Abstract
Bioassay-guided isolation and subsequent structure elucidation of a Bael tree Aegle marmelos lipid extract yielded two unstable acylated geranyloxycoumarin mixtures (1–2), six geranyloxycoumarins (3–8), (+)-9′-isovaleroxylariciresinol (9), and dehydromarmeline (10). In a T47D cell-based reporter assay, 1 and 2 potently inhibited hypoxia-induced HIF-1 activation (IC50 values 0.18 and 1.10 μg mL−1, respectively). Insufficient material and chemical instability prevented full delineation of the fatty acyl side chain olefin substitution patterns in 1 and 2. Therefore, five fatty acyl geranyloxycoumarin ester derivatives (11–15) were prepared from marmin (3) and commercial fatty acyl chlorides by semisynthesis. The unsaturated C-6′ linoleic acid ester derivative 14 that was structurally most similar to 1 and 2, inhibited HIF-1 activation with comparable potency (IC50 0.92 μM). The octanoyl (11) and undecanoyl (12) ester derivatives also suppressed HIF-1 activation (IC50 values 3.1 and 0.87 μM, respectively). Mechanistic studies revealed that these geranyloxycoumarin derivatives disrupt mitochondrial respiration, primarily at complex I. Thus, these compounds may inhibit HIF-1 activation by suppressing mitochondria-mediated hypoxic signaling. One surprising observation was that, while less potent, the purported cancer chemopreventive agent auraptene (8) was found to act as a mitochondrial poison that disrupts HIF-1 signaling in tumors.

Keywords: Botanical Dietary Supplements, Mitochondrial Poisons, Geranyloxycoumarin, Auraptene, Hypoxia-Inducible Factor-1 (HIF-1)
1. Introduction
Aegle marmelos (L.) Corrêa ex Roxb. (Rutaceae) is a medicinal plant used in Indian Ayurvedic medicine to treat an array of ailments.1 Phytochemical studies of A. marmelos (common name: Bael tree) have led to the isolation and identification of over 100 compounds that include amide alkaloids,2–4 lignan glucosides,5 coumarins,6,7 protolimonoids,8 and anthraquinones.9,10 While A. marmelos extracts have been demonstrated to be pharmacologically active in various animal models,1 the molecular target(s) and/or mechanism(s) of action for the majority of the purified A. marmelos metabolites remain unknown.
The transcription factor hypoxia-inducible factor-1 (HIF-1) regulates cellular oxygen homeostasis and represents an important molecular target for anticancer drug discovery.11 As part of a natural product drug discovery program aimed at the identification of small-molecule HIF-1 inhibitors, a lipid extract of A. marmelos trunk bark was found to suppress HIF-1 activation in a T47D cell-based reporter assay.
Protolimonoids skimmiarepin A and C isolated from Bael tree extracts were recently shown to act as mitochondrial poisons and inhibited HIF-1 activation.12 Upon further examination, Bael tree extracts were found to contain structurally unrelated compounds with similar bioactivities. Subsequent bioassay-guided chromatographic separation yielded two inseparable mixtures of potent lipophilic HIF-1 inhibitors. Analysis of the NMR spectral data suggested that these two active mixtures were structurally related geranyloxycoumarins with similar polyunsaturated fatty acyl chains substituted at the C-6′ position of the geranyloxycoumarin skeleton. Biogenetically, these compounds are unusual in that they appear to derive from the incorporation of shikimate, terpene, and fatty acid-derived precursors. Components from three separate biogenetic sources are uniquely assembled to construct these structurally novel acylated geranyloxycoumarins. The scarcity and the instability of these active materials prohibited further isolation and structure elucidation of the lead compounds. Therefore, less potent acylated geranyloxycoumarin analogues were isolated and a semisynthetic approach was employed to generate geranyloxycoumarin derivatives conjugated to fatty acyl chains at C-6′. Both natural and semisynthetic geranyloxycoumarins were evaluated in human breast tumor cell-based in vitro models for their effects on hypoxic signaling and mitochondrial function. Herein, we report the isolation of geranyloxycoumarins from A. marmelos, semisynthesis of acylated derivatives, and in vitro biological evaluation of these unique metabolites that appear to derive from a mixed biogenetic origin.
2. Results and discussion
2.1. Bioassay-guided isolation and identification of geranyloxycoumarins
In a human breast tumor T47D cell-based reporter assay,13 a sample of trunk bark lipid extract from A. marmelos inhibited hypoxia (1% O2, 16 h)-induced HIF-1 activation by 93% at the concentration of 5 μg mL−1. Bioassay-guided chromatographic separation yielded two active mixtures 1 and 2. Following repeated attempts at chromatographic separation, the ESIMS of 1 revealed two protonated molecular ions m/z 661.3 and 663.4. This observation suggested that 1 was an inseparable mixture of two related compounds, differing only in the presence of a single olefin unit. The 1H and 13C NMR spectra of 1 exhibited a set of proton resonances attributable to the 7-(6′,7′-dihydroxygeranyloxycoumarin) [δH 7.63 (1H, d, J = 9.2 Hz, H-4), 7.36 (1H, d, J = 8.4 Hz, H-5), 6.85 (1H, dd, J = 8.4, 2.4 Hz, H-6), 6.82 (1H, d, J = 2.4 Hz, H-8), 6.25 (1H, d, J = 9.2 Hz, H-3), 5.47 (1H, t, J = 6.4 Hz, H-2′), 4.82 (1H, dd, J = 10.4, 2.4 Hz, H-6′], 4.59 (2H, d, J = 6.4 Hz, H2-1′), 2.07 (2H, overlapped, H2-4′), 1.80 (2H, overlapped, H2-5′), 1.76 (3H, s, H3-10′), 1.20 (6H, s, H3-8′,9′); δC 162.1 (C-7), 161.3 (C-2), 155.9 (C-8a), 143.4 (C-4), 141.4 (C-3′), 128.7 (C-5), 118.9 (C-2′), 113.2 (C-6), 113.0 (C-3), 112.5 (C-4a), 101.6 (C-8), 79.1 (C-6′), 72.4 (C-7′), 65.3 (C-1′), 35.9 (C-4′), 27.5 (C-5′), 26.6 (-C-10′), 25.1 (C-8′), 16.8 (C-9′)].14 The NMR spectra also contained resonances for long methylene chains [δH 2.37, 2.06, 1.26, 1.32; δC 34.5, 31.9, 29.1–29.9, 27.2, 25.6, 25.5, 22.7, 20.5], two terminal methyl groups [δH 0.978, 0.884; δC 14.3, 14.1], double bonds [δH 5.37 (6H, m); δC 132.0, 130.2, 128.3, 128.2, 127.7, 127.1], and at least one carbonyl carbon (δC 173.9). Substance 1 was an inseparable mixture of acylated 7-(6′,7′-dihydroxygeranyloxycoumarins), with a geranyloxycoumarins-core motif analogous to the previously reported compound marmin (3). However, in this case, these geranyloxycoumarins were esterified with long polyunsaturated fatty acyl side chains. Comparison of the ESIMS data with that of 3 revealed that the polyunsaturated side chains in the two mixed compounds in 1 could be assigned as C23H37O and C23H39O, respectively. However, the positions of the double bonds could not be definitively assigned. The downfield shift of C-6′ (δC 79.1) and H-6′ (δH 4.82, dd, J = 10.4, 2.4 Hz) indicated that the hydroxy group at C-6′ was acylated. This was further confirmed by the HMBC correlations between H-6′ (δH 4.82) and the carbonyl carbon(s) (δC 173.9) of long-chain polyunsaturated fatty acyl groups. The mixture in 1 was determined to be two marmin (3)-like derivatives with long-chain polyunsaturated fatty acyl esters at C-6′.
The ESIMS of 2 showed two protonated molecular ions m/z 647.4 and 649.4, indicating that, like 1, substance 2 was a mixture of two related compounds that differed only in the presence of a single double bond. The 1H NMR spectrum of 2 was comparable to that of 1. As in the spectrum of 1, both a 7-(6′,7′-dihydroxygeranyloxycoumarin) and polyunsaturated fatty acyl groups were observable. The ESIMS indicated that the two compounds in 2 were 14 mass units (one CH2) less than those of 1. Therefore, the long-chain polyunsaturated fatty acyl side chains of the two compounds in 2 could be assigned as C22H35O and C22H37O, respectively. Similarly, the downfield shift of H-6′ (δH 4.82, dd, J = 10.4, 2.4 Hz) indicated that the fatty acyl side chains were esterified at C-6′.
2.2. Effects of natural and semisynthetic geranyloxycoumarins on HIF activation
Acylated geranyloxycoumarins 1 and 2 were found to be unstable. The original TLC spots were lost upon storage and attempts to isolate the compounds from the samples by HPLC were unsuccessful. Due to their low yields and tendency to decompose, insufficient material was purified for hydrolysis to fully define the olefin substitution patterns in their fatty acyl side chains. However, the mixtures were examined for their effects on hypoxia (1% O2)- and iron chelator (10 μM 1,10-phenanthroline)-induced HIF-1 activation in a T47D cell-based reporter assay. The assays were performed with freshly isolated samples to reduce the potential loss of activity associated with chemical degradation. Freshly prepared 1 and 2 were assayed for purity prior to, and immediately following, the bioassays. The samples of 1 and 2 used in the bioassays were determined to be 97.8% and 98.2% pure, respectively. The purity of 1 and 2 recovered from the stock solutions were found to be 89% and 91.2% pure, respectively. Both 1 and 2 selectively inhibited hypoxia-induced HIF-1 activation in a concentration-dependent manner (IC50 values 0.18 and 1.10 μg mL−1, respectively, Fig. 1A and 1B). At similar concentrations, neither mixture significantly inhibited iron chelator-induced HIF-1 activation (IC50 > 6.5 μg mL−1) (1, Fig. 1A; and 2, Fig. 1B). The low yield, difficulty in separation, chemical instability, and the potent HIF-1 inhibitory activities of 1 and 2 prompted the further isolation of simple geranyloxycoumarins from A. marmelos and the semisynthesis of a small panel of acylated geranyloxycoumarin prototypes for biological evaluation. Upon further purification, coumarin-rich fractions of A. marmelos extract yielded six previously reported geranyloxycoumarins (3–8),6,14–16 and two other known compounds (+)-9′-isovaleroxylariciresinol (9)16 and dehydromarmeline (10).17 Using marmin (3)14 and commercially available fatty acyl chlorides (octanoyl chloride, undecanoyl chloride, palmitoyl chloride, linoleoyl chloride and cis-vaccenoyl chloride) as starting materials, five fatty acyl marmin ester derivatives (11–15) were prepared. The fatty acyl chlorides were selected to provide a variety of chain lengths and different unsaturation patterns to the esterified products. The structures of the semisynthetic esters (11–15) were confirmed by interpretation of 1H and 13C-NMR spectra and analysis of HRESIMS data.
Figure 1. Acylated geranyloxycoumarins 1 and 2 inhibit hypoxia-induced HIF-1 activation.
T47D cells transfected with the pHRE-luc construct were exposed to hypoxia (1% O2, 16 h, solid bar) and chemical hypoxia (10 μM 1,10-phenanthroline, 16 h, open bar) in the presence of 1 (A) and 2 (B) at the specified concentrations. The protein translation inhibitor cycloheximide (CHX, 10 μM) was used as a positive control. Data are presented as “% Inhibition” of the induced control (average + standard deviation, n = 3, representative of two independent experiments).
Both the natural products 3–10 and the semisynthetic esters 11–15 were evaluated for their effects on HIF-1 activation in a T47D cell-based concentration-response study (IC50 values summarized in Table 1). The conditions applied to induce HIF-1 are 1% oxygen (hypoxia), and 10 μM 1,10-phenanthroline (chemical hypoxia). The simple geranyloxycoumarins 4–8, (+)-9′-isovaleroxylariciresinol (9), and dehydromarmeline (10) displayed moderate inhibitory activity against hypoxia-induced HIF-1 activation (IC50 values range from 9 to 19 μM). Three semisynthetic fatty acyl marmin esters 11, 12 and 14 exhibited significantly more potent hypoxia-activated HIF-1 inhibitory activity (IC50 values of 3.08, 0.87, and 0. 92 μM, respectively). Neither marmin (3) nor the two semisynthetic esters 13 and 15 significantly inhibited HIF-1 activation by either hypoxia or chemical hypoxia (< 50% inhibition at 30 μM). Preliminary structure-activity relationships were observed among these geranyloxycoumarins. Auraptene (8, AUR), the natural geranyloxycoumarin with the most lipophilic side chain, was among the most potent non-acylated HIF-1 inhibitors (IC50 10.2 μM against hypoxia-activated HIF-1). However, the presence of a hydroxy group at C-6′ of the geranyl side chain (5) did not affect the activity (IC50 10.4 μM against hypoxia-activated HIF-1). The presence of a methoxy group at C-6 of the coumarin backbone appears to reduce the activity (although the differences were not statistically significant, p >0.05) of 6-methoxyauraptene (7, IC50 17.5 μM), suggesting that C-6 substitution may interfere with the bioactivity associated with the coumarin backbone. Additional geranyl side chain substitution [i.e., hydroxy (3), methoxy (4), and chloride (6) groups] at C-7′ dramatically decreased the activity (IC50 values of >30, 19.3, and 16.7 μM for 3, 4, and 6, respectively). This observation would tend to indicate that the substitution pattern at C-7′ of the geranyl side chain is critical for the HIF-1 inhibitory activity. Although marmin (3) exhibited very weak HIF-1 inhibitory activity under either hypoxia-inducing or chemical hypoxia-inducing conditions (IC50 values >30 μM), the introduction of a long-chain fatty acid ester [octanoyl (11), undecanoyl (12) esters] at C-6′ of the geranyl side chain markedly enhanced the potency by at least one order of magnitude. While the fatty acyl chain length did not directly correlate with the potency, the unsaturation index was important. The 9″,12″-unsaturated C-6′ linoleic acid ester derivative 14, that was structurally most similar to the original inseparable mixtures 1 and 2, exhibited HIF-1 inhibitory activity with comparable potency to that of 1 and 2; in contrast, the semisynthetic analogue 15 with one less double bond, did not significantly inhibit HIF-1 activation at the highest concentration tested (20% inhibition at 30 μM). It appears that conformational changes produced by the conjugated fatty acyl side chain may contribute to the increased potency of 14.
Table 1.
IC50 values of 3–15 suppressing HIF-1 activation in a T47D cell-based reporter assay. The IC50 and 95% confidence interval (CI) values were determined from one representative experiment performed in triplicate.
| Compound | 1% O2, 16 h | 10 μM 1,10-phen, 16 h | ||
|---|---|---|---|---|
|
| ||||
| IC50 μM | (95% CI) μM | IC50 μM | (95% CI) μM | |
| 3 | > 30 | – | > 30 | – |
| 4 | 19.3 | (14.9 – 25.0) | > 30 | – |
| 5 | 10.4 | (8.3 – 13.1) | 21.8 | – |
| 6 | 16.7 | (11.3 – 24.8) | 11.1 | (8.1 – 15.1) |
| 7 | 17.5 | (12.3 – 24.8) | 24.2 | – |
| 8 | 10.2 | (7.5 – 13.9) | 18.8 | – |
| 9 | 9.1 | (5.9 – 14.0) | 4.9 | (3.6 – 6.8) |
| 10 | 15.1 | (10.3 – 22.3) | > 30 | – |
| 11 | 3.08 | (2.4 – 3.9) | 3.71 | (2.95 – 4.67) |
| 12 | 0.87 | (0.73 – 1.05) | 3.67 | (2.53 – 5.33) |
| 13 | > 30 | – | > 30 | – |
| 14 | 0.92 | (0.83 – 1.02) | 2.35 | (1.86 – 2.96) |
| 15 | > 30 | – | > 30 | – |
The positive control cycloheximide (10 μM) inhibited HIF-1 activation by 97% under both conditions.
2.3. Geranyloxycoumarins suppress cellular respiration by targeting the mitochondrial electron transport chain
Geranyloxycoumarins and their semisynthetic fatty acyl ester derivatives exert more pronounced inhibitory effects on hypoxia (1% O2)-induced HIF-1 activation, relative to their effects on chemical hypoxia (1,10-phenanthroline)-activated HIF-1 (Table 1). This “hypoxia-selective” profile is typical for small-molecule HIF-1 inhibitors that disrupt the function of mitochondrial electron transport chain (ETC).18 Concentration-response studies were performed in a T47D cell-based respiration assay19 to investigate if these HIF-1 inhibitors disrupt mitochondrial function (results shown in Fig. 2A) at concentrations comparable to those which inhibit HIF-1. The mitochondrial ETC complex I (NADH-ubiquinone oxidoreductase) inhibitor rotenone was used as a positive control. Natural geranyloxycoumarins 4–8 and semisynthetic marmin ester derivatives 11 and 12 inhibited T47D cell respiration in a concentration-dependent manner (30–70% inhibition at 30 μM). Marmin (3) and the other three semisynthetic ester derivatives 13–15 weakly inhibited T47D cell respiration (20–30% inhibition at 30 μM). Dehydromarmeline (10) inhibited cellular respiration (70% inhibition at 30 μM), while (+)-9′-isovaleroxylariciresinol (9) did not affect respiration. Compounds 4–8 and 10 suppressed cellular respiration at concentrations comparable to those required to inhibit hypoxia-induced HIF-1 activation (Table 1). However, 11 and 12 required higher concentrations to disrupt cellular respiration, relative to the concentrations that inhibited HIF-1 activation. Additionally, 14 potently suppressed HIF-1 activation (IC50 0.92 μM, Table 1), but only weakly inhibited cellular respiration (25% inhibition at 30 μM). The potency differences suggest that the effects on mitochondria may not be directly involved in the inhibition of HIF. However, because mitochondrial respiration is assayed after acute (5–10 min) rather than chronic (16 h, as in the HIF assay) exposure to each compound, slow membrane penetration and/or metabolic processing to the active compounds could also explain the differences in potency. Therefore, T47D cells were treated with structurally related representative compounds 5, 11, 14, and 15 for 2 h and the rate of cellular respiration measured. The compounds were tested at 10 μM, a concentration that was found to exert little to no acute inhibitory effects on cellular respiration (Fig. 2A). After 2 h, 14 exhibited the most pronounced increase in potency (from 6% to 62% inhibition of respiration at 10 min vs. 2 h, relative to the untreated controls), followed by the complex I inhibitor rotenone (21% to 68% inhibition, at 0.01 μM) and 11 (28% to 39% inhibition). No significant increase in respiration inhibitory activity was observed with either 5 (27% to 31% inhibition) or 15 (5% to −6% inhibition) upon prolonged incubation. Thus, the substantially increased potency in the respiration assay after the 2 h pretreatment suggests that 14 may indeed suppress HIF-1 by inhibiting mitochondrial electron transport.
Figure 2. Effects of natural and semisynthetic geranyloxycoumarin derivatives on cellular respiration.
A) Compounds 3–15 were evaluated at the concentrations of 3 μM (black bar), 10 μM (open bar), and 30 μM (striped bar) for their effects on cellular respiration in T47D cells. Rotenone (rot) was included as a positive control at the concentrations of 1 nM (black bar), 10 nM (open bar), and 100 nM (striped bar). Data shown are “% Inhibition” of the untreated control (average + deviation from average of two independent experiments). B) T47D cells were pretreated with rotenone (0.01 μM) and 14 (10 μM) for 2 h, the cells detached, and the rate of oxygen consumption monitored upon the addition of specified substrates and inhibitors. Changes in the slopes following substrate and/or inhibitor addition reflect alterations in the observed cellular respiration rates. C) Similar to those described in B), except that succinate was added to initiate respiration at complex II. D) Rates of oxygen consumption initiated by a mixture of malate/pyruvate (malate/pyruvate) and succinate (succinate) in T47D cells pretreated for 2 h with rotenone (0.01 μM, rot), 5 (10 μM), 11 (10 μM), and 14 (10 μM). Data shown are normalized to the rate of the untreated control (con).
To discern the specific site(s) within the mitochondrial ETC affected by geranyloxycoumarin pretreatment, a panel of substrates and inhibitors were added to digitonin-permeabilized T47D cells (untreated control and compound treated) in a sequential order to determine the activity of each ETC complex. Briefly, mitochondrial respiration was initiated at complex I or complex II by the addition of malate and pyruvate or succinate, respectively. Antimycin A was added to inhibit respiration by blocking electron transfer through complex III, and a mixture of TMPD/ascorbate (complex IV substrates) were added to re-initiate antimycin A-stalled respiration [untreated control, rotenone (0.01 μM)-, and 14 (10 μM)-treated T47D cells, Fig. 2B, Fig. 2C; 5 (10 μM)- and 11 (10 μM)-treated T47D cells, not shown]. The relative rates of malate/pyruvate- and succinate-initiated respiration are summarized in Fig. 2D. Compound 14 treatment not only drastically decreased malate/pyruvate-initiated respiration (68% inhibition, relative to the untreated control), but also impaired succinate-initiated respiration (41% inhibition). The structurally similar 11 exhibited a profile comparable to that observed with 14, while the less potent analogue 5 modestly affected respiration (16–17% inhibition). As anticipated for a mitochondrial complex I inhibitor, rotenone preferentially decreased malate/pyruvate-initiated respiration (75% inhibition), relative to the inhibitory effect on succinate-initiated respiration (18% inhibition). The rates of oxygen consumption in the presence of complex IV substrates ascorbate/TMPD were comparable between untreated control, 5-, 11-, 14-, and rotenone-treated cells (control, rotenone, and 14, Fig. 2B and 2C; 5 and 11, data not shown). These observations suggest that geranyloxycoumarin derivative such as 11 and 14 may interfere with the function of ETC complex II and/or complex III, in addition to their inhibitory effects on complex I.
Similar studies were performed to identify the specific site(s) within the mitochondrial ETC affected by natural geranyloxycoumarins. Representative prenylated coumarins 6, 8, and dehydromarmeline (10) were examined at 30 μM. Substrates and inhibitors were added to digitonin-permeabilized T47D cells in a sequential order, and the rate of oxygen consumption was measured, as described in the pretreatment study. In rotenone-inhibited mitochondria oxidizing the complex II substrate succinate, 6, 8, and 10 failed to inhibit respiration suggesting that these compounds do not inhibit complex II, III, or IV (Fig. 3A, B, and C). To ensure that the mitochondrial ETC remained functional under these experimental conditions, antimycin A was added to suppress respiration, and a mixture of ascorbate/TMPD was applied to rescue cellular antimycin A-suppressed respiration (Fig. 3A, B, C). As observed in the case of the complex I inhibitor rotenone, 6, 8, and 10 each inhibited mitochondrial respiration initiated by malate/pyruvate in T47D cells, and succinate overcame this inhibition (Fig. 3D, E, and F). These results indicate that 6, 8, and 10 inhibit electron transfer through complex I.
Figure 3. Compounds 6, 8, and 10 inhibit mitochondrial respiration at ETC complex I.
A panel of substrates and inhibitors were added to digitonin-permeabilized T47D cells in a sequential manner and the rates of oxygen consumption monitored. Changes in the slopes following substrate and/or inhibitor addition reflect alterations in the observed cellular respiration rates. Compounds 6 (30 μM, A), 8 (30 μM, B), and 10 (30 μM, C) failed to suppress mitochondrial respiration in the presence of succinate. All three compounds decreased mitochondrial respiration initiated by a mixture of malate/pyruvate at complex I (6, D; 8, E; and 10, F; respectively).
2.4. Geranyloxycoumarins inhibit breast tumor cell proliferation/viability in a time- and cell line-dependent manner
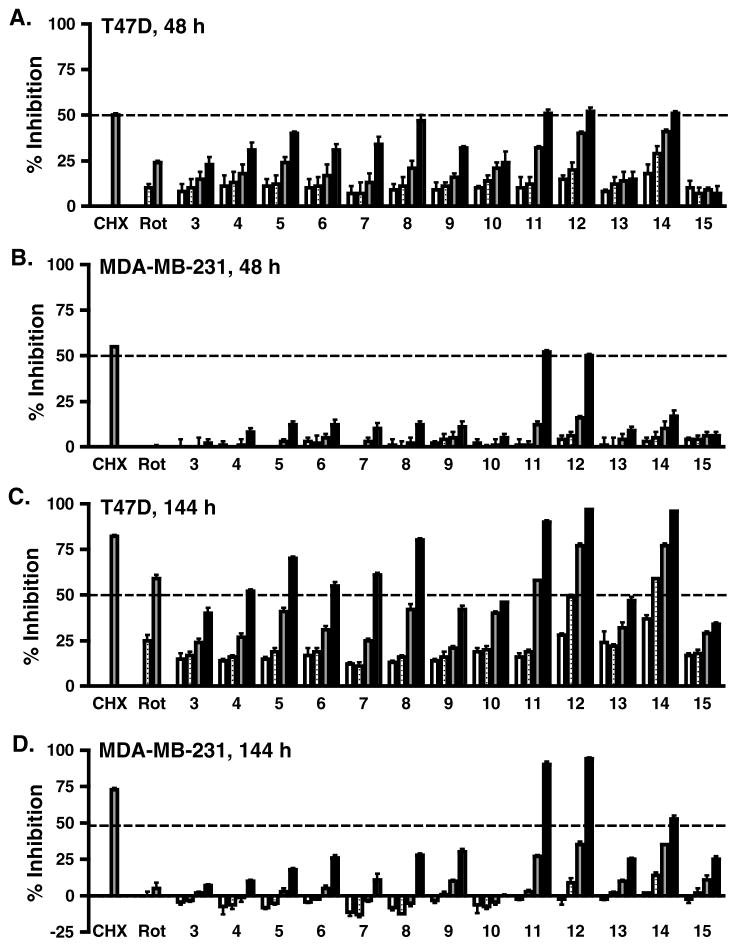
The effects of 3–15 on tumor cell proliferation/viability were examined in T47D and MDA-MB-231 human breast tumor cells. Compounds were tested at the concentrations of 1, 3, 10, and 30 μM. Rotenone (1 and 10 nM) and cycloheximide (10 μM) were included as mitochondrial respiration inhibitor and protein translation inhibitor controls, respectively. Results from the commonly applied 48 h exposure study revealed that T47D cells are more sensitive to the cytostatic/cytotoxic effects of geranyloxycoumarins than MDA-MB-231 cells (T47D, Fig. 4A; MDA-MB-231, Fig. 4B). However, the semisynthetic derivatives 11 and 12 suppressed the proliferation/viability of both cell lines at the highest concentration tested (30 μM), to the same extent as the cycloheximide control. An extended exposure time (6-day) study was performed to further examine the effects of mitochondrial inhibitors on cell proliferation/viability.19 When subjected to longer compound treatment time (144 h), the antiproliferative effects of geranyloxycoumarins (Fig. 4C) correlated more closely with their HIF-1 inhibitory activities in T47D cells (Table 1). In the extended incubation study, derivatives 11, 12, and 14 also suppressed the proliferation/viability of MDA-MB-231 cells (Fig. 4D).
Figure 4. Concentration-response of 3–15 on breast tumor cell proliferation/viability.
Cultured T47D (48 h, A; 144 h, C) and MDA-MB-231 (48 h, B; 144 h, D) cells were exposed to 3–15 at the concentrations of 1 μM (open bar), 3 μM (striped bar), 10 μM (gray bar), and 30 μM (black bar) for 48 h and 144 h (6 d). At the end of treatment, cell viability was determined by the sulforhodamine B method and presented as % Inhibition of the untreated control. For the 6 d exposure study, conditioned medium were replaced with fresh medium with compounds after 3 days. Cycloheximide (CHX, 10 μM) and rotenone (Rot; 1 nM, open bar; 10 nM, striped bar) were included as positive controls. Data shown are average + standard deviation (n = 3).
2.5. Discussion
Geranyloxycoumarins are produced by plants that belong to the families Rutaceae and Umbelliferae.20 Biological activities that range from cancer chemoprevention to antiulcer have been reported for geranyloxycoumarins.20 The geranyloxycoumarin auraptene (8), commonly found in Citrus fruit, has been intensively evaluated as a potential chemopreventive agent.21 In various animal models, auraptene suppressed both tumor initiation and progression.22–24 Our results suggest that the inhibition of HIF-1-mediated hypoxic signaling may represent a molecular mechanism that contributes to the antiproliferative/antitumor activity of 8 and other geranyloxycoumarins. The HIF-1 inhibitory activity of geranyloxycoumarin derivatives appears to be highly dependent upon the substitution pattern of both the coumarin backbone and the C-7-substituted geranyl side chain. Addition of certain fatty acyl chains to the basic geranyloxycoumarin system can markedly increase the HIF-1 inhibitory activity. Mechanistic studies revealed that 8 and related geranyloxycoumarins inhibited cellular respiration, which may subsequently suppressed mitochondria-mediated HIF-1 activation by hypoxia. Based on the observation that 1, 2, and the semisynthetic fatty acyl marmin esters 11, 12, and 14 exhibited more potent bioactivities than the non-acylated geranyloxycoumarins, one can hypothesize that natural products such as 3 or auraptene (8) may be less active precursors of potent metabolically activated acylated derivatives. Alternatively, acylated geranyloxycoumarins such as 1 and 2 may act like “prodrugs” that display potent activity in cell-based assays because their lipophilic ester side chains facilitate membrane penetration and these ester derivatives are subsequently hydrolyzed by esterases within the cell to produce potentially active geranyloxycoumarins, that may otherwise not readily penetrate the cell membrane. The mechanism(s) responsible for the chemical labiality of the natural acylated geranyloxycoumarins was not determined. However, 1 and 2 may have been relatively stable in crude extracts, but less stable when purified and exposed to air and solvents.
3. Conclusion
Coumarin-derived phytochemicals have been shown to disrupt mitochondrial function. Farnesylated coumarins surangin B25 and ferulenol26 inhibited mitochondrial ETC complexes, while mammea E/BB and related prenylated coumarins uncoupled mitochondrial respiration.27,28 Coumarin derivatives that disrupt mitochondrial function all possess a bicyclic coumarin component and a prenylated (or polyprenylated) side chain. Both structural features appear to be essential for the mitochondria-disrupting activity. Structural resemblance to the electron carrier ubiquinone suggests that these highly lipophilic geranyloxycoumarin derivatives may act as ubiquinone antagonists that interfere with the electron flow in the mitochondrial ETC. Given the critical role of mitochondria in maintaining cellular homeostasis, it is possible that many of the pharmacological activities attributed to auraptene and other geranyloxycoumarins may stem from the disruption of mitochondrial function.
From this same active A. marmelos extract, we previously isolated the protolimonoids skimmiarepin A and C that suppressed mitochondrial respiration at ETC complex I.12 The protolimonoid mitochondrial respiration inhibitors induced hyperphosphorylation and inactivation of translation initiation factor eIF2α and elongation factor eEF2, signaling an ER stress response that blocks protein translation. One possible scenario is that these compounds disrupt mitochondrial respiration, which then transmits the stress signal to the ER to stall protein translation and the subsequent downstream events. While these findings suggest that geranyloxycoumarins potentially exert antitumor effects by inhibiting hypoxia-induced HIF-1 activation, these results also imply that these relatively common phytochemicals may have the potential to induce mitochondrial poison-associated toxicities, such as liver damage29 and neurodegeneration30.
4. Experimental section
4.1. General procedure and plant material
Optical rotations were obtained on an AP IV/589-546 digital polarimeter. The NMR spectra were recorded in CDCl3 on AMX-NMR spectrometers (Bruker) operating at 400 MHz for 1H and 100 MHz for 13C, respectively. Residual solvent resonances (δ7.26 for 1H and δ77.16 for 13C) were used as internal references for the NMR spectra recorded running gradients. The HRESIMS were determined on a Bruker Daltonic micro TOF fitted with an Agilent 1100 series HPLC and an electrospray ionization source. HPLC was performed on a Waters system, equipped with a 600 controller and a 2998 photodiode array detector (DAD). Semi-preparative HPLC (Phenomenex Luna C18, 5 μm, 250 × 10.00 mm) was employed for isolation. The acyl halides octanoyl, undecanoyl, palmitoyl, linoleoyl and cis-vaccenoyl chloride were purchased from Sigma (USA). The TLCs were performed using Merck Si60F254 plates, sprayed with a 10% H2SO4 solution in EtOH, heated, and/or visualized under UV at 254 and 365 nm. The purities of 11–15 (samples that were submitted for bioassay) were determined to be more than 95% by normalization of peak areas detected by HPLC-DAD.
4.1.1. Plant material
The bark of A. marmelos was collected from Sumba, eastern Indonesia (February, 1994). The plant material was identified by A. McDonald and a voucher specimen (collection/extract number N085633) was deposited at the Smithsonian Institution National Museum of Natural History, Washington, D.C.
4.2. Chemistry
4.2.1. Extraction and isolation
Dried plant material was extracted with CH2Cl2-MeOH (1:1), residual solvents removed under vacuum, and the extract (NCI Open Repository collection/extract number N085633) stored at −20 °C in the NCI Open Repository at the Frederick Cancer Research and Development Center (Frederick, Maryland). The A. marmelos extract (4.19 g sample from NCI stock supply, 93% HIF-1 inhibition at 5 μg mL−1) was separated into six fractions by Sephadex LH-20 column chromatography eluted with CH2Cl2-MeOH (1:1). The second fraction (1.23 g, inhibited HIF-1 activation by 84% at 5 μg mL−1) was further separated into ten subfractions by a C18 vacuum column chromatography (eluted with step gradients of 10% to 100% MeOH in H2O). The eighth subfraction (66 mg, eluted with 80% MeOH in H2O, inhibited HIF-1 activation by 92% at 0.5 μg mL−1) was further separated by HPLC [Luna 5 μm, C18 (2) 100 Å, 250 × 10.0 mm, isocratic 95% MeOH in H2O, 4.0 mL min−1] to produce mixtures 1 (2.0 mg, 0.04% yield, tR 12 min, inhibited HIF-1 activation by 73% at 0.5 μg mL−1) and 2 (1.8 mg, 0.04% yield, tR 15 min, inhibited HIF-1 activation by 79% at 5 μg mL−1). TLC analysis revealed the fourth fraction (1.8 g) from the original Sephadex LH-20 column was rich in coumarins. This fraction was separated into nine subfractions by vacuum column chromatography (C18, eluted with step gradients of 20% to 100% MeOH in H2O). The third subfraction (150 mg, eluted with 40% MeOH in H2O) was further separated by HPLC [Luna 5 μm, C18 (2) 100 Å, 250 × 10.0 mm, isocratic 57% MeOH in H2O, 4.0 mL min−1] to produce marmin14 (3, 35 mg, tR 19 min, 0.84% yield). The fourth subfraction (84 mg, eluted with 50% MeOH in H2O) was separated by HPLC [Luna 5 μm, C18 (2) 100 Å, 250 × 10.0 mm, isocratic 65% MeOH in H2O, 4.0 mL min−1] to produce (+)-9′-isovaleroxylariciresinol16 (9, 0.8 mg, tR 13 min, 0.02% yield), R-(+)-7′-methoxymarmin15 (4, 6.5 mg, tR 15 min, 0.16% yield), and 515 (1.5 mg, tR 20 min, 0.04% yield). The fifth subfraction (25 mg, eluted with 60% MeOH in H2O) was separated by HPLC [Luna 5 μm, C18 (2) 100 Å, 250 × 10.0 mm, isocratic 75% MeOH in H2O, 4.0 mL min−1] to produce chloromarmin4 (6, 3.0 mg, tR 9 min, 0.07%), dehydromarmeline21 (10, 0.8 mg, tR 15 min, 0.02% yield), and 6-methoxyauraptene31 (7, 1.8 mg, tR 20 min, 0.04% yield). The sixth subfraction (78 mg, eluted with 70% MeOH in H2O) was separated by HPLC [Luna 5 μm, C18 (2) 100 Å, 250 × 10.0 mm, isocratic 82% MeOH in H2O, 4.0 mL min−1] to produce auraptene14 (8, 0.8 mg, tR 9 min, 0.02% yield). The structures of the known geranyloxycoumarin derivatives (3–10) were identified based on comparison of their 1H and 13C NMR spectra and specific rotation values with those previously reported. Additional details are available in the Supplementary Data.
4.2.2. Preparation of geranyloxycoumarin fatty acyl derivatives
To each of five separate R-(+)-marmin (3) solutions (4.0 mg in 1 mL CH2Cl2, cooled on ice), 2 to 3 molar equivalents of the fatty acyl halides octanoyl chloride (4 μL), undecanoyl chloride (6 mg), palmitoyl chloride (8 μL), linoleoyl chloride (10 mg), and cis-vaccenoyl chloride (10 μL) were added, respectively, under a nitrogen gas stream. To each solution, a catalytic amount of 4-dimethylaminopyridine (DMAP, 3.5 mg) was added. The reaction mixtures were stirred at room temperature and monitored by TLC. When completed, each of the reactions was quenched with water (200 μL). The following workup procedure was performed for each reaction: Saturated NaHCO3 solution (10 mL) was added and the product extracted with CH2Cl2 (5 mL, 2x). The combined CH2Cl2 extract was washed with HCl (2 N, 10 mL). The CH2Cl2 layer was washed with saturated NaHCO3 solution (10 mL) and then washed with saturated NaCl solution (10 mL, 2x). The CH2Cl2 layer was initially dehydrated by addition of anhydrous Na2SO4 (15 g) and dried under vacuum. The residue was dissolved in MeOH, the crude products were purified by semi-preparative HPLC (Luna 5 μm, C18 (2) 100 Å, 250 × 10.0 mm) to give R-(+)-marmin 6′-octanoate (11) (2.1 mg, 39% yield), R-(+)-marmin 6′-undecanoate (12) (3.7 mg, 64% yield), R(+)-marmin 6′-palmitate (13) (3.5 mg, 51% yield), R-(+)-marmin 6′-linoleate (14) (3.0 mg, 41% yield) and R-(+)-marmin 6′-cis-vaccenoate (15) (1.5 mg, 13% yield), respectively.
4.2.3
R-(+)-marmin 6′-octanoate (11): colorless oil; [α]25D + 23.1 (c 0.13, MeOH); 1H NMR (CDCl3, 400 MHz): δ7.63 (1H, d, J = 9.6 Hz, H-4), 7.36 (1H, d, J = 8.4 Hz, H-5), 6.84 (1H, dd, J = 8.4, 2.4 Hz, H-6), 6.81 (1H, d, J = 2.4 Hz, H-8), 6.24 (1H, d, J = 9.6 Hz, H-3), 5.48 (1H, t, J = 6.8 Hz, H-2′), 4.81 (1H, dd, J = 10.4, 2.4 Hz, H-6′], 4.59 (2H, d, J = 6.8 Hz, H2-1′), 2.36 (2H, t, J = 7.6 Hz, H2-2″), 2.07 (2H, overlapped, H2-4′), 1.80 (2H, overlapped, H2-5′), 1.76 (3H, s, H3-10′), 1.65 (2H, m, H2-3″), 1.28~1.32 (8H, overlapped, H2-4″~7″), 1.20 (6H, s, H3-8′,9′), 0.88 (3H, t, J = 7.2 Hz, H3-8″); 13C NMR (CDCl3, 100 MHz): δ 174.0 (C-1″), 162.1 (C-7), 161.3 (C-2), 155.9 (C-8a), 143.5 (C-4), 141.4 (C-3′), 128.7 (C-5), 118.9 (C-2′), 113.2 (C-6), 113.0 (C-3), 112.5 (C-4a), 101.6 (C-8), 79.0 (C-6′), 72.4 (C-7′), 65.4 (C-1′), 35.9 (C-4′), 34.5 (C-2″), 31.6 (C-6″), 29.2 (C-5″), 28.9 (C-4″), 27.5 (C-5′), 26.6 (-C-10′), 25.1 (C-8′), 25.1 (C-3″), 22.6 (C-7″), 16.8 (C-9′), 14.0 (C-8″); HRESIMS m/z 481.2559 [M + Na]+ (calcd for C27H38O6Na 481.2566).
4.2.4
R-(+)-marmin 6′-undecanoate (12): colorless oil; [α]2D + 28.6 (c 0.25, MeOH); 1H NMR (CDCl3, 400 MHz): δ7.63 (1H, d, J = 9.2 Hz, H-4), 7.36 (1H, d, J = 8.4 Hz, H-5), 6.84 (1H, dd, J = 8.4, 2.4 Hz, H-6), 6.82 (1H, d, J = 2.4 Hz, H-8), 6.24 (1H, d, J = 9.2 Hz, H-3), 5.47 (1H, t, J = 6.8 Hz, H-2′), 4.81 (1H, dd, J = 10.4, 2.4 Hz, H-6′], 4.59 (2H, d, J = 6.4 Hz, H2-1′), 2.36 (2H, t, J = 7.6 Hz, H2-2″), 2.07 (2H, overlapped, H2-4′), 1.82 (2H, overlapped, H2-5′), 1.76 (3H, s, H3-10′), 1.65 (2H, m, H2-3″), 1.26 ~1.31 (14H, overlapped, H2-4″~10″), 1.20 (6H, s, H3-8′,9′), 0.87 (3H, t, J = 6.8 Hz, H3-11″); 13C NMR (CDCl3, 100 MHz): δ173.9 (C-1″), 162.1 (C-7), 161.3 (C-2), 155.9 (C-8a), 143.4 (C-4), 141.4 (C-3′), 128.7 (C-5), 118.9 (C-2′), 113.2 (C-6), 113.0 (C-3), 112.5 (C-4a), 101.6 (C-8), 79.0 (C-6′), 72.4 (C-7′), 65.4 (C-1′), 35.9 (C-4′), 34.5 (C-2″), 31.9 (C-9″), 29.5~29.2 (C-4″~8″), 27.5 (C-5′), 26.6 (-C-10′), 25.1 (C-8′), 25.1 (C-3″), 22.7 (C-10″), 16.8 (C-9′), 14.1 (C-11″); HRESIMS [M + Na]+ m/z 523.3047 (calcd for C30H44O6Na 523.3036).
4.2.5
R-(+)-marmin 6′-palmitate (13): colorless oil; [α]25D + 35.3 (c 0.24, MeOH); 1H NMR (CDCl3, 400 MHz): δ7.63 (1H, d, J = 9.2 Hz, H-4), 7.36 (1H, d, J = 8.4 Hz, H-5), 6.84 (1H, dd, J = 8.4, 2.0 Hz, H-6), 6.81 (1H, d, J = 2.0 Hz, H-8), 6.24 (1H, d, J = 9.2 Hz, H-3), 5.46 (1H, t, J = 6.4 Hz, H-2′), 4.81 (1H, dd, J = 10.4, 2.4 Hz, H-6′], 4.58 (2H, d, J = 6.4 Hz, H2-1′), 2.35 (2H, t, J = 7.6 Hz, H2-2″), 2.06 (2H, overlapped, H2-4′), 1.81 (2H, overlapped, H2-5′), 1.75 (3H, s, H3-10′), 1.64 (2H, m, H2-3″), 1.24 ~1.31 (22H, overlapped, H2-4″~14″), 1.19 (6H, s, H3-8′,9′), 0.87 (3H, t, J = 6.8 Hz, H3-15″); 13C NMR (CDCl3, 100 MHz): δ 173.9 (C-1″), 162.1 (C-7), 161.3 (C-2), 155.9 (C-8a), 143.4 (C-4), 141.4 (C-3′), 128.7 (C-5), 118.9 (C-2′), 113.2 (C-6), 113.0 (C-3), 112.5 (C-4a), 101.6 (C-8), 79.0 (C-6′), 72.4 (C-7′), 65.4 (C-1′), 35.9 (C-4′), 34.5 (C-2″), 31.9 (C-13″), 29.7~29.2 (C-4″~12″), 27.5 (C-5′), 26.6 (-C-10′), 25.1 (C-8′), 25.1 (C-3″), 22.7 (C-14″), 16.8 (C-9′), 14.1 (C-15″); HRESIMS [M + Na]+ m/z 593.3840 (calcd for C35H54O6Na 593.3818).
4.2.6
R-(+)-marmin 6′-linoleate (14): colorless oil; [α]25D + 18.6 (c 0.18, MeOH); 1H NMR (CDCl3, 400 MHz): δ7.63 (1H, d, J = 9.6 Hz, H-4), 7.35 (1H, d, J = 8.4 Hz, H-5), 6.83 (1H, dd, J = 8.4, 2.4 Hz, H-6), 6.81 (1H, d, J = 2.4 Hz, H-8), 6.24 (1H, d, J = 9.6 Hz, H-3), 5.46 (1H, t, J = 6.4 Hz, H-2′), 5.35 (4H, m, H-9″, 10″, 12″, 13″), 4.81 (1H, dd, J = 10.4, 2.4 Hz, H-6′], 4.58 (2H, d, J = 6.4 Hz, H2-1′), 2.76, (2H, m, H2-11″), 2.35 (2H, t, J = 7.6 Hz, H2-2″), 2.06 (6H, overlapped, H2-4′, 8″, 14″), 1.82 (2H, overlapped, H2-5′), 1.75 (3H, s, H3-10′), 1.64 (2H, m, H2-3″), 1.26 ~1.38 (14H, overlapped, H2-4″, 5″, 6″, 7″, 15″, 16″, 17″), 1.19 (6H, s, H3-8′,9′), 0.88 (3H, t, J = 6.8 Hz, H3-18″); 13C NMR (CDCl3, 100 MHz): δ 173.9 (C-1″), 162.1 (C-7), 161.3 (C-2), 155.9 (C-8a), 143.4 (C-4), 141.4 (C-3′), 130.2 (C-10″), 130.0 (C-9″), 128.7 (C-5), 128.1 (C-13″), 127.9 (C-12″), 118.9 (C-2′), 113.2 (C-6), 113.0 (C-3), 112.5 (C-4a), 101.6 (C-8), 79.1 (C-6′), 72.4 (C-7′), 65.4 (C-1′), 35.9 (C-4′), 34.5 (C-2″), 31.5 (C-16″), 29.6~29.1 (C-4″, 5″, 6″, 8″, 15″), 27.5 (C-5′), 27.2 (C-7″), 27.2 (C-14″), 26.6 (-C-10′), 25.6 (C-11″), 25.1 (C-8′), 25.1 (C-3″), 22.6 (C-17″), 16.8 (C-9′), 14.1 (C-18″); HRESIMS [M + Na]+ m/z 617.3829 (calcd for C37H54O6Na 617.3818).
4.2.7
R-(+)-marmin 6′-cis-vaccenoate (15): colorless oil; [α]25D + 23.1 (c 0.13, MeOH); 1H NMR (CDCl3, 400 MHz): δ7.63 (1H, d, J = 9.2 Hz, H-4), 7.36 (1H, d, J = 8.8 Hz, H-5), 6.83 (1H, dd, J = 8.8, 2.0 Hz, H-6), 6.81 (1H, d, J = 2.0 Hz, H-8), 6.24 (1H, d, J = 9.2 Hz, H-3), 5.46 (1H, t, J = 6.4 Hz, H-2′), 5.34 (4H, m, H-11″, 12″), 4.81 (1H, dd, J = 10.4, 2.4 Hz, H-6′], 4.58 (2H, d, J = 6.4 Hz, H2-1′), 2.76, (2H, m, H2-11″), 2.36 (2H, t, J = 7.6 Hz, H2-2″), 2.08 (2H, m, H2-4′), 2.01 (4H, overlapped, H2-10″, 13″), 1.81 (2H, overlapped, H2-5′), 1.75 (3H, s, H3-10′), 1.64 (2H, m, H2-3″), 1.25 ~1.35 (20H, overlapped, H2-4″~9″, 14″~17″), 1.19 (6H, s, H3-8′,9′), 0.88 (3H, t, J = 6.8 Hz, H3-18″); HRESIMS [M + Na]+ m/z 619.3977 (calcd for C37H56O6Na 619.3975).
4.3. Biological studies
4.3.1. Cell-based reporter and viability assays
Human breast tumor T47D and MDA-MB-231 cells were obtained from ATCC and maintained as described previously.19 The T47D cell-based reporter assay for HIF-1 activity was performed as described.13 Cell viability was determined using the sulforhodamine B method.19 Samples of extract and fractions were dissolved in DMSO and the final solvent concentration in cell-based assays was less than 0.5%. Pure compounds were prepared as 4 mM stock solutions in 2-propanol and stored at −20 °C. The final solvent concentration was less than 0.75%. Data are presented as “% Inhibition” of the control, calculated using the formula:
4.3.2. Mitochondrial respiration assay
The rate of oxygen consumption in T47D cells was measured in an Oxytherm Clarke-type electrode System (Hansatech). A cell-based respiration assay was used to determine the effects of compounds on cellular respiration.19 Data are presented as “% Inhibition” of the control, calculated using the formula:
Mechanistic studies were conducted in digitonin-permeabilized T47D cells to identify the specific site(s) within the mitochondrial ETC affected by active compounds as previously described.19 For the pretreatment study, T47D cells plated in 10 cm tissue culture plates (CytoOne) were treated with test compounds for 2 h, the cells detached, washed, and the rate of oxygen consumption determined.
4.3.3. Statistical analysis
Data analyses were performed with GraphPad Prism 5. Differences between data sets were considered statistically significant when p < 0.05.
Supplementary Material
Acknowledgments
The authors thank Dr. D. J. Newman and Ms. E. C. Brown (Natural Products Branch Repository Program, NCI - Frederick) for providing plant extracts and collection information, Dr. S. L. McKnight (UT Southwestern Medical Center at Dallas) for the pHRE-TK-luc construct, and Dr. D. Ferreira (University of Mississippi) for his input on the manuscript. This research was supported in part by NIH grant CA98787. The work at the University of Mississippi was conducted in a facility constructed with NIH Research Facilities Improvement grant C06 RR-14503-01.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bmc.XXXXXXXXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Maity P, Hansda D, Bandyopadhyay U, Mishra DK. Indian J Exp Biol. 2009;47:849. [PubMed] [Google Scholar]
- 2.Sharma BR, Rattan RK, Sharma P. Phytochemistry. 1981;20:2606. [Google Scholar]
- 3.Narender T, Shweta S, Tiwari P, Reddy KP, Khaliq T, Prathipati P, Puri A, Srivastava AK, Chander R, Agarwal SC, Raj K. Bioorg Med Chem Lett. 2007;17:1808. doi: 10.1016/j.bmcl.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 4.Phuwapraisirisan P, Puksasook T, Jong-Aramruang J, Kokpol U. Bioorg Med Chem Lett. 2008;18:4956. doi: 10.1016/j.bmcl.2008.08.024. [DOI] [PubMed] [Google Scholar]
- 5.Ohashi K, Watanabe H. Chem Pharm Bull. 1994;42:1924. [Google Scholar]
- 6.Ohashi K, Watanabe H, Ohi K, Arimoto H, Okumura Y. Chem Lett. 1995;24:881. [Google Scholar]
- 7.Ali MS, Pervez MK. Nat Prod Res. 2004;18:141. doi: 10.1080/14786410310001608037. [DOI] [PubMed] [Google Scholar]
- 8.Samarasekera JKRR, Khambay BPS, Hemalal KP. Nat Prod Res. 2004;18:117. doi: 10.1080/1478641031000149858. [DOI] [PubMed] [Google Scholar]
- 9.Mishra BB, Kishore N, Tiwari VK, Singh DD, Tripathi V. Fitoterapia. 2010;81:104. doi: 10.1016/j.fitote.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Mishra BB, Singh DD, Kishore N, Tiwari VK, Tripathi V. Phytochemistry. 2010;71:230. doi: 10.1016/j.phytochem.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Semenza GL. Oncogene. 2010;29:625. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Mahdi F, Du L, Datta S, Nagle DG, Zhou Y-D. J Nat Prod. 2011;74:1894. doi: 10.1021/np200370z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodges TW, Hossain CF, Kim YP, Zhou Y-D, Nagle DG. J Nat Prod. 2004;67:767. doi: 10.1021/np030514m. [DOI] [PubMed] [Google Scholar]
- 14.Yamada Y, Nakatani N, Fuwa H. Agric Biol Chem. 1987;51:1105. [Google Scholar]
- 15.Masuda T, Muroya Y, Nakatani N. Biosci Biotech Biochem. 1992;56:1257. [Google Scholar]
- 16.Lin S, Chen T, Liu XH, Shen YH, Shan L, Liu RH, Xu XK, Zhang WD, Wang H. J Nat Prod. 2010;73:632. doi: 10.1021/np900795c. [DOI] [PubMed] [Google Scholar]
- 17.Govindachari TR, Premila MS. Phytochemistry. 1983;22:755. [Google Scholar]
- 18.Nagle DG, Zhou Y-D. Phytochem Rev. 2009;8:415. doi: 10.1007/s11101-009-9120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Veena CK, Morgan JB, Mohammed KA, Jekabsons MB, Nagle DG, Zhou YD. J Biol Chem. 2009;284:5859. doi: 10.1074/jbc.M806744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curini M, Cravotto G, Epifano F, Giannone G. Curr Med Chem. 2006;13:199. doi: 10.2174/092986706775197890. [DOI] [PubMed] [Google Scholar]
- 21.Genovese S, Epifano F. Curr Drug Targets. 2011;12:381. doi: 10.2174/138945011794815248. [DOI] [PubMed] [Google Scholar]
- 22.Murakami A, Kuki W, Takahashi Y, Yonei H, Nakamura Y, Ohto Y, Ohihashi H, Koshimizu K. Jpn J Cancer Res. 1997;88:443. doi: 10.1111/j.1349-7006.1997.tb00402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayashi K, Suzuki R, Miyamoto S, Shin-Ichiroh Y, Kohno H, Sugie S, Takashima S, Tanaka T. Nutr Cancer. 2007;58:75. doi: 10.1080/01635580701308216. [DOI] [PubMed] [Google Scholar]
- 24.Krishnan P, Yan KJ, Windler D, Tubbs J, Grand R, Li BD, Aldaz CM, McLarty J, Kleiner-Hancock HE. BMC Cancer. 2009;9:259. doi: 10.1186/1471-2407-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng Y, Nicholson RA. Pestic Biochem Physiol. 2005;81:39. [Google Scholar]
- 26.Lahouel M, Zini R, Zellagui A, Rhouati S, Carrupt PA, Morin D. Biochem Biophys Res Comm. 2007;355:252. doi: 10.1016/j.bbrc.2007.01.145. [DOI] [PubMed] [Google Scholar]
- 27.Du L, Mahdi F, Jekabsons MB, Nagle DG, Zhou Y-D. J Nat Prod. 2010;73:1868. doi: 10.1021/np100501n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du L, Mahdi F, Jekabsons MB, Nagle DG, Zhou Y-D. J Nat Prod. 2011;74:240. doi: 10.1021/np100762s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones DP, Lemasters JJ, Han D, Boelsterli UA, Kaplowitz N. Mol Interv. 2010;10:98. doi: 10.1124/mi.10.2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayala A, Venero JL, Cano J, Machado A. Front Biosci. 2007;12:986. doi: 10.2741/2119. [DOI] [PubMed] [Google Scholar]
- 31.Larsen PK, Sandberg F. Acta Chem Scand. 1970;24:1113. doi: 10.3891/acta.chem.scand.24-1113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.