Abstract
Purpose
The receptor tyrosine kinase (RTK) c-MET and its ligand hepatocyte growth factor (HGF) are deregulated and promote malignancy in cancer and brain tumors. Consequently, clinically applicable c-MET inhibitors have been developed. The purpose of this study was to investigate the not well known molecular determinants that predict responsiveness to c-MET inhibitors, and to explore new strategies for improving inhibitor efficacy in brain tumors.
Experimental design
We investigated the molecular factors and pathway activation signatures that determine sensitivity to c-MET inhibitors in a panel of glioblastoma and medulloblastoma cells, glioblastoma stem cells (GSCs), and established cell line-derived xenografts using functional assays, reverse protein microarrays, and in vivo tumor volume measurements, but validation with animal survival analyses remains to be done. We also explored new approaches for improving the efficacy of the inhibitors in vitro and in vivo.
Results
We found that HGF co-expression is a key predictor of response to c-MET inhibition among the examined factors, and identified an ERK/JAK/p53 pathway activation signature that differentiates c-MET inhibition in responsive and non-responsive cells. Surprisingly, we also found that short pre-treatment of cells and tumors with exogenous HGF moderately but statistically significantly enhanced the anti-tumor effects of c-MET inhibition. We observed a similar ligand-induced sensitization effect to an EGFR small molecule kinase inhibitor.
Conclusions
These findings allow the identification of a subset of patients that will be responsive to c-MET inhibition, and propose ligand pre-treatment as a potential new strategy for improving the anti-cancer efficacy of RTK inhibitors.
Keywords: Brain tumors, Receptor tyrosine kinases inhibitors, c-MET, Hepatocyte growth factor, Oncogene addiction
INTRODUCTION
The receptor tyrosine kinase (RTK) c-MET and its ligand hepatocyte growth factor (HGF) are key determinants of malignancy in human cancers including brain tumors. c-MET is aberrantly activated in high grade gliomas and embryonal brain tumors and activation is associated with poor clinical outcomes (1, 2). c-MET activation enhances malignancy by inducing cell proliferation, survival, migration, invasion, promoting tumor angiogenesis, and supporting a stem cell phenotype (3–7). Inhibiting endogenous c-MET and/or HGF in experimental tumors leads to growth inhibition and tumor regression (8–14). The oncogenic effects of c-MET are mediated by signaling networks that include the Ras/MAPK and PI3K/Akt pathways (15–18). c-MET pathway activation is regulated by several factors including ligand and receptor expressions, cross talk with the epidermal growth factor receptor (EGFR) (19–22), and modulation of c-MET-dependent signaling by the tumor suppressor PTEN (23, 24).
Clinically translatable c-MET inhibitors have been developed (13, 25–27). Prominent among these are small-molecules that target the catalytic activity of the kinase. Four selective kinase inhibitors (PF-02341066, ARQ197, JNJ-38877605, PF-04217903) and four broad-spectrum kinase inhibitors (XL184, PM470, MGCD265, MK-2461) have entered initial clinical evaluations (28). PF-02341066, also known as Crizotinib (METi) (Pfizer), is a clinically applicable, potent and selective ATP competitive small molecule kinase inhibitor of c-MET (13, 29). The sensitivity of cancer cells to c-MET inhibition varies and the factors that determine this sensitivity have not been systematically studied to date (30). However, the efficient use of c-MET inhibitors requires an understanding of these factors in order to identify responsive patients.
In this study, we used a panel of brain tumor cell lines, primary cells, glioblastoma stem cells and xenografts to investigate the factors that determine sensitivity to c-MET inhibition. We found that HGF co-expression is a key predictor of sensitivity to METi among the tested factors and identified an ERK/JAK/p53 pathway activation signature that differentiates responsive from non-responsive tumor cells. Furthermore, we discovered for the first time that short-term exogenous HGF treatment of tumor cells and xenografts sensitizes them to METi. Similarly, short-term exogenous EGF treatment of sensitized tumor cells to an EGFR kinase inhibitor. These findings identify a subset of tumors that are more likely to respond to c-MET inhibition, and uncover ligand pre-treatment as a potential new strategy for improving the efficacy of RTK inhibitors.
MATERIALS AND METHODS
Reagents and cells
The c-MET kinase inhibitor METi (PF-2341066) [(R)-3-[1-(2,6-dichloro-3-fluoro-phenyl)-ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)-pyridin-2-ylamine] was from Pfizer. The EGFR inhibitor, Erlotinib, was from Sigma (St Louis, MO). Human glioblastoma cell lines (U87, A172, U373, T98G, U1242, SF-767), primary cells (GBM-6, GBM-10), glioblastoma stem cells (GSCs) (1228, 0308) and medulloblastoma cell lines (DAOY, PFSK, D425, ONS-76), and xenografts were used in this study. U87, A172, U373, T98G, and DAOY were from American Type Culture Collection (Manassas, VA). U1242 and SF-767 were kind gifts from Dr. Isa Hussaini (University of Virginia), Dr. Jasti Rao (University of Illinois), and Dr. Russel Pieper (UCSF), respectively. Primary glioblastoma cells (GBM-6 and GMB-10 at 5–10 passages), a gift from Dr. Jann Sarkaria (Mayo Clinic), were isolated from patients who underwent surgery at the Mayo clinic and were molecularly and functionally characterized (31, 32). GSCs 1228 and 0308 were a gift from Dr. Howard Fine (National Institutes of Health) (33). PFSK, D425, and ONS-76 cells were a gift from Dr. Charles Eberhart (Johns Hopkins University). The cells were cultured as described in the supplemental Methods.
Treatment with METi and Erlotinib
The cells were grown overnight in low-serum media and treated with METi (30–300 nM) or Erlotinib (2 µM) for 1 hr prior to treatment with or without 20 ng/ml HGF or 100 ng/ml EGF, respectively. For in vivo sensitivity studies, mice with established intracranial glioblastoma xenografts were randomly separated into control and experimental groups. Treatment groups consisted of vehicle water or 25 mg/kg METi administered by daily oral gavage from day 7 – 28 post-tumor implantation. For METi in vivo sensitization studies, subcutaneous GSC xenografts were injected with either PBS or HGF (400 ng/cm3) for 2 hrs prior to treatment with METi (20 µl/cm3 tumor volume). The animals were treated for 2 weeks before tumor removal and measurement of tumor weight.
Immunoblotting
Quantitative immunoblotting was performed to measure protein expression and phosphorylation as previously described (3). Antibodies were used that are specific for c-MET, phospho-MET (p-MET), HGF, EGFR, p-EGFR (Santa Cruz Biotechnologies, Santa Cruz, CA) and PTEN (Cell Signaling Technology, Danvers, MA). All blots were stripped and re-probed with β-actin antibody (Santa Cruz Biotechnologies, Santa Cruz, CA) as loading control. All antibodies were used at a 1:1000 dilution. Quantitative analysis was performed by densitoMETry on film (BioRad GS 800) and normalized to the β-actin measured on the same blots. Two independent immunoblotting experiments were performed for all cell lines and quantified protein levels were averaged.
Cell death and cell proliferation assays
Cell death and apoptosis were assessed by AnnexinV-PE/7AAD flow cytometry as previously described (3). Cell samples were analyzed on a FACsan and dead cell and apoptotic fractions were determined. Cell proliferation was assessed by cell counting for five days as previously described (3). All experiments were performed three times using triplicate samples.
Reverse Phase Protein Microarray construction and analysis
Pathway activation mapping was performed by reverse phase protein microrray as previously described (34–36). The cells were treated with METi (300nM) or vehicle control in triplicates for 24 hrs and lysed (more details in Supplemental methods). Protein signaling analytes were chosen for analysis based on their previously described involvement in key aspects of c-MET-associated tumor biology. Detection was performed using a fluorescence-based tyramide signal amplification strategy using Streptavidin-conjugated IRDye680 (LI-COR Biosciences, Lincoln NE) as detection reagent. All antibodies were validated for single band specificity and for ligand-induction (for phospho-specific antibodies) by immunoblotting prior to use on the arrays as previously described (34–36). Each array was scanned using a TECAN LS (Vidar Systems Corporation, Herndon VA). After scanning, spot intensity was analyzed, data were normalized to total protein and a standardized, single data value was generated for each sample on the array by MicroVigene software V2.999 (VigeneTech, North Billerica, MA).
In vivo experiments
In vivo sensitivity of glioblastoma cells to METi was assessed using an orthotopic xenograft mouse model. U87 (3 × 105) and T98G (6 × 105) were stereotactically implanted into the right striatum of immunodeficient SCID mice (n=6 per treatment group). One week after implantation, the animals were treated with oral gavage of METi or control every day for three weeks at 25 mg/kg body weight. Four weeks after tumor implantation, the maximal tumor cross-sectional area was determined by computer-assisted image analysis of H&E stained brain cross sections, and the tumor volume was calculated using the formula: volume = (square root of maximal tumor cross-sectional area)3 (37).
Flank xenografts were used to determine the effects bolus HGF injections on responses to METi. GSCs 1228 (3×105) in 50% matrigel were injected into the flanks of SCID mice (n=6 per treatment group). When the tumors reached ~ 5 mm diameter, they were treated with intratumoral injections of HGF (400 ng/cm3 tumor volume) or control for 2 hrs prior to treatment with METi (5 µM at 20 µl/cm3 tumor volume). The animals were treated for 2 weeks. Tumor diameters were measured every other day and tumors were weighted at the end of the experiment.
Statistical analyses
The statistical association between apoptosis (or proliferation) and protein expression were evaluated both with regression and Spearman (rank-based) correlation analyses. The latter analysis was performed to robustly evaluate statistical significance of such association even though the protein expression values were normally distributed for the regression analysis. To evaluate the statistical significance of the difference between each treated and control animal groups in vivo, we used both two-sample t-test and non-parametric Wilcoxen rank-sum test.
The continuous variable reverse phase protein microrray data generated were subjected to both unsupervised and supervised statistical analyses. Statistical analyses were performed on final microarray intensity values obtained using R version 2.9.2 software (The R Foundation for Statistical Computing). Quartile determination was performed to determine which cell lines and treatment groups fell into the highest (75th percentile) and lowest (25th percentile) of HGF expression. If the distribution of variables for the analyzed groups were normal, a two-sample t-test was performed. If the variances of two groups were equal, two-sample t-test with a pooled variance procedure was used to compare the means of intensity between two groups. Otherwise, two-sample t-test without a pooled variance procedure was adopted. For non-normally distributed variables, the Wilcoxon rank sum test was used. Significance levels were set at p < 0.05.
RESULTS
c-MET kinase inhibitor METi completely inhibits c-MET activation in all brain tumor cells
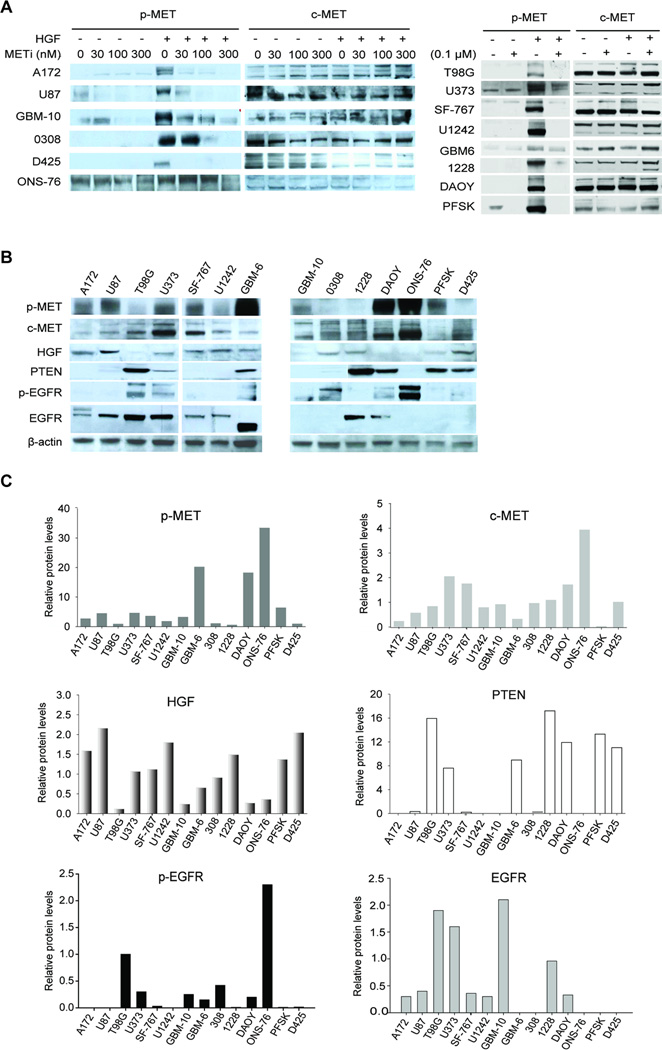
We tested the efficacy of the c-MET kinase inhibitor METi in inhibiting c-MET activation to ensure that differences in anti-tumor response are not caused by differences in c-MET inhibition levels. We treated fourteen different brain tumor cells with METi and measured c-MET phosphorylation relative to total c-MET protein using quantitative immunoblotting. METi was applied at different concentrations (30 nM to 300 nM) for 1 hour prior to treatment of the cells with (or without) HGF (20 ng/ml) for 5 min. The cells included glioblastoma cells (U87, A172, T98G, U373, SF-767, and U1242), GSCs (0308 and 1228), glioblastoma primary cells (GBM-6 and GBM-10) and medulloblastoma cells (DAOY, ONS-76, D425, and PFSK). METi completely or almost completely inhibited both basal and HGF-induced c-MET phosphorylation at 300 nM in all tested cells (Fig. 1A). We therefore used this concentration in all subsequent experiments.
Figure 1. METi effects on c-MET activation, and molecular backgrounds of brain tumor cells.
A The glioblastoma cell lines U87, A172, T98G, U373, SF-767 and U1242, primary cells GBM-10 and GBM-6, GSCs 0308 and 1228, and the medulloblastoma cell lines D425, ONS-76, DAOY and PFSK were treated with METi prior to treatment with HGF. Protein lysates were immunoblotted for phosphorylated (p-MET) or total c-MET. The results show that METi inhibits basal and HGF-induced c-MET phosphorylation in all cells. B Protein lysates from the above cells were subjected to immunoblotting for phospho-c-MET (p-MET), c-MET, HGF, PTEN, phosphor-EGFR (p-EGFR), EGFR and β-Actin using specific antibodies. C Protein levels from the blots described in (B) were quantified by densitometry on film. The data show substantial variability in the expressions of the different proteins in the different cells.
The expressions of c-MET, p-MET, HGF, EGFR, p-EGFR and PTEN vary in human glioblastoma and medulloblastoma cells
Based on published literature on c-MET functions, regulation, signaling and interactions, we focused on c-MET, p-MET, HGF, PTEN, EGFR and p-EGFR levels as the most likely predictors of anti-tumor responses to c-MET inhibition. We first used quantitative immunoblotting to measure the levels of the above proteins in all cells. Expression levels of the proteins varied significantly between the different cells (Fig. 1B & 1C). We also analyzed all cells for c-MET gene amplification, another potential predictor of responsiveness to c-MET inhibitors. No gene amplification was detected in any of the cells (data not shown). Overall, the cells exhibited significant variation in the factors that are most likely to predict responsiveness to c-MET inhibitors.
The effects of METi on death and apoptosis vary between the different brain tumor cells
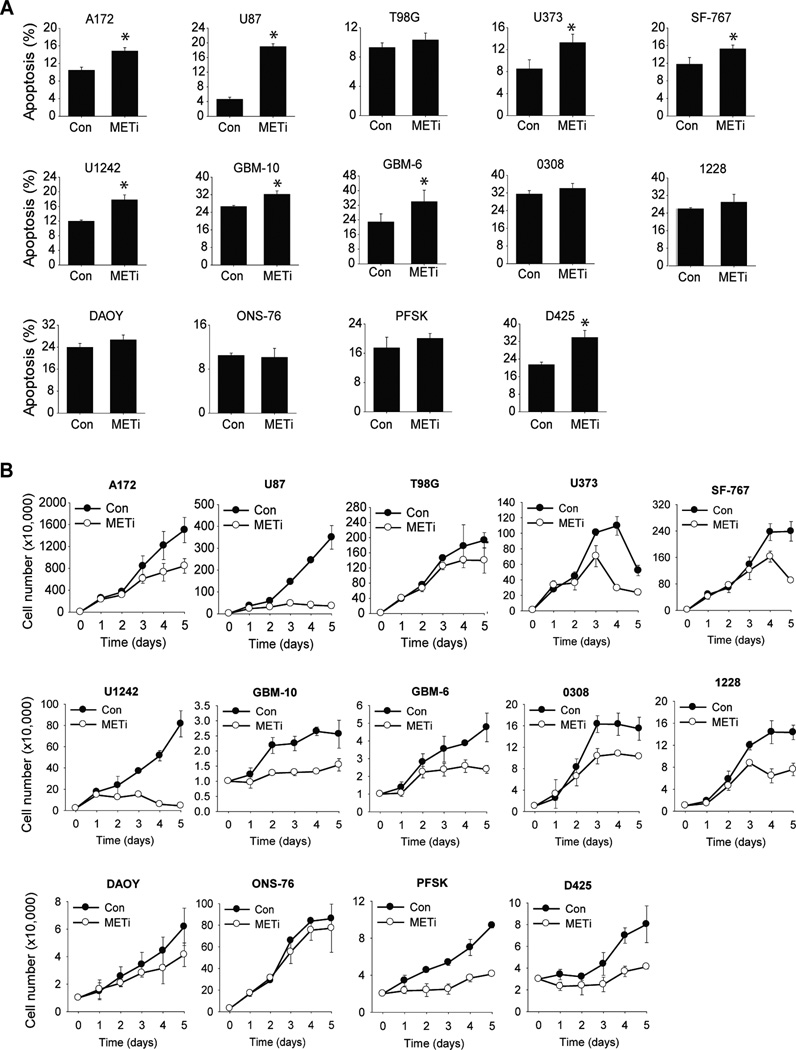
To investigate the effects of METi on brain tumor cell death and apoptosis, cells were treated with METi (300 nM) for 48 hours and assessed for cell death and apoptosis by AnnexinV/7AAD flow cytometry (n=3). Total cell death and apoptosis levels induced by METi varied between the different cells. METi induced strong apoptosis in some cells such as U87, GBM-6 and D425 but had no effect on other cells such as ONS-76 (Fig. 2A). These results indicate that METi induces brain tumor cell death at various degrees in the different cells.
Figure 2. The effects of METi on cell death / apoptosis and proliferation vary between the different brain tumor cells.
A The glioblastoma cell lines U87, A172, T98G, U373, SF-767 and U1242, primary cells GBM-10 and GBM-6, GSCs 0308 and 1228, and the medulloblastoma cell lines D425, ONS-76, DAOY and PFSK were treated with METi or control. The cells were subsequently assessed for apoptosis and cell death by AnnexinV/7AAD flow cytometry and the percentage of dead cells was determined. B The same brain tumor cells as in (A) were treated with METi or control. The cells were subsequently assessed for proliferation by cell counting over a period of 5 days and growth curves were established. All experiments were performed in triplicates and repeated three times.
The effects of METi on proliferation vary between the different brain tumor cells
To investigate the effects of METi on brain tumor cell proliferation, cells were incubated with METi (300 nM) for five days in 0.01% FBS and counted every day. The experiments were repeated three times and growth curves were established. The inhibitory effect of METi on proliferation varied between the different tumor cell lines. METi strongly inhibited the proliferation of some cells such as U87, GBM-6 and D425 but had modest or no effects on other cells such as ONS-76 (Fig. 2B). Overall, the differential effects of METi on tumor cell proliferation were stronger than those on cell death but similar in their differential effects between the different cell lines. These results indicate that METi exerts differential inhibitory effects on cell proliferation in different cells.
HGF co-expression determines sensitivity to c-MET kinase inhibitor
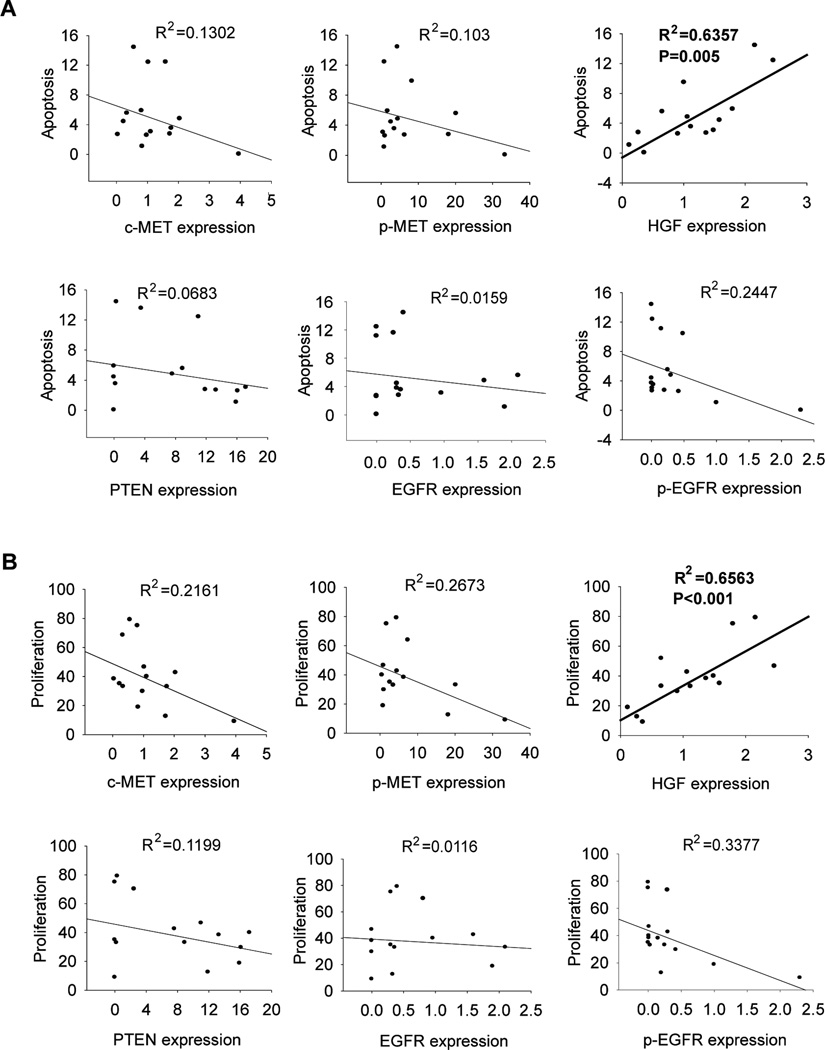
To identify the factor(s) that determine responsiveness of brain tumor cells to METi-induced cell death and inhibition of cell proliferation, we analyzed the correlations between p-MET, c-MET, HGF, PTEN, EGFR, p-EGFR and responsiveness to METi in all 14 brain tumor cell lines using both regression and Spearman correlation analyses as described in the METhods. Responsiveness of cells to METi anti-survival or anti-proliferative effects did not correlate with c-MET, p-MET, PTEN, EFGR or p-EGFR expression levels (Fig. 3A & 3B). However, there was a statistically significant correlation between HGF expression levels and both METi-induced cell apoptosis and death (Adjust R square = 0.6357, p<0.005; Spearman correlation p = 0.005) and METi-induced inhibition of cell proliferation (Adjust R square = 0.6563, p-value < 0.001; Spearman correlation p-value < 0.001) (Fig. 3, Supplemental Table 1). These data show that HGF co-expression is an important determinant of responsiveness of brain tumor cells to c-MET inhibition.
Figure 3. HGF co-expression determines sensitivity to c-MET kinase inhibitor.
Analysis of correlations between the levels of METi-induced inhibition of cell survival A and proliferation B with the expressions of c-MET, p-MET, HGF, PTEN, p-EGFR and EGFR using both regression and Spearman correlation analyses. The results show that METi-induced inhibition of proliferation and survival statistically significantly correlates with HGF expression but not with c-MET, p-MET, PTEN, p-EGFR or EGFR.
Pathway activation mapping of METi effects in high HGF and low HGF expressing cells
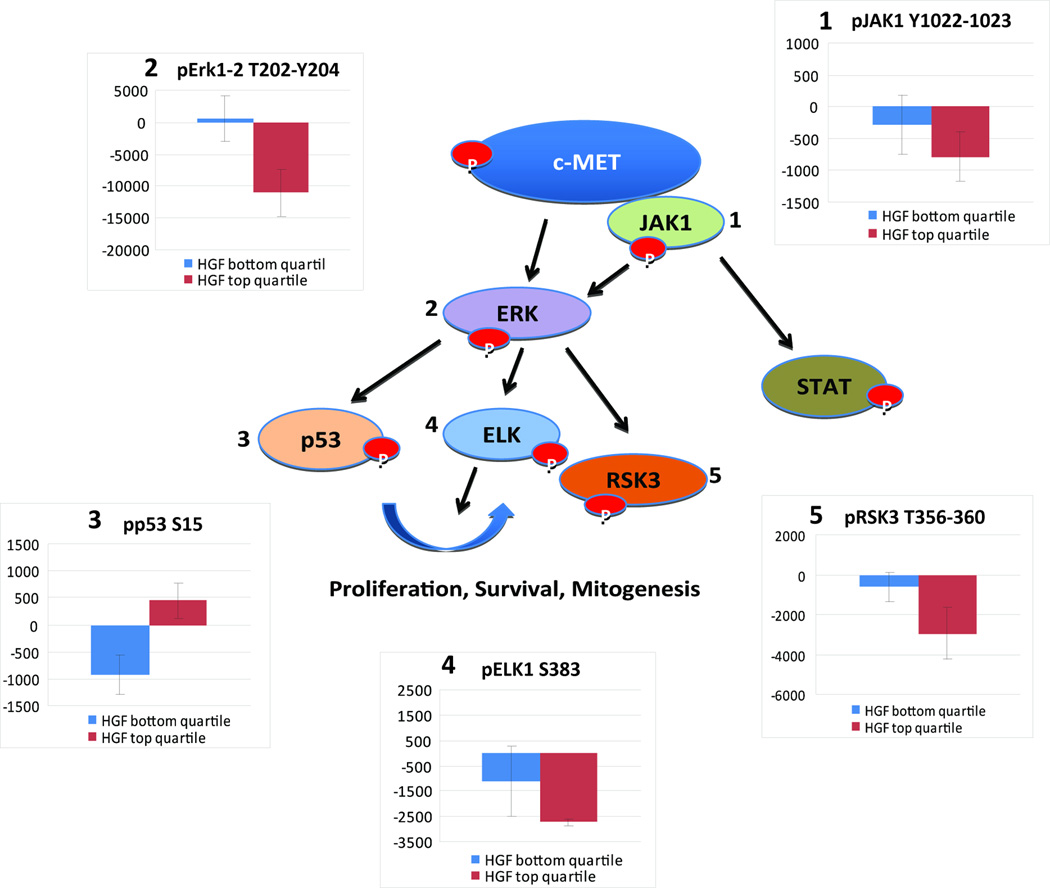
To investigate the cell signaling pathway(s) that might predict and mediate the responses to METi, we conducted a pathway activation mapping analysis using Reverse Phase Protein Microarrays. Phosphorylation/activation mapping by Reverse Phase Protein Microarray was obtained after 24 hr treatment with METi (300 nM) or vehicle on three cell lines that comprised the top quartile of the highest HGF expression (U1242, U87, A172) and on four cell lines that comprised the bottom quartile of HGF expression (ONS76, DAOY, GBM-10 and T98G). Of the key signaling proteins measured, statistically significant differences between the effects of c-MET inhibitor treatment (calculated as the difference between triplicate independent experiments of the treatment – control/untreated) were found for 5 biochemically interconnected ERK pathway-linked signaling proteins that are found downstream of the c-MET receptor: JAK (Y1022/Y1023) p=0.04; ERK 1,2 (T202/Y204) p= 0.01; p53 (S15) p=0.06; ELK (S383) p=0.03; and RSK3 (T356/S360) p=0.05 (Fig. 4, Supplemental Table 2). These data reveal a molecular activation signature that differentiates the responses to METi of high-HGF from low HGF cells and provide new insights into the pathways that mediate the effects of c-MET inhibition in METi-responsive tumors.
Figure 4. Pathway activation mapping of c-MET inhibitor effects in high HGF and low HGF expressing cell lines.
Cells were treated with METi or control and phosphorylation/activation mapping by Reverse Phase Protein Microarray (RPMA) was obtained on three cell lines that comprised the top quartile of the highest HGF expression (U1242, U87, A172) and on four cell lines that comprised the bottom quartile of HGF expression (ONS-76, DAOY, GFM-10 and T98G). Statistically significant differences between the effects of c-MET inhibitor treatment (calculated as the difference between triplicate independent experiments of the treatment – control/untreated) were found for 5 biochemically interconnected ERK pathway-linked signaling proteins that are found downstream of c-MET: JAK (Y1022/Y1023) p=0.04; ERK 1,2 (T202/Y204) p=0.01; p53 (S15) p=0.06; ELK (S383) p=0.03; and RSK3 (T356/S360) p=0.05. The figure reveals the biochemically interlinked signature of differential treatment response in the context of high vs. low HGF production along with the RPMA data results as a histogram for each of the proteins.
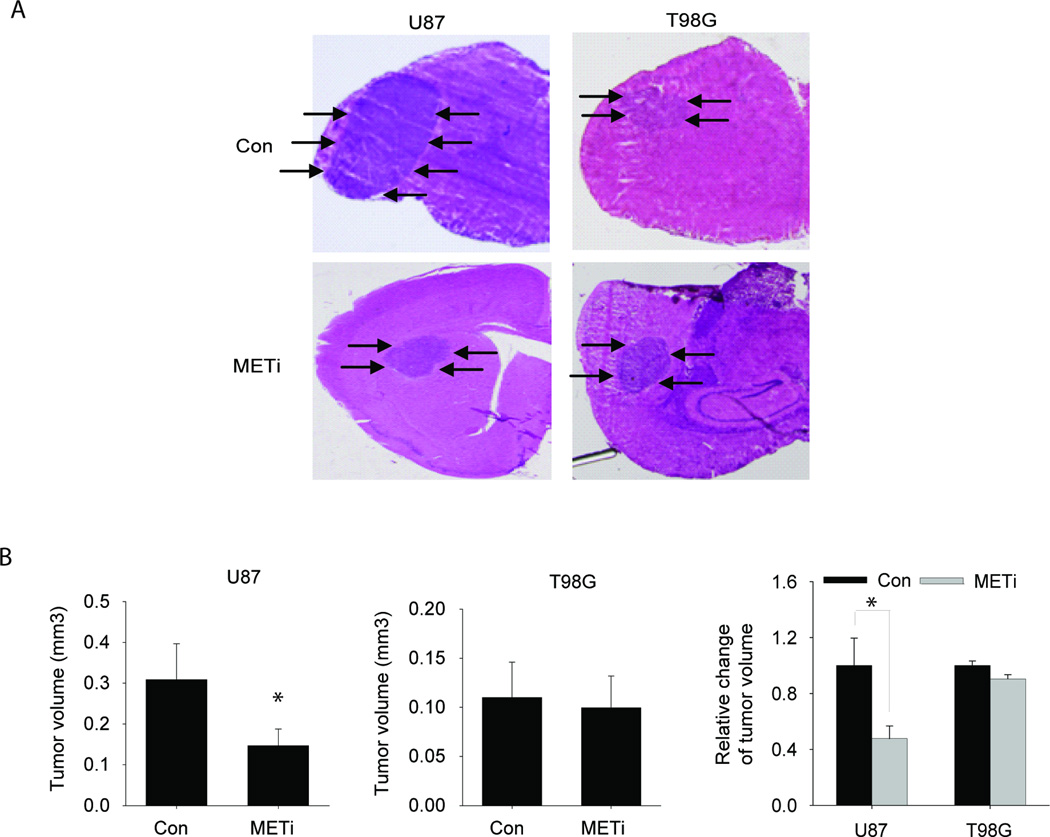
The in vivo anti-tumor effects of METi are significantly greater in high HGF-expressing glioblastoma xenografts than in low- HGF-expressing xenografts
To determine if HGF expression also predicts sensitivity to METi in vivo, we selected two cell lines that are known to generate in vivo glioblastoma xenografts in immunodeficient mice. One of the cell lines expresses high HGF (U87) and the other expresses low HGF (T98G). We generated xenografts from the two cell lines. One week after implantation, tumor formation was verified by MRI (not shown) and the animals were treated with oral gavage of METi (25 mg/kg body weight) or vehicle control everyday for three weeks. The mice were euthanized 4 weeks after tumor implantation. Tumor sizes were measured on H&E-stained maximal brain cross-sectional areas using computer-assisted image analysis. METi significantly inhibited the in vivo growth of high HGF-expressing U87-derived xenografts, but did not affect the growth of low HGF-expressing T98G-derived xenografts (Fig. 5A). Oral delivery of METi led to a decrease of 53% of U87 tumor volume (p<0.01) but did not significantly alter the volume of T98G tumors (p=0.64) (Fig. 5B). These data show that HGF expression also predicts in vivo responsiveness of brain tumor xenografts to c-MET inhibition by METi. However, the application of the tumor volume measurements to clinically relevant animal survival endpoints is yet to be determined.
Figure 5. The in vivo anti-tumor effects of METi are significantly greater in high HGF-expressing glioblastoma xenografts than in low-expressing xenografts.
Glioblastoma cells U87 with high HGF-expression and T98G with low HGF-expression were stereotactically implanted in the striatum of immunodeficient mice (n=6 for each treatment group). METi or vehicle control was administered by daily oral gavage starting one week post-tumor implantation. The animals were euthanized 4 weeks after tumor implantation and the volumes of all tumors were measured. The results show that METi significantly inhibited the growth of U87 xenografts but not the growth of T98G xenografts. A Representative brain cross sections with implanted xenografts (arrows). B Quantification of tumor sizes. *, p<0.05.
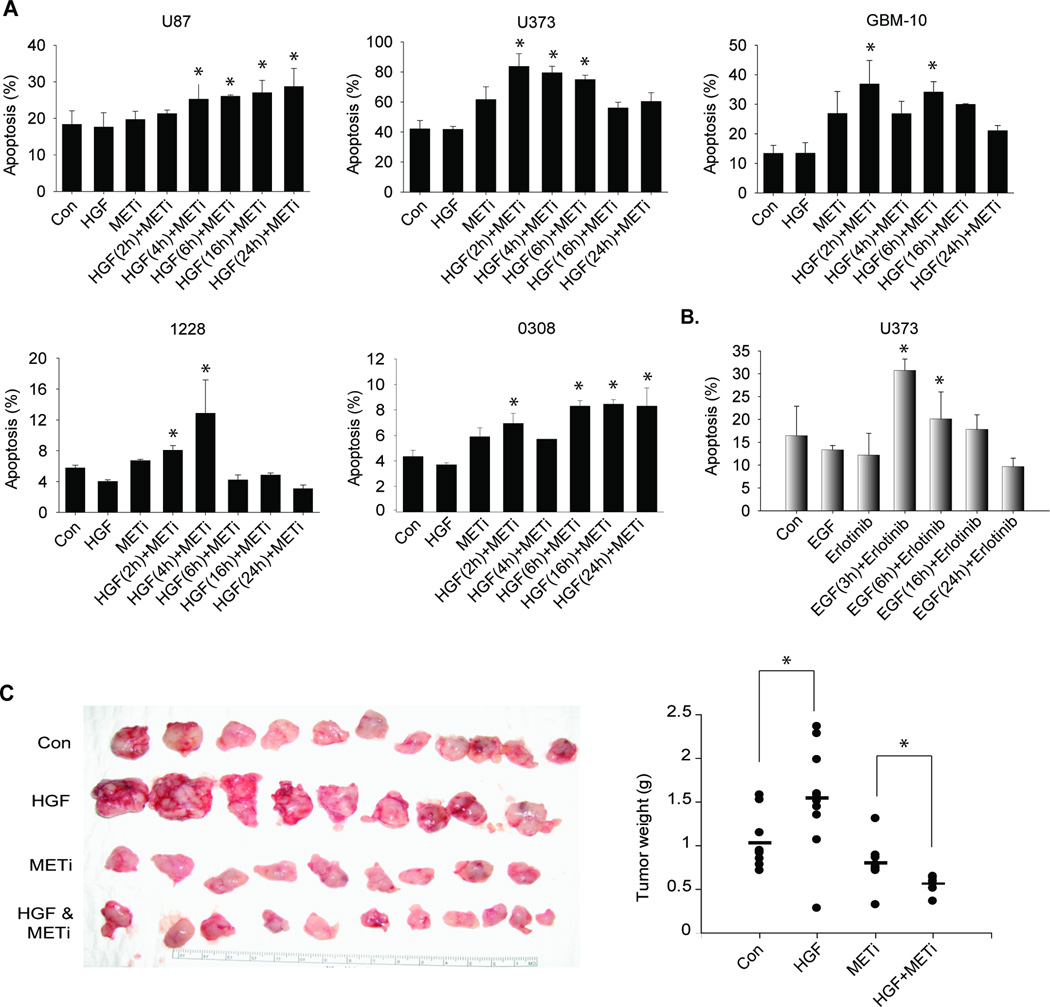
Brief exogenous HGF/EGF treatment sensitizes glioblastoma cells and GSCs to RTK inhibitor-induced apoptosis
While investigating the effects of METi on glioblastoma cell and GSCs malignancy, we surprisingly noticed that METi induced significantly greater cell death when the cells were pre-treated with HGF. We therefore systematically investigated this intriguing observation, also in response to EGFR kinase inhibition. Glioblastoma cells (U87, U373), primary glioblastoma cells (GBM10) and GSCs (1228, 0308) were pre-treated with 20 ng/ml HGF for various times (2–24 hrs) prior to treatment with METi for 24 hrs. The cells were subsequently assessed for apoptosis using annexin-V/7AAD flow cytometry. The results showed that METi induced significantly greater (34–79%; two-sample t-test p-value < 0.05) cell death in HGF pre-treated cells than in control (Fig. 6A). The optimal time of pre-activation that achieved greatest sensitization to METi-induced cell death was 2–6 hrs. We observed a similar intriguing result when we pretreated glioma cells with EGF for 3–24 hrs before treatment with the EGFR kinase inhibitor erlotinib. Erlotinib induced significantly greater cell death (45–151%; two-sample t-test p < 0.05) in EGF pre-treated cells than in control (Fig. 6B). The optimal time of pre-activation that achieved greatest sensitization of the receptor to erlotinib was 3 hours. These data suggest that cancer cells can be sensitized to c-MET and possibly also other RTK inhibitions by brief activation with their respective ligands.
Figure 6. Short-time exogenous HGF treatment sensitizes glioblastoma cells and GSCs to RTK inhibitor-induced apoptosis in vitro and in vivo.
A Glioblastoma cell lines U87 and U373, primary cells GBM-10, and GSCs 1228 and 0308 were treated with HGF for 2–24 hours and then treated with METi. The cells were assessed for apoptosis and death by Annexin V/7AAD flow cytometry. METi induces greater apoptosis in cells pre-treated with HGF for 2–6 hr prior METi treatment. B The glioblastoma cells U373 were treated with EGF (100 ng/ml) for 3–24 hours and then treated with the EGFR inhibitor Erlotinib (2 µM). The results show that Erlotinib induces greater apoptosis in cells pre-treated with EGF for 3 hrs before Erlotinib treatment. C GSCs 1228-derived flank xenografts were pre-treated with HGF intratumoral injections 2 hr prior to treatment with METi and tumor sizes were measured after 3 weeks. The results show that HGF-pre-treated tumors were more inhibited by METi than non-pretreated tumors (n=6 for each treatment group). *, p<0.05
Brief exogenous HGF treatment sensitizes glioblastoma xenografts to METi in vivo
Based on the above findings, we hypothesized that short-term exogenous activation of c-MET by HGF might sensitize glioblastoma in vivo xenograft growth to inhibition by METi. To test this hypothesis, we established GSC-derived xenografts in the flanks of immunodeficient mice. Flank xenografts were chosen to facilitate HGF delivery to the tumors as HGF protein might not penetrate the blood brain barrier. The tumor-bearing animals were randomized and intratumorally injected with HGF at 400 ng/cm3 tumor volume or control for 2 hours prior to intratumoral injection of METi at 50 µM/cm3 tumor volume. The animals were treated every day for 2 weeks and tumor sizes and weights were measured. The results showed that METi induced moderately but statistically significantly greater inhibition of growth in HGF pre-treated tumors than control (two-sample t-test p-value = 0.002; Wilcoxen test p-value = 0.004) (Fig. 6C and Supplementary Figure). These data suggest that short-term bolus pre-treatment of tumors with HGF could be used as a novel strategy to improve the efficacy of c-MET kinase inhibitors.
DISCUSSION
PF-2341066 (METi) is an orally bioavailable small-molecule kinase inhibitor of c-MET that has been tested in few experimental cancers (13, 38, 39), but not in brain tumors before. The factors that determine sensitivity of cancers to this drug have not been determined to date. We show that while METi strongly inhibits c-MET phosphorylation in brain tumor cells, the METi effects on cell proliferation and apoptosis vary tremendously between the different cells. We identify HGF as one key determinant of responsiveness to METi. Our data therefore suggest that c-MET kinase inhibitors are likely to achieve greater clinical benefits in patients with tumors expressing higher levels of HGF, though these findings remain to be validated using animal survival studies to complement the tumor volume measurements. Surprisingly, responsiveness to METi did not correlate with p-MET which is usually induced by HGF. A probable explanation for this apparent inconsistency (illustrated by the findings in the ONS-76 cell line) is the previously described HGF-independent c-MET activation via mechanisms other than HGF and including basal c-MET kinase activity that increases with c-MET expression (40) and ligand-independent activation via dimerization with EGFR (41). While our manuscript was being prepared for submission, a publication by Vande Woude and colleagues showed that c-MET autocrine loop predicts responsiveness to another c-MET kinase inhibitor (SGX523) (42). These findings, which were derived from fewer cell lines and which used an inhibitor that was retracted from a phase I clinical trial due to toxicity, are in line with our findings. On the other hand, a previous publication concluded that amplification of c-MET may identify a subset of rare gastric cancers with sensitivity to the selective tyrosine kinase inhibitor PHA-665752 (43). However, c-MET amplifications are relatively rare in brain tumors and were not detected in our cell lines. The c-MET inhibitor METi (PF-2341066) also has affinity to the anaplastic lymphoma kinase (ALK). However, while c-MET is a well characterized oncogene in brain tumors (1, 2), ALK has not been described as a major regulator of these tumors. Therefore, the effects of METi in brain tumors are most likely due to inhibition of c-MET.
We also investigated the molecular signaling events that underlie responsiveness to c-MET inhibition in high HGF vs. low HGF-expressing cells. We found that p-p53 is significantly increased while p-JAK, p-ERK, p-ELK and p-RSK3 are significantly inhibited by METi in high-HGF expressing cells, but not in low-HGF cells. These data suggest that autocrine-activated c-MET depends on these molecules to maintain tumor cell proliferation and survival. The activation and stabilization of p53 are mediated by specific protein modifications, with phosphorylation at serine Ser-15 representing a critical event (44). JAK, ERK, ELK, RSK3 are activated by c-MET, and mediate its effects on proliferation, survival and mitogenesis (45–48). The data provide insights into the molecular basis of responsiveness to c-MET inhibition in cancer cells.
Surprisingly, we also found that when glioblastoma cells and xenografts were pretreated with a pulse of exogenous HGF, METi induced greater cell apoptosis and tumor growth inhibition than in non-pretreated cells. We also observed this phenomenon for EGF/EGFR, suggesting that it might also occur in other RTKs. A possible explanation for this intriguing phenomenon is that short-term exogenous activation of c-MET by HGF triggers fast oncogene addiction that renders tumors dependent on c-MET (49). Another potential explanation is that ligand binding to the receptor induces conformational changes that render the receptor more susceptible to the inhibitor. A third possible explanation is based on the finding that the extracellular domain of c-MET interacts with the death receptor Fas to prevent Fas ligand (FasL) binding, Fas activation and apoptosis through the extrinsic pathway (50). Exogenous HGF might therefore disrupt the Fas/MET aggregation leading to Fas activation and apoptosis. Irrespective of the underlying mechanism(s), our findings suggest that short-term delivery of ligands prior to treatment with RTK inhibitors might be developed as a new strategy for improving the efficacy of RTK inhibitors. Unlike long-term treatment, short-term delivery of ligands is not likely to enhance tumor growth as evidenced by our in vivo findings. Delivery of the ligand to brain tumors would probably be local due to questionable penetration of the blood brain barrier by HGF.
Altogether, our findings suggests that c-MET activation by either long-term autocrine HGF/c-MET loop formation or short-term exogenous HGF application sensitizes cancer cells to c-MET kinase inhibition and that this might also apply for other RTKs.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Our study identifies HGF co-expression as one key predictor of brain tumor sensitivity to a clinically applicable small molecule kinase inhibitor (METi) of the receptor tyrosine kinase (RTK) c-MET and uncovers an ERK/JAK/p53 pathway activation signature that differentiates responsive from non-responsive tumor cells. The study also shows for the first time that short-term exogenous HGF treatment of tumor cells and xenografts sensitizes them to METi. Similarly, short-term exogenous EGF treatment sensitized tumor cells to the anti-tumor effects of the EGFR kinase inhibitor erlotinib. These findings have important practical implications for the clinical use of recently developed c-MET (and possibly also other RTK) inhibitors in cancer and brain tumors. They identify a subset of patients that are more likely to respond to c-MET inhibition and that therefore could be pre-selected for such therapies. They also uncover ligand pre-treatment as a potential new strategy for improving the efficacy of RTK inhibitors.
Acknowledgments
Financial support: NIH grants RO1 NS045209 and RO1 CA134843 (R. Abounader).
Footnotes
Conflict of interest: Dr. Christensen is an employee and shareholder of Pfizer, Inc. and patent holder on PF2341066.
REFERENCES
- 1.Arrieta O, Garcia E, Guevara P, Garcia-Navarrete R, Ondarza R, Rembao D, et al. Hepatocyte growth factor is associated with poor prognosis of malignant gliomas and is a predictor for recurrence of meningioma. Cancer. 2002;94:3210–3218. doi: 10.1002/cncr.10594. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Rumayor A, Arrieta O, Guevara P, Escobar E, Rembao D, Salina C, et al. Coexpression of hepatocyte growth factor/scatter factor (HGF/SF) and its receptor cMET predict recurrence of meningiomas. Cancer Lett. 2004;213:117–124. doi: 10.1016/j.canlet.2004.04.026. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Lal B, Kwon S, Fan X, Saldanha U, Reznik TE, et al. The scatter factor/hepatocyte growth factor: c-met pathway in human embryonal central nervous system tumor malignancy. Cancer Res. 2005;65:9355–9362. doi: 10.1158/0008-5472.CAN-05-1946. [DOI] [PubMed] [Google Scholar]
- 4.Fan S, Meng Q, Laterra JJ, Rosen EM. Ras effector pathways modulate scatter factor-stimulated NF-kappaB signaling and protection against DNA damage. Oncogene. 2007;26:4774–4796. doi: 10.1038/sj.onc.1210271. [DOI] [PubMed] [Google Scholar]
- 5.Li Y, Guessous F, Johnson EB, Eberhart CG, Li XN, Shu Q, et al. Functional and molecular interactions between the HGF/c-Met pathway and c-Myc in large-cell medulloblastoma. Lab Invest. 2008;88:98–111. doi: 10.1038/labinvest.3700702. [DOI] [PubMed] [Google Scholar]
- 6.Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, et al. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391:285–288. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, et al. c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype. Proc Natl Acad Sci U S A. 2011;108:9951–9956. doi: 10.1073/pnas.1016912108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockmann MA, Papadimitriou A, Brandt M, Fillbrandt R, Westphal M, Lamszus K. Inhibition of intracerebral glioblastoma growth by local treatment with the scatter factor/hepatocyte growth factor-antagonist NK4. Clin Cancer Res. 2003;9:4578–4585. [PubMed] [Google Scholar]
- 9.Abounader R, Lal B, Luddy C, Koe G, Davidson B, Rosen EM, et al. In vivo targeting of SF/HGF and c-met expression via U1snRNA/ribozymes inhibits glioma growth and angiogenesis and promotes apoptosis. Faseb J. 2002;16:108–110. doi: 10.1096/fj.01-0421fje. [DOI] [PubMed] [Google Scholar]
- 10.Abounader R, Ranganathan S, Lal B, Fielding K, Book A, Dietz H, et al. Reversion of human glioblastoma malignancy by U1 small nuclear RNA/ribozyme targeting of scatter factor/hepatocyte growth factor and c- met expression. J Natl Cancer Inst. 1999;91:1548–1556. doi: 10.1093/jnci/91.18.1548. [DOI] [PubMed] [Google Scholar]
- 11.Abounader R, Montgomery R, Dietz H, Laterra J. Design and expression of chimeric U1/ribozyme transgenes. Methods Mol Biol. 2004;252:209–219. doi: 10.1385/1-59259-746-7:209. [DOI] [PubMed] [Google Scholar]
- 12.Christensen JG, Schreck R, Burrows J, Kuruganti P, Chan E, Le P, et al. A selective small molecule inhibitor of c-Met kinase inhibits c-Met-dependent phenotypes in vitro and exhibits cytoreductive antitumor activity in vivo. Cancer Res. 2003;63:7345–7355. [PubMed] [Google Scholar]
- 13.Zou HY, Li Q, Lee JH, Arango ME, McDonnell SR, Yamazaki S, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007;67:4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- 14.Guessous F, Zhang Y, diPierro C, Marcinkiewicz L, Sarkaria J, Schiff D, et al. An orally bioavailable c-Met kinase inhibitor potently inhibits brain tumor malignancy and growth. Anticancer Agents Med Chem. 2010;10:28–35. doi: 10.2174/1871520611009010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abounader R, Ranganathan S, Kim BY, Nichols C, Laterra J. Signaling pathways in the induction of c-met receptor expression by its ligand scatter factor/hepatocyte growth factor in human glioblastoma. J Neurochem. 2001;76:1497–1508. doi: 10.1046/j.1471-4159.2001.00158.x. [DOI] [PubMed] [Google Scholar]
- 16.Ponzetto C, Bardelli A, Zhen Z, Maina F, Zonca P, Giordano S, et al. A multifunctional docking site mediates signalling and transformation by the HGF/SF receptor family. Cell. 1994;77:261–271. doi: 10.1016/0092-8674(94)90318-2. [DOI] [PubMed] [Google Scholar]
- 17.Weidner KM, Di Cesare S, Sachs M, Brinkmann V, Behrens J, Birchmeier W. Interaction between Gab1 and the c-Met receptor tyrosine kinase is responsible for epithelial morphogenesis. Nature. 1996;384:173–176. doi: 10.1038/384173a0. [DOI] [PubMed] [Google Scholar]
- 18.Lock LS, Royal I, Naujokas MA, Park M. Identification of an atypical Grb2 carboxyl-terminal SH3 domain binding site in Gab docking proteins reveals Grb2-dependent and -independent recruitment of Gab1 to receptor tyrosine kinases. J Biol Chem. 2000;275:31536–31545. doi: 10.1074/jbc.M003597200. [DOI] [PubMed] [Google Scholar]
- 19.Reznik TE, Sang Y, Ma Y, Abounader R, Rosen EM, Xia S, et al. Transcription-dependent epidermal growth factor receptor activation by hepatocyte growth factor. Mol Cancer Res. 2008;6:139–150. doi: 10.1158/1541-7786.MCR-07-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi J, McTigue MA, Rogers A, Lifshits E, Christensen JG, Janne PA, et al. Multiple mutations and bypass mechanisms can contribute to development of acquired resistance to MET inhibitors. Cancer Res. 2011;71:1081–1091. doi: 10.1158/0008-5472.CAN-10-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- 22.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Guessous F, DiPierro C, Zhang Y, Mudrick T, Fuller L, et al. Interactions between PTEN and the c-Met pathway in glioblastoma and implications for therapy. Mol Cancer Ther. 2009;8:376–385. doi: 10.1158/1535-7163.MCT-08-0627. [DOI] [PubMed] [Google Scholar]
- 24.Abounader R. Interactions between PTEN and receptor tyrosine kinase pathways and their implications for glioma therapy. Expert Rev Anticancer Ther. 2009;9:235–245. doi: 10.1586/14737140.9.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen PY, Schiff D, Cloughesy TF, Raizer JJ, Laterra J, Smitt M, et al. A phase II study evaluating the efficacy and safety of AMG 102 (rilotumumab) in patients with recurrent glioblastoma. Neuro Oncol. 2011;13:437–446. doi: 10.1093/neuonc/noq198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto K, Nakamura T. NK4 (HGF-antagonist/angiogenesis inhibitor) in cancer biology and therapeutics. Cancer Sci. 2003;94:321–327. doi: 10.1111/j.1349-7006.2003.tb01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov. 2008;7:504–516. doi: 10.1038/nrd2530. [DOI] [PubMed] [Google Scholar]
- 28.Cecchi F, Rabe DC, Bottaro DP. Targeting the HGF/Met signalling pathway in cancer. Eur J Cancer. 2010;46:1260–1270. doi: 10.1016/j.ejca.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen JG, Burrows J, Salgia R. c-Met as a target for human cancer and characterization of inhibitors for therapeutic intervention. Cancer Lett. 2005;225:1–26. doi: 10.1016/j.canlet.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 30.McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarkaria JN, Yang L, Grogan PT, Kitange GJ, Carlson BL, Schroeder MA, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6:1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 32.Sarkaria JN, Carlson BL, Schroeder MA, Grogan P, Brown PD, Giannini C, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12:2264–2271. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 33.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 34.Einspahr JG, Calvert V, Alberts DS, Curiel-Lewandrowski C, Warneke J, Krouse R, et al. Functional protein pathway activation mapping of the progression of normal skin to squamous cell carcinoma. Cancer Prev Res (Phila) 2012;5:403–413. doi: 10.1158/1940-6207.CAPR-11-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierobon M, Vanmeter AJ, Moroni N, Galdi F, Petricoin EF., 3rd Reverse-phase protein microarrays. Methods Mol Biol. 2012;823:215–235. doi: 10.1007/978-1-60327-216-2_14. [DOI] [PubMed] [Google Scholar]
- 36.Paweletz CP, Charboneau L, Bichsel VE, Simone NL, Chen T, Gillespie JW, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 37.Gunther B. On theories of biological similarity. Fortschr Exp Theoret Biophys. 1975;19:9–28. [Google Scholar]
- 38.McDermott U, Pusapati RV, Christensen JG, Gray NS, Settleman J. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer Res. 2010;70:1625–1634. doi: 10.1158/0008-5472.CAN-09-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zillhardt M, Christensen JG, Lengyel E. An orally available small-molecule inhibitor of c-Met, PF-2341066, reduces tumor burden and metastasis in a preclinical model of ovarian cancer metastasis. Neoplasia. 2010;12:1–10. doi: 10.1593/neo.09948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dulak AM, Gubish CT, Stabile LP, Henry C, Siegfried JM. HGF-independent potentiation of EGFR action by c-Met. Oncogene. 2011;30:3625–3635. doi: 10.1038/onc.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pillay V, Allaf L, Wilding AL, Donoghue JF, Court NW, Greenall SA, et al. The plasticity of oncogene addiction: implications for targeted therapies directed to receptor tyrosine kinases. Neoplasia. 2009;11:448–458. doi: 10.1593/neo.09230. 2 p following 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Q, Bradley R, Kang L, Koeman J, Ascierto ML, Worschech A, et al. Hepatocyte growth factor (HGF) autocrine activation predicts sensitivity to MET inhibition in glioblastoma. Proc Natl Acad Sci U S A. 2012;109:570–575. doi: 10.1073/pnas.1119059109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smolen GA, Sordella R, Muir B, Mohapatra G, Barmettler A, Archibald H, et al. Amplification of MET may identify a subset of cancers with extreme sensitivity to the selective tyrosine kinase inhibitor PHA-665752. Proc Natl Acad Sci U S A. 2006;103:2316–2321. doi: 10.1073/pnas.0508776103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kastan MB, Zhan Q, el-Deiry WS, Carrier F, Jacks T, Walsh WV, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 45.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 46.Lai AZ, Abella JV, Park M. Crosstalk in Met receptor oncogenesis. Trends Cell Biol. 2009;19:542–551. doi: 10.1016/j.tcb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Sipeki S, Bander E, Buday L, Farkas G, Bacsy E, Ways DK, et al. Phosphatidylinositol 3-kinase contributes to Erk1/Erk2 MAP kinase activation associated with hepatocyte growth factor-induced cell scattering. Cell Signal. 1999;11:885–890. doi: 10.1016/s0898-6568(99)00060-1. [DOI] [PubMed] [Google Scholar]
- 48.Zhang YW, Wang LM, Jove R, Vande Woude GF. Requirement of Stat3 signaling for HGF/SF-Met mediated tumorigenesis. Oncogene. 2002;21:217–226. doi: 10.1038/sj.onc.1205004. [DOI] [PubMed] [Google Scholar]
- 49.Sharma SV, Settleman J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev. 2007;21:3214–3231. doi: 10.1101/gad.1609907. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, DeFrances MC, Dai Y, Pediaditakis P, Johnson C, Bell A, et al. A mechanism of cell survival: sequestration of Fas by the HGF receptor Met. Mol Cell. 2002;9:411–421. doi: 10.1016/s1097-2765(02)00439-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.