except the lowest tested, S534N showed significantly higher enzyme activity than that of wt (minimum P=7.1×10−4). The activity of I560S was intermediate between wt and *2A (minimum P=1.8×10−6 and P=8.3×10−5, respectively). Enzyme activity in lysate from cells expressing the *2A variant showed no difference in activity compared to lysate from cells transfected with empty vector. All variants were equally expressed (Fig. 2D). Additionally, the rank order of variations was not altered by changing the amount of DNA transfected (Supplementary Fig. S1A-B) or when expressed in a second cell line (HCT116 cells, data not shown).
DPYD variants determine 5-FU sensitivity
DPYD variant-expressing cells were continuously monitored for changes in cell proliferation using RTCA to closely measure the cellular responses following 5-FU treatment. The culture plates that are used for RTCA are lined with gold electrodes (Fig. 3A). A small electric current is passed through the culture and the impedance recorded. These data are expressed in cell index (CI) units. Changes in CI can indicate a change in cell number, morphology, and/or attachment. Cell death reduces the impedance between electrodes and results in an overall downward trend in the CI profile (Fig. 3B). Conversely, cell proliferation is indicated by an increase in CI.
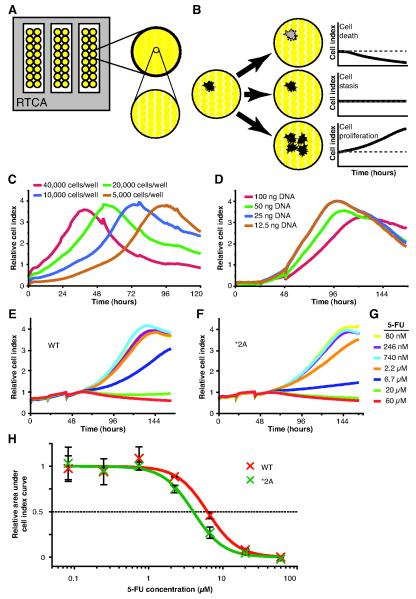
Figure 3. Real-time cellular analysis of variant-expressing cells treated with 5-FU.
A, the surface area of RTCA plates are ~80% covered with gold microelectrodes used for measuring impedance. B, cells attached to the plate surface disrupt conductivity at the electrode-solution interface enabling the kinetic monitoring of changes in the cell population. C, HEK293T/c17 cells were seeded at varying densities and CI monitored. D, cells were transfected with varying amounts of plasmid DNA (empty pIRES-neo3 vector) using a 3:1 ratio of FuGene HD to plasmid. Cells were cultured on RTCA plates for 20 hours and transfected with vectors encoding wt DPYD (WT, E) and the catalytically inactive *2A variant (F). 20 hours after transfection, cells were treated with the concentrations of 5-FU indicated (G). H, area under the curve was determined for each CI profile using an analysis window defined as 8 hours to 96 hours after 5-FU treatment and the mean inhibitory concentration (IC50) of 5-FU was calculated. Data was rescaled relative to the maximum and minimum asymptotes for ease of visualization and is presented +/− the standard deviation of 3 technical replicates. Cell index profiles in C-F represent the average of 3 technical replicates. Data in E and F was normalized to 8 hours post 5-FU treatment using the delta method.
To determine an appropriate number of cells to use in subsequent experiments, various quantities of HEK293T/c17 cells were seeded into the wells of an RTCA plate (Fig. 3C). A greater starting density of cells resulted in an earlier CI plateau, indicative of culture confluency and/or nutrient limitation. Based on these results, a seeding density of 5,000 cells per well was chosen for subsequent experiments. To determine the amount of vector that could be transfected without inducing a change in the CI profile, varying amounts of empty pIRES-neo3 vector were transfected in parallel (Fig. 3D). Cells receiving 100 ng, and to a lesser extent 50 ng, showed altered CI profiles relative to cells transfected with either 25 or 12.5 ng plasmid (Fig. 3D). Based on these results, subsequent experiments were performed by transfecting 25 ng of plasmid per well.
Using the optimized parameters detailed above, cells were transfected with vectors encoding wt DPYD or *2A and subsequently treated with varying concentrations of 5-FU to assess sensitivity to the drug (Fig. 3E-G). The greatest difference CI profiles between wt and *2A was noted following treatment with 6.7 μM 5-FU. At this drug concentration, the CI profile demonstrated less of an upward trend for *2A-expressing cells than for cells expressing wt (Fig. 3E-F). Similar differences were noted between wt and *2A when treated with 2.2 or 20 μM 5-FU. To determine relevant treatment concentrations for subsequent studies, the IC50 concentrations for 5-FU were determined. IC50 concentrations for wt and *2A were 6.9 and 3.9 μM 5-FU, respectively (Fig. 3G). These values were similar to those obtained using a conventional means of measuring cellular sensitivity to 5-FU (Fig. 1D).
RTCA classification of selected DPYD variants by 5-FU sensitivity
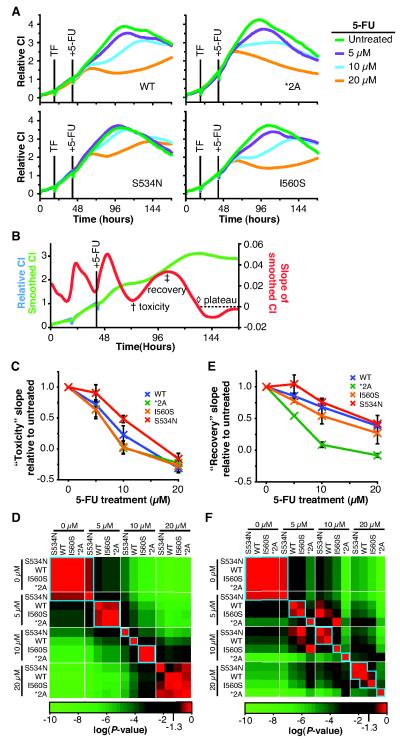
Given the unexpected hyperactivity that resulted from changing amino acid 534 of DPD from serine to asparagine, we decided to more closely study this variation using RTCA. HEK293T/c17 cells were again plated on RTCA plates and transfected as described for Figure 3, except in addition to wt DPYD and *2A, vectors encoding S534N and I560S were also transfected in parallel (Fig. 4A). Based on the IC50 values determined in Figure 3, cells were treated with three concentrations of 5-FU (5, 10, and 20 μM) and monitored for an extended period of time (Fig. 4A). For each variant, an untreated control was included. Experiments were performed as three independent biological replicates, each consisting of three technical replicates. A representative biological replicate is presented in Figure 4A. Relative to untreated cells, the CI profiles for S534N-expressing cells were higher than those for wt, *2A, or I560S at each 5-FU treatment concentration (Fig. 4A). Cells expressing *2A showed the greatest reduction in CI following 5-FU treatment relative to untreated. Overall, CI profiles for wt and I560S were similar, although CI values tended to be slightly higher for wt following treatment with 5, 10, or 20 μM 5-FU.
Figure 4. Classification of functional variants using RTCA to measure relative sensitivity to 5-FU.
A, HEK293T/c17 cells were plated on RTCA plates, cultured for 20 hours, and transfected with expression plasmids encoding wt DPYD (WT), *2A, S534N, or I560S. After 20 hours, media was replaced and cells were treated with the indicated concentrations of 5-FU. Experiments were conducted as three independent biological replicates, each consisting of three technical replicates. A representative biological replicate is presented in A. B, to objectively analyze the kinetic data produced, profiles were smoothed and the CI slope was determined for each data point collected. The relative minimum and maximum CI slopes, following the initial increase due to changing of the culture media, were determined as detailed in Supplementary Figure S3 and were termed the “toxicity” (†) and “recovery” (‡) timepoints, respectively. C, a summary of CI slopes at the toxicity time point is presented normalized to the untreated data for a given variant. Error bars represent the standard deviation of 3 biological replicates. D, P values comparing differences between individual samples were determined using the least squares means differences Student’s t-test and are presented as a heat map. E, the recovery slope was compared for different treatments. Data were normalized and results are presented in the same manner as for panel C. F, a heat map of P values determined for (E) is presented.
The separation between CI profiles was not always evident when using a single window of analysis defined by the kinetics of the untreated samples (Supplementary Fig. S2). To better utilize the time-course data generated during RTCA, we developed a series of data analysis methods in which relevant measurement times were objectively determined for each sample using defined characteristics of the CI profile. The developed methods are detailed in the materials and methods section and Supplementary Figure S3. Briefly, a smoothing algorithm was used to remove irregularities between adjacent points within a CI profile without altering the overall shape of a CI profile (Fig. 4B). The slope of the smoothed CI profile at each time point collected was plotted and yielded two features common to all profiles following drug treatment (Fig. 4B). Based on the location along the CI profile for these two features, they were termed “toxicity” and “recovery” time points. The slope of the CI profile at these time points was measured as a statistical endpoint.
The greatest differences in CI slopes between treatments were noted for the analysis of the “toxicity” time point (Supplementary Fig. S4, summarized in Fig. 4C). When treated with 5 μM 5-FU, cells expressing S534N had a significantly higher slope than those expressing wt, I560S, or *2A (Fig. 4C-D). For cells treated with 10 μM 5-FU, the toxicity slope for S534N remained significantly higher than that of wt, which in turn was significantly greater than that of I560S or *2A. Cells treated with 20 μM 5-FU showed largely similar responses for this endpoint regardless of the DPYD variant expressed.
Comparing the CI slope at the “recovery” time point, significant differences were also noted for drug treatment levels and DPYD variants (Supplementary Fig. S5, summarized in Fig. 4E-F). At the 5 μM treatment level, the recovery slope for S534N was significantly higher than that for wt. Wt was not significantly different than I560S; however the slope was significantly higher than that of *2A. For cultures treated with 10 μM 5-FU, wt and S534N were not significantly different from each other; however both slopes were significantly higher than those for I560S and *2A. The recovery slopes were similar for S534N, wt, and I560S when treated with 20 μM 5-FU. For each of the three treatments with 5-FU, the recovery slope for *2A-expressing cells was significantly lower than that for any other constructs tested.
Evaluating variants expressed as heterozygous alleles
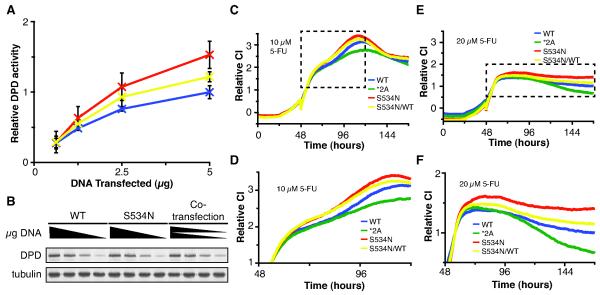
Based on the reported allele frequencies, all of the variants studied are most often detected as heterozygotes. Therefor, we sought to determine if the developed system could be used to determine if variants significantly altered cellular sensitivity to 5-FU when co-expressed with wt DPYD to mimic the heterozygous state. Co-transfection of plasmids encoding wt and S534N resulted in enzyme activity mid way between single transfections (Fig. 5A-B). Similar results were obtained for cellular sensitivity to 5-FU by RTCA (Fig. 5C-F).
Figure 5. Co-transfection studies to evaluate variants present as heterozygous alleles.
A, the indicated quantities of expression plasmids for wt and S534N were transfected into HEK293T/c17 cells and DPD enzyme activity determined for lysates. For co-transfections (S534N/WT), equal quantities of wt and S534N expression plasmids were mixed, and the indicated amount of total plasmid was transfected. Error bars indicate standard deviation of three independent biological replicates. B, expression of DPD and alpha-tubulin was determined. A representative immunoblot is presented. C-F, HEK293T/c17 cells were seeded onto RTCA plates and cultured for 24 hours, at which time they were transfected with the indicated expression plasmids or mixture of plasmids (S534N/WT). After an additional 24 hours, cells were treated with 10 μM (C) or 20 μM (E) 5-FU. The boxed regions in C and E correspond to the enlarged areas presented in D and F, respectively. CI data represents the average values for three technical replicates performed in parallel and normalized to 6 hours after 5-FU treatment using the delta method.
Discussion
In this report, we demonstrate a robust phenotypic assay to functionally classify nonsynonymous coding variations in the context of chemotherapy drug toxicity. The most striking result in this study was obtained for the S534N variant, which possessed greater enzymatic activity than wt in vitro and was more protective against 5-FU toxicity when expressed in cells. To a lesser extent, the more common C29R variation also exhibited increased enzymatic activity. These findings support a model in which hyperactive forms of DPD reduce mean circulating levels of 5-FU by increased drug catabolism. Circulating levels of 5-FU have been correlated with clinical toxicity (36, 37). Consistent with our model, clinical studies by Seck et al. (32) and Kleibl et al. (18) have suggested that C29R is protective against 5-FU toxicity. To our knowledge, the less common S534N variation has not been specifically evaluated as a hyperactive allele in clinical trials; however, based on our results, we speculate that S534N may also be protective against 5-FU toxicity. As a consequence of increased drug clearance, hyperactive alleles may also reduce the effectiveness of 5-FU as an anti-tumor drug.
The mildly hyperactive phenotype we observed for C29R contradicts previous reports that have suggested that the variant is catalytically inactive when expressed in E. coli (35, 38, 39). Bacteria lack many factors necessary for efficient folding and post-translational modification of large multi-domain proteins such as DPD. Additionally, eukaryotic proteins produced in high levels in E. coli have been shown to be subject to inactivation through aggregation into inclusion bodies (40). The lack of enzyme activity for C29R previously reported may have been an artifact of the bacterial expression system used (35, 38, 39).
Structurally, amino acids 534 and 543 of DPD lie at opposite ends of the loop-structure that covers the opening of the 5-FU catabolic domain (Supplementary Fig. S6 and ref. 41). This domain is a member of the dihydroorotate dehydrogenase (PyrD) conserved domain family (42). Based on the conservation at the position corresponding to S534N within the PyrD family, this variation is predicted to impact enzyme function. The PolyPhen-2 (43) prediction for this amino acid change is “probably damaging” with a dScore of +2.13 based on alignment with 106 homologs. The S534N substitution may alter the localized protein structure through hydrogen bond interactions between the asparagine side chain and the peptide backbone. In contrast, the I543V variation is not predicted to impact function since isoleucine and valine are chemically and structurally similar. Given the location within the DPD structure and the increased rate of 5-FU catabolism for the S534N variant, we postulate that this variation affects the structure of the loop at the active site opening, and as a consequence, increases substrate turnover.
To develop this phenotypic assay as a useful tool for genotype/phenotype analysis, we studied two DPYD variations known to be clinically associated with severe toxicity to 5-FU, *2A and I560S (8). In our study, cells expressing either of these variants showed significantly reduced proliferation following treatment with 5-FU. These results are consistent with the numerous clinical reports demonstrating that *2A is a null allele (15, 20, 44). The less common I560S variant has only been reported in a few published cases (21, 22). In one case, I560S was present in a compound heterozygous state with *2A, and resulted in markedly reduced DPD activity and severe toxicity to 5-FU (22). Enzyme function was also severely reduced in a family member carrying I560S but not *2A. Morel et al. (21) detected I560S in a single patient who had experienced grade 3-4 toxicity following chemotherapy containing 5-FU. Taken together with these previous reports, our data suggest that dose reduction, or avoiding the use of 5-FU in favor of other treatment options, may be warranted for carriers of either *2A or I560S.
It has long been recognized that genetics have a major role in the variability of drug metabolism and hence contribute greatly to the drugs efficacy and toxicity. Personalized medicine initiatives strive to tailor individual treatment regimens based on a number of factors, including genetic factors. The results presented in this manuscript support 5-FU dose reduction in patients that carry either the *2A or I560S variants to minimize risk of severe adverse toxicity. Our findings do not suggest that C29R, S534N, I543V, or V732I contribute to DPD deficiency individually. Additionally, the in vivo cellular model presented in this manuscript could be employed to rapidly identify functional alleles of interest in a given pathway and use that information to better individualize therapies. Additional studies currently underway suggest that this model can be expanded to evaluate multiple variations within a gene present in both cis and trans combinations.
Supplementary Material
Acknowledgments
Grant Support
This study was funded by NIH grant CA116964.
Footnotes
Author Contributions
S.M.O. conceived and developed the hypothesis, designed the experimental approach, performed the experimental work, analyzed and interpreted the data, coordinated the project, and wrote the manuscript. N.J.W. performed cell culture studies, interpreted the data, discussed the hypothesis and wrote the manuscript. C.F. and K.W. performed HPLC studies. R.B.D. developed the hypothesis, led the project, and assisted in writing and editing the manuscript.
References
- 1.Rich TA, Shepard RC, Mosley ST. Four decades of continuing innovation with fluorouracil: current and future approaches to fluorouracil chemoradiation therapy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2004;22:2214–32. doi: 10.1200/JCO.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 2.MacMillan WE, Wolberg WH, Welling PG. Pharmacokinetics of fluorouracil in humans. Cancer Res. 1978;38:3479–82. [PubMed] [Google Scholar]
- 3.Heggie GD, Sommadossi JP, Cross DS, Huster WJ, Diasio RB. Clinical pharmacokinetics of 5-fluorouracil and its metabolites in plasma, urine, and bile. Cancer Res. 1987;47:2203–6. [PubMed] [Google Scholar]
- 4.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–8. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 5.Harris BE, Song R, Soong SJ, Diasio RB. Relationship between dihydropyrimidine dehydrogenase activity and plasma 5-fluorouracil levels with evidence for circadian variation of enzyme activity and plasma drug levels in cancer patients receiving 5-fluorouracil by protracted continuous infusion. Cancer Res. 1990;50:197–201. [PubMed] [Google Scholar]
- 6.Fleming RA, Milano G, Thyss A, Etienne MC, Renee N, Schneider M, et al. Correlation between dihydropyrimidine dehydrogenase activity in peripheral mononuclear cells and systemic clearance of fluorouracil in cancer patients. Cancer Res. 1992;52:2899–902. [PubMed] [Google Scholar]
- 7.Etienne MC, Lagrange JL, Dassonville O, Fleming R, Thyss A, Renee N, et al. Population study of dihydropyrimidine dehydrogenase in cancer patients. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1994;12:2248–53. doi: 10.1200/JCO.1994.12.11.2248. [DOI] [PubMed] [Google Scholar]
- 8.Diasio RB, Beavers TL, Carpenter JT. Familial deficiency of dihydropyrimidine dehydrogenase. Biochemical basis for familial pyrimidinemia and severe 5-fluorouracil-induced toxicity. The Journal of clinical investigation. 1988;81:47–51. doi: 10.1172/JCI113308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris BE, Carpenter JT, Diasio RB. Severe 5-fluorouracil toxicity secondary to dihydropyrimidine dehydrogenase deficiency. A potentially more common pharmacogenetic syndrome. Cancer. 1991;68:499–501. doi: 10.1002/1097-0142(19910801)68:3<499::aid-cncr2820680309>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 10.Tuchman M, Ramnaraine ML, O’Dea RF. Effects of uridine and thymidine on the degradation of 5-fluorouracil, uracil, and thymine by rat liver dihydropyrimidine dehydrogenase. Cancer Res. 1985;45:5553–6. [PubMed] [Google Scholar]
- 11.Wadman SK, Berger R, Duran M, de Bree PK, Stoker-de Vries SA, Beemer FA, et al. Dihydropyrimidine dehydrogenase deficiency leading to thymine-uraciluria. An inborn error of pyrimidine metabolism. Journal of inherited metabolic disease. 1985;8(Suppl 2):113–4. doi: 10.1007/BF01811484. [DOI] [PubMed] [Google Scholar]
- 12.Braakhekke JP, Renier WO, Gabreels FJ, De Abreu RA, Bakkeren JA, Sengers RC. Dihydropyrimidine dehydrogenase deficiency. Neurological aspects. J Neurol Sci. 1987;78:71–7. doi: 10.1016/0022-510x(87)90079-7. [DOI] [PubMed] [Google Scholar]
- 13.Wilcken B, Hammond J, Berger R, Wise G, James C. Dihydropyrimidine dehydrogenase deficiency--a further case. Journal of inherited metabolic disease. 1985;8(Suppl 2):115–6. doi: 10.1007/BF01811485. [DOI] [PubMed] [Google Scholar]
- 14.Boisdron-Celle M, Remaud G, Traore S, Poirier AL, Gamelin L, Morel A, et al. 5-Fluorouracil-related severe toxicity: a comparison of different methods for the pretherapeutic detection of dihydropyrimidine dehydrogenase deficiency. Cancer letters. 2007;249:271–82. doi: 10.1016/j.canlet.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Meinsma R, Fernandez-Salguero P, Van Kuilenburg AB, Van Gennip AH, Gonzalez FJ. Human polymorphism in drug metabolism: mutation in the dihydropyrimidine dehydrogenase gene results in exon skipping and thymine uracilurea. DNA Cell Biol. 1995;14:1–6. doi: 10.1089/dna.1995.14.1. [DOI] [PubMed] [Google Scholar]
- 16.van Kuilenburg AB, Hausler P, Schalhorn A, Tanck MW, Proost JH, Terborg C, et al. Evaluation of 5-fluorouracil pharmacokinetics in cancer patients with a c.1905+1G>A mutation in DPYD by means of a Bayesian limited sampling strategy. Clinical pharmacokinetics. 2012;51:163–74. doi: 10.1007/BF03257473. [DOI] [PubMed] [Google Scholar]
- 17.The Single Nucleotide Polymorphism Database (dbSNP) of Nucleotide Sequence Variation. 2012 Aug 10; cited. Available from: http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?showRare=on&chooseRs=coding&go=Go&locusId=1806.
- 18.Kleibl Z, Fidlerova J, Kleiblova P, Kormunda S, Bilek M, Bouskova K, et al. Influence of dihydropyrimidine dehydrogenase gene (DPYD) coding sequence variants on the development of fluoropyrimidine-related toxicity in patients with high-grade toxicity and patients with excellent tolerance of fluoropyrimidine-based chemotherapy. Neoplasma. 2009;56:303–16. doi: 10.4149/neo_2009_04_303. [DOI] [PubMed] [Google Scholar]
- 19.Irelan JT, Wu MJ, Morgan J, Ke N, Xi B, Wang X, et al. Rapid and quantitative assessment of cell quality, identity, and functionality for cell-based assays using real-time cellular analysis. J Biomol Screen. 2011;16:313–22. doi: 10.1177/1087057110397359. [DOI] [PubMed] [Google Scholar]
- 20.Johnson MR, Hageboutros A, Wang K, High L, Smith JB, Diasio RB. Life-threatening toxicity in a dihydropyrimidine dehydrogenase-deficient patient after treatment with topical 5-fluorouracil. Clin Cancer Res. 1999;5:2006–11. [PubMed] [Google Scholar]
- 21.Morel A, Boisdron-Celle M, Fey L, Soulie P, Craipeau MC, Traore S, et al. Clinical relevance of different dihydropyrimidine dehydrogenase gene single nucleotide polymorphisms on 5-fluorouracil tolerance. Mol Cancer Ther. 2006;5:2895–904. doi: 10.1158/1535-7163.MCT-06-0327. [DOI] [PubMed] [Google Scholar]
- 22.Johnson MR, Wang K, Diasio RB. Profound dihydropyrimidine dehydrogenase deficiency resulting from a novel compound heterozygote genotype. Clin Cancer Res. 2002;8:768–74. [PubMed] [Google Scholar]
- 23.van Kuilenburg AB, Dobritzsch D, Meinsma R, Haasjes J, Waterham HR, Nowaczyk MJ, et al. Novel disease-causing mutations in the dihydropyrimidine dehydrogenase gene interpreted by analysis of the three-dimensional protein structure. Biochem J. 2002;364:157–63. doi: 10.1042/bj3640157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loganayagam A, Arenas-Hernandez M, Fairbanks L, Ross P, Sanderson JD, Marinaki AM. The contribution of deleterious DPYD gene sequence variants to fluoropyrimidine toxicity in British cancer patients. Cancer chemotherapy and pharmacology. 2010;65:403–6. doi: 10.1007/s00280-009-1147-x. [DOI] [PubMed] [Google Scholar]
- 25.Amstutz U, Farese S, Aebi S, Largiader CR. Dihydropyrimidine dehydrogenase gene variation and severe 5-fluorouracil toxicity: a haplotype assessment. Pharmacogenomics. 2009;10:931–44. doi: 10.2217/pgs.09.28. [DOI] [PubMed] [Google Scholar]
- 26.Schwab M, Zanger UM, Marx C, Schaeffeler E, Klein K, Dippon J, et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: a prospective clinical trial by the German 5-FU Toxicity Study Group. J Clin Oncol. 2008;26:2131–8. doi: 10.1200/JCO.2006.10.4182. [DOI] [PubMed] [Google Scholar]
- 27.Gross E, Busse B, Riemenschneider M, Neubauer S, Seck K, Klein HG, et al. Strong association of a common dihydropyrimidine dehydrogenase gene polymorphism with fluoropyrimidine-related toxicity in cancer patients. PloS one. 2008;3:e4003. doi: 10.1371/journal.pone.0004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Li YM, Jin X. DPYD*5 gene mutation contributes to the reduced DPYD enzyme activity and chemotherapeutic toxicity of 5-FU: results from genotyping study on 75 gastric carcinoma and colon carcinoma patients. Med Oncol. 2007;24:251–8. doi: 10.1007/BF02698048. [DOI] [PubMed] [Google Scholar]
- 29.Zhang XP, Bai ZB, Chen BA, Feng JF, Yan F, Jiang Z, et al. Polymorphisms of dihydropyrimidine dehydrogenase gene and clinical outcomes of gastric cancer patients treated with fluorouracil-based adjuvant chemotherapy in Chinese population. Chin Med J (Engl) 2012;125:741–6. [PubMed] [Google Scholar]
- 30.Lazar A, Mau-Holzmann UA, Kolb H, Reichenmiller HE, Riess O, Schomig E. Multiple organ failure due to 5-fluorouracil chemotherapy in a patient with a rare dihydropyrimidine dehydrogenase gene variant. Onkologie. 2004;27:559–62. doi: 10.1159/000081338. [DOI] [PubMed] [Google Scholar]
- 31.Collie-Duguid ES, Etienne MC, Milano G, McLeod HL. Known variant DPYD alleles do not explain DPD deficiency in cancer patients. Pharmacogenetics. 2000;10:217–23. doi: 10.1097/00008571-200004000-00002. [DOI] [PubMed] [Google Scholar]
- 32.Seck K, Riemer S, Kates R, Ullrich T, Lutz V, Harbeck N, et al. Analysis of the DPYD gene implicated in 5-fluorouracil catabolism in a cohort of Caucasian individuals. Clinical cancer research: an official journal of the American Association for Cancer Research. 2005;11:5886–92. doi: 10.1158/1078-0432.CCR-04-1784. [DOI] [PubMed] [Google Scholar]
- 33.Gross E, Ullrich T, Seck K, Mueller V, de Wit M, von Schilling C, et al. Detailed analysis of five mutations in dihydropyrimidine dehydrogenase detected in cancer patients with 5-fluorouracil-related side effects. Human mutation. 2003;22:498. doi: 10.1002/humu.9201. [DOI] [PubMed] [Google Scholar]
- 34.Vreken P, Van Kuilenburg AB, Meinsma R, van Gennip AH. Identification of novel point mutations in the dihydropyrimidine dehydrogenase gene. J Inherit Metab Dis. 1997;20:335–8. doi: 10.1023/a:1005357307122. [DOI] [PubMed] [Google Scholar]
- 35.Vreken P, Van Kuilenburg AB, Meinsma R, van Gennip AH. Dihydropyrimidine dehydrogenase (DPD) deficiency: identification and expression of missense mutations C29R, R886H and R235W. Hum Genet. 1997;101:333–8. doi: 10.1007/s004390050637. [DOI] [PubMed] [Google Scholar]
- 36.Grem JL, McAtee N, Murphy RF, Balis FM, Steinberg SM, Hamilton JM, et al. A pilot study of interferon alfa-2a in combination with fluorouracil plus high-dose leucovorin in metastatic gastrointestinal carcinoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1991;9:1811–20. doi: 10.1200/JCO.1991.9.10.1811. [DOI] [PubMed] [Google Scholar]
- 37.Fety R, Rolland F, Barberi-Heyob M, Hardouin A, Campion L, Conroy T, et al. Clinical impact of pharmacokinetically-guided dose adaptation of 5-fluorouracil: results from a multicentric randomized trial in patients with locally advanced head and neck carcinomas. Clinical cancer research: an official journal of the American Association for Cancer Research. 1998;4:2039–45. [PubMed] [Google Scholar]
- 38.Vreken P, van Kuilenburg AB, Meinsma R, van Gennip AH. Dihydropyrimidine dehydrogenase deficiency. Identification of two novel mutations and expression of missense mutations in E. coli. Adv Exp Med Biol. 1998;431:341–6. [PubMed] [Google Scholar]
- 39.Vreken P, van Kuilenburg AB, Meinsma R, Beemer FA, Duran M, van Gennip AH. Dihydropyrimidine dehydrogenase deficiency: a novel mutation and expression of missense mutations in E. coli. Journal of inherited metabolic disease. 1998;21:276–9. doi: 10.1023/a:1005380525218. [DOI] [PubMed] [Google Scholar]
- 40.Georgiou G, Valax P. Expression of correctly folded proteins in Escherichia coli. Curr Opin Biotechnol. 1996;7:190–7. doi: 10.1016/s0958-1669(96)80012-7. [DOI] [PubMed] [Google Scholar]
- 41.Dobritzsch D, Schneider G, Schnackerz KD, Lindqvist Y. Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J. 2001;20:650–60. doi: 10.1093/emboj/20.4.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic acids research. 2011;39:D225–9. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei X, McLeod HL, McMurrough J, Gonzalez FJ, Fernandez-Salguero P. Molecular basis of the human dihydropyrimidine dehydrogenase deficiency and 5-fluorouracil toxicity. J Clin Invest. 1996;98:610–5. doi: 10.1172/JCI118830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.