Abstract
The transcriptional repressor Bcl6 is a critical regulator of T helper cell fate, and inhibits Th2-type inflammation. We have found that microRNA-21 (miR-21) is a novel target gene for Bcl6 in Treg cells. Bcl6 represses miR-21 transcription through a Stat3 binding element in the promoter, indicating opposing regulation of miR-21 by the two transcription factors. Ectopic expression of miR-21 promoted Th2 differentiation in non-polarized T cells. The pro-Th2 activity of miR-21 was associated with increased Gata3 expression and decreased expression of the miR-21 target Sprouty1. Increased miR-21 promoted Th2 and Treg gene expression in wild-type Tregs. MiR-21 could thus help promote the Th2 bias of Bcl6-deficient conventional T cells and Treg cells. MiR21 expression is increased in Th2-type inflammation, and our results define miR-21 as a critical target of Bcl6, thus providing a new link between Bcl6 and Th2 inflammation. Finally, our results reveal a novel T cell autonomous role for miR-21 in promoting Th2 differentiation.
Keywords: MicroRNA, Bcl6, Th2 differentiation, Regulatory T cells
Introduction
Bcl6 is a potent sequence-specific transcriptional repressor originally identified as an oncogene in B cell lymphoma (Dalla-Favera et al. 1999). Bcl6-deficient (Bcl6−/−) mice develop severe Th2-mediated myocarditis and pulmonary vasculitis, thus revealing a key role for Bcl6 in inhibiting Th2-type inflammatory disease (Dent et al. 1997; Dent et al. 1998). The spontaneous nature of the inflammatory disease in Bcl6−/− mice suggests defects in the regulatory T cell (Treg) lineage, however, little is known about the role of Bcl6 in Treg function. Bcl6 is a lineage-defining transcription factor for follicular helper T cells (Tfh) (Yu et al. 2009). One mechanism for how Bcl6 acts as a master regulator of the Tfh lineage is that Bcl6 represses micro-RNAs that normally repress Tfh cell development (Yu et al. 2009).
Micro-RNAs (miRs) are important inhibitors of gene expression in multiple biological systems, and function by binding to 3’-UTRs of specific target mRNAs, mediating either mRNA degradation or translational inhibition (Bartel 2004). MiRs are critical for normal cell development and function but are also dysregulated in diseases such as cancer, autoimmunity and inflammation (Cho 2007; Jeker and Bluestone 2010; O'Connell et al. 2012). Individual miRs have been identified that regulate T helper cell differentiation: miR-155 promotes Th1 and Th17 differentiation (O'Connell et al. 2010), miR-326 promotes Th17 differentiation (Du et al. 2009), and miR-29 inhibits Th1 differentiation (Steiner et al. 2011). Mice with a conditional deletion of Dicer in the Treg lineage develop fatal multi-organ autoimmunity similar to Foxp3-deficient mice, revealing a key function for miRs in Treg stability and function (Liston et al. 2008; Zhou et al. 2008). Indeed, specific miRs are differentially expressed in Tregs and regulate Treg cell biology, for instance, miR-155 is required for Treg development (Kohlhaas et al. 2009), miR-142-3p regulates cAMP production in Tregs (Huang et al. 2009), miR-146a regulates ability of Tregs to control Th1-inflammation (Lu et al. 2011), and miR-10a helps stabilize FoxP3 expression in Tregs (Jeker et al. 2012; Takahashi et al. 2012).
MicroRNA-21 (miR21) is the most commonly up-regulated microRNA in a variety of cancers (Jung and Calin 2010). MiR-21 is also increased in allergic disease in both mouse and human (Lu et al. 2009; Lu et al. 2012). MiR-21 can promote Th2 responses by inhibiting IL-12 in myeloid cells (Lu et al. 2009; Lu et al. 2011). While Stat3 and AP-1 can promote miR-21 expression (Loffler et al. 2007; Fujita et al. 2008; Iliopoulos et al. 2010), the signals that regulate miR-21 expression in Th2 inflammation are not understood. MiR-21 is expressed at higher levels in Tregs compared to conventional CD4+CD25− T cells (Cobb et al. 2006), but the functional significance of Treg-specific expression of miR-21 has not been ascertained. Here we report a novel pathway of gene regulation in Treg cells, involving repression of miR-21 by Bcl6, and a novel pathway for promoting Th2 responses via miR-21.
Results
Bcl6 directly represses miR-21 and shares a binding site in the miR-21 promoter with Stat3
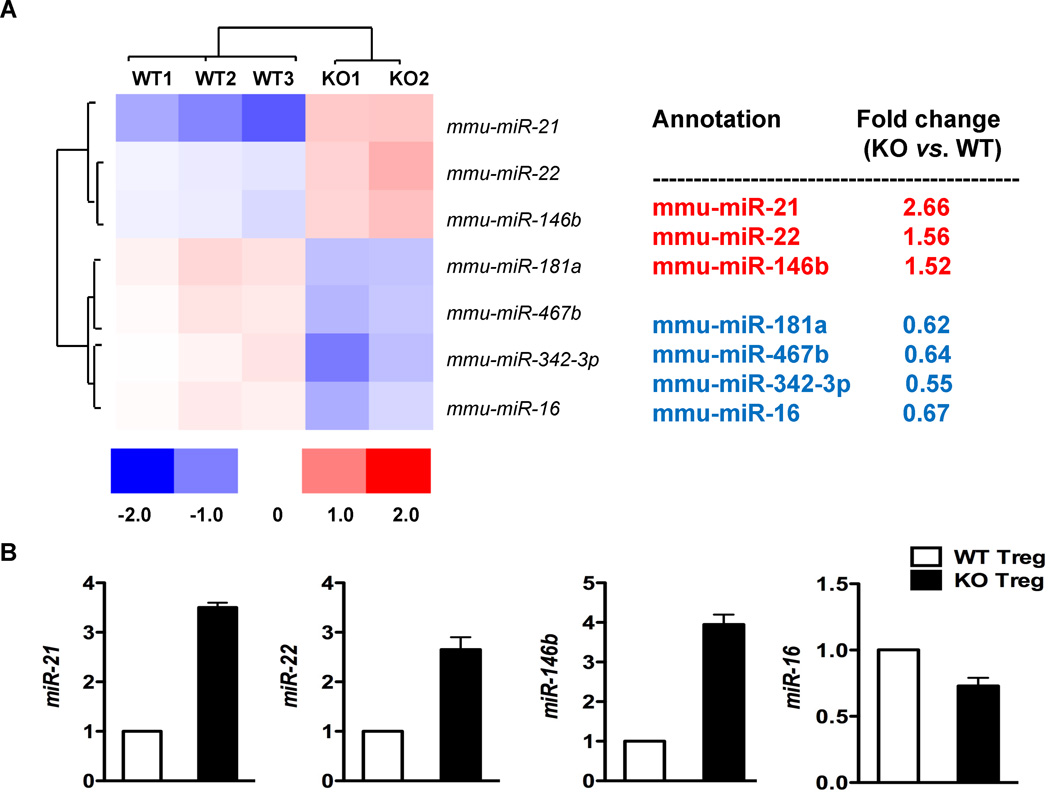
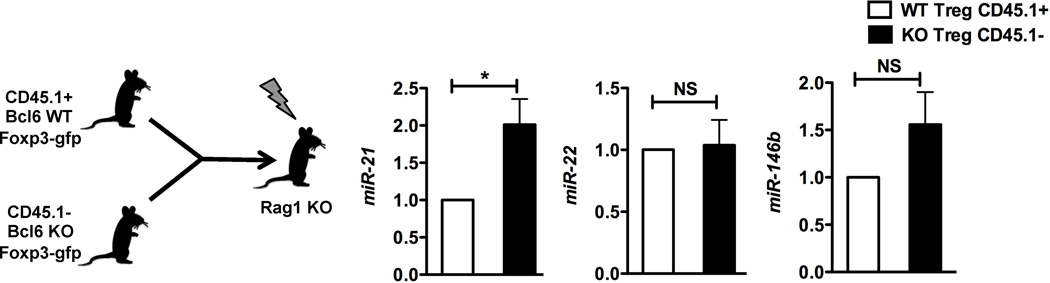
MiRs are important regulators of Treg lineage stability under inflammatory contexts (Liston et al. 2008; Zhou et al. 2008), and Bcl6 represses the expression of a large number of miRs in T cells (Yu et al. 2009). We found that Bcl6−/− Foxp3+ Tregs express elevated Th2 genes (Gata3 and Th2 cytokines) and fail to suppress Th2 inflammatory responses in vivo (Sawant et al. 2012). We thus wondered if Bcl6 regulated Treg lineage stability by repressing miRs in the Treg lineage. MiR profiling of wild-type and Bcl6−/− CD4+CD25+Foxp3+ Tregs identified 3 miRs increased significantly in Bcl6−/− Tregs relative to wild-type: miR-21, miR-22 and miR-146b (Fig. 1A). Increased expression of these miRs was verified by QPCR analysis (Fig. 1B). To test whether these miRs were regulated intrinsically in Tregs by Bcl6 or extrinsically by inflammatory signals in the Bcl6−/− mice, mixed chimeras were generated with bone marrow from CD45.1+ wild-type Foxp3-gfp mice and CD45.1− Bcl6−/− Foxp3-gfp mice, using Rag1−/− recipient mice. As shown in Fig. 2, only miR-21 was increased significantly in chimeric Bcl6−/− Tregs compared to chimeric wild-type Tregs, indicating that only miR-21 is an intrinsic target of Bcl6 in Tregs, and that miR-22 and miR-146b are not likely to be directly regulated by Bcl6 in Tregs.
Figure 1. Bcl6 represses miR-21 in Tregs.
A. The heat map shows miRs differentially expressed with statistical significance between sorted Bcl6−/− and wild-type Tregs, analyzed by expression microarrays. The color scale shown at the bottom illustrates the relative expression level of a miR across all samples: red color represents an expression level above mean, blue color represents expression level lower than the mean. N = Tregs from 3 wild-type mice and 2 Bcl6−/− mice.
B. Validation of miR expression in Bcl6−/− and wild-type Tregs by QPCR assay. N= Tregs from 2 wild-type mice and 2 Bcl6−/− mice.
Figure 2. Intrinsic regulation of miR-21 by Bcl6 in Tregs.
QPCR analysis of miR transcripts in sorted bone marrow chimera-derived Bcl6−/− (CD45.1−) and wild-type Tregs (CD45.1+), following activation for 16 hrs with anti-CD3 and anti-CD28. N= 6 mice per group.
*p<0.05 (Student’s t-test) (error bars, s.e.m.)
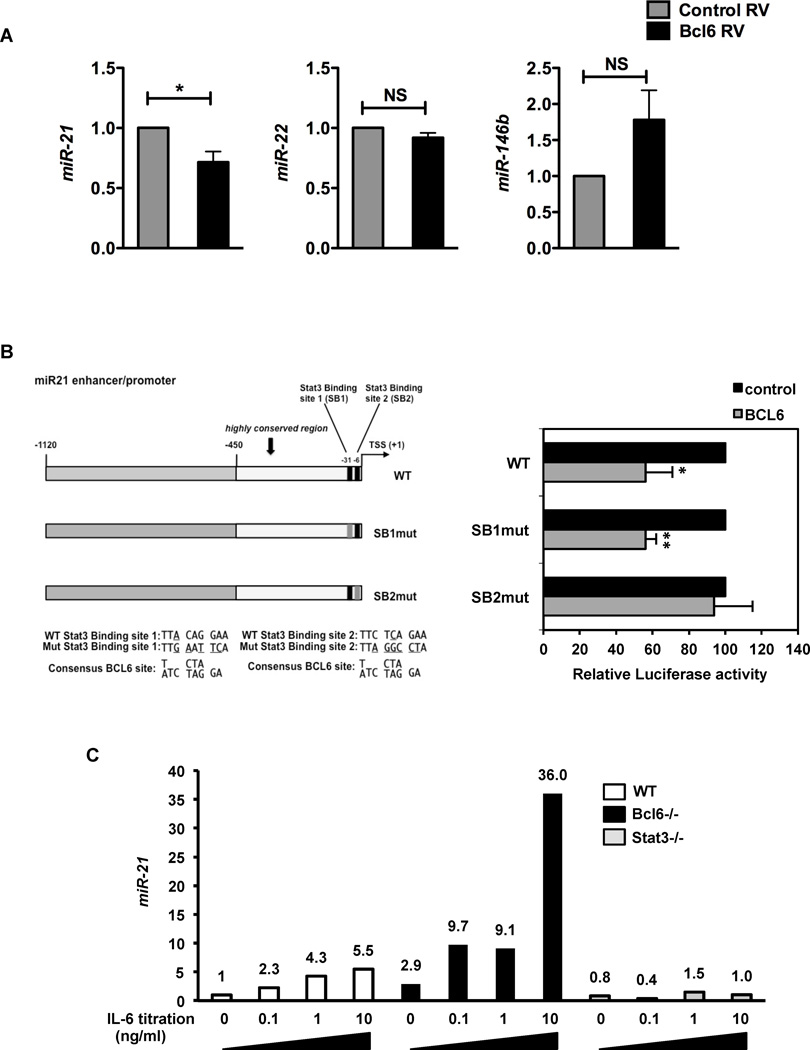
To further test the regulation of miR-21 by Bcl6, we infected naïve CD4+ T cells with retrovirus (RV) expressing Bcl6. Ectopic Bcl6 expression resulted in specific repression of miR-21, but not miR-22 or miR-146b relative to control RV (Fig. 3A), consistent with a study by Yu et al (Yu et al. 2009), which also noted miR-21 as one of the most strongly down-regulated miRNA by Bcl6 in T cells (~3.9 fold), while miR-146a was down-regulated (~1.1 fold) and miR-22 was up-regulated (~1.2 fold). Thus, this result confirms miR-21 as a novel Bcl6 target gene in T cells. Next, we analyzed the miR-21 promoter sequence and found two potential Bcl6 binding sites near the start site, which overlap with previously characterized binding sites for the transcription factor, Stat3 (Fig. 3B) (Loffler et al. 2007; Fujita et al. 2008; Iliopoulos et al. 2010; van der Fits et al. 2011; Barnes et al. 2012). Multiple studies have shown that IL-6 or IL-21, acting on Stat3, can positively regulate miR-21 transcription (Loffler et al. 2007; Iliopoulos et al. 2010; van der Fits et al. 2011; Barnes et al. 2012). We found that Bcl6 could repress the wild-type miR-21 promoter significantly in a transient transfection assay in Jurkat T cells (Fig. 3B). Mutation of the 5’ Stat3-binding site (SB1) had no effect on repression by Bcl6, whereas mutation of the 3’ Stat3-binding site (SB2), completely abolished repression by Bcl6, consistent with SB2 having a better core Bcl6 recognition motif. These data suggested that Bcl6 and Stat3 may compete for regulation of the miR-21 promoter through a common site, with Bcl6 repressing and Stat3 activating miR-21 transcription. To test this further, we analyzed miR-21 induction by IL-6 in CD4 T cells from wild-type, Bcl6−/− or Stat3−/− mice (Fig. 3C). IL-6 induced miR-21 up to 5.5 fold, whereas in Bcl6−/− T cells, basal miR-21 was increased almost 3-fold and induction of miR-21 by IL-6 was augmented over 30-fold. In contrast, miR-21 induction by IL-6 was completely abolished in Stat3−/− T cells. Thus, Bcl6 and Stat3 have opposing effects on the transcription of miR-21 in T cells.
Figure 3. Opposing regulation of miR-21 by Bcl6 and Stat3.
A. QPCR analysis of expression of miRs – 21, 22 and 146b following ectopic expression of Bcl6 in naïve T cells. Sorted RV+ T cells were re-stimulated for 6 hrs to assess its effect on miR expression. Data are averaged from at least 3 different experiments.
B. Luciferase activity in Jurkat T cells co-transfected with full-length or SB1 and SB2 mutated miR-21 promoter driven luciferase reporters and expression constructs for CXN and CXN-Bcl6. Cells were stimulated with PMA and Ionomycin for 24 hrs prior to harvest and luciferase measurement. Results are averaged from 5 independent experiments, where the basal luciferase activity of each promoter construct was set to 100%.
C. QPCR analysis of miR-21 expression following culture of WT, Bcl6−/− and Stat3−/− naïve T cells (CD4+CD62L+) in response to increasing doses of IL-6 (0, 0.1, 1, 10 ng/ml) for 24 hrs, with expression normalized to U6. Data representative of 2 independent experiments.
(A–B) *p<0.05, **p<0.01 (Student’s t-test) (error bars=s.e.m.)
MiR-21 promotes Th2 differentiation in a T cell intrinsic manner
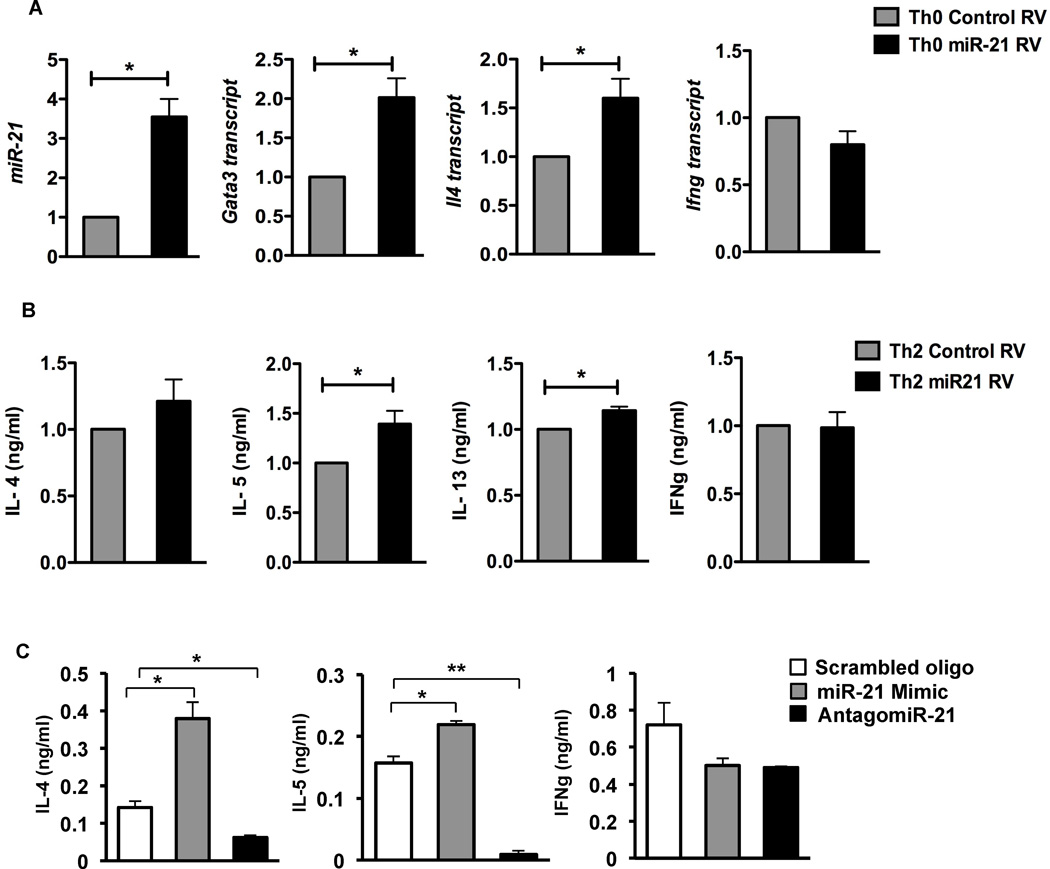
MiR-21 can inhibit Th1 responses and promote Th2 responses by repressing IL-12 expression in myeloid cells (Lu et al. 2009; Lu et al. 2011). To assess the functional relevance of miR-21 within T cells, we tested whether miR-21 regulates T cell differentiation in a T cell intrinsic manner. We therefore constructed a miR-21-expressing RV, and infected naïve CD4 T cells from wild-type mice, to test whether over-expression of miR-21 in T cells affected helper cell differentiation activated under non-polarized “Th0” conditions. The miR-21 RV promoted a 3- to 4-fold increase in miR-21 in differentiated T cells (Fig. 4A), which was comparable to the relative increase in miR-21 between wild-type and Bcl6−/− Tregs (Fig. 1). In T cells differentiated in the presence of increased miR-21, we observed increases in the Th2 genes, Gata3 and Il4, and a slight decrease in Ifng compared to T cells infected with control retrovirus (Fig. 4A). Under Th2 differentiation conditions, miR-21 RV augmented Th2 cytokines but not IFNγ production (Fig. 4B). To further assess miR-21 activity in promoting Th2 differentiation, we tested a synthetic miR-21 “mimic” and a miR-21 “antagomir” (inhibitor) in Th0 differentiation cultures, using a scrambled oligo as control. As shown in Figure 4C, the mimic significantly promoted Th2 cytokine production but not IFNγ production, whereas the antagomir potently inhibited Th2 cytokine production. Thus, miR-21 can specifically promote Th2 differentiation by a T cell autonomous mechanism.
Figure 4. MiR-21 promotes Th2 differentiation in a T cell-intrinsic manner.
A. QPCR analysis of miR-21 and Gata3, Il4 and Ifng following ectopic expression of miR-21 RV in non-polarized (Th0) T cells, relative to control RV transduced cells. Sorted RV+ cells were re-stimulated with anti-CD3 and anti-CD28 for 6 hrs for gene expression analysis. Data are averaged from at least 3 independent experiments.
B. ELISA for cytokines assayed from supernatants of miR-21 transduced Th2 differentiated T cells, relative to control RV transduced cells. Cells were re-stimulated with anti-CD3 and anti-CD28 for 24 hrs following sorting of RV+ Th2 cells for cytokine measurements.
C. ELISA for cytokines assayed from supernatants of scrambled control, miR-21 mimic and antagomiR-21 treated Th0 differentiated T cells. Cells were cultured with the oligos (1 µ M) over a 5-day period, then re-stimulated with anti-CD3 and anti-CD28 overnight for cytokine measurements. Data representative of 3 independent experiments.
(A–C) *p<0.05, **p<0.01(two-tailed Student’s t-test) (error bars, s.e.m.)
Consistent with the promotion of Th2 differentiation by increased miR-21 expression, miR-21 is expressed higher in Th2 cells compared to Th1 cells (Suppl. Fig. 1). Stat3 is activated during Th2 but not Th1 differentiation (Stritesky et al. 2011), and this Stat3 activity likely promotes the elevated miR-21 in Th2 cells. Furthermore, Stat3−/− T cells have defective Th2 differentiation (Stritesky et al. 2011), and a lack of miR-21 up-regulation can account for the reduced Th2 responses of Stat3−/− T cells.
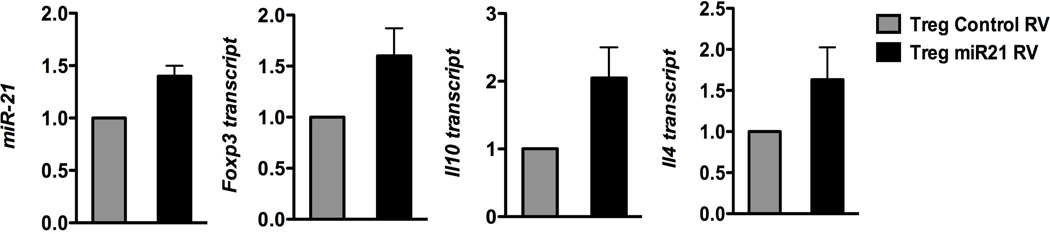
MiR-21 can positively regulate Foxp3 expression in human Tregs, although the exact mechanism has not been elucidated (Rouas et al. 2009). Consistent with that observation, we also noted increased Treg genes (Foxp3 and IL-10) following miR-21 over-expression in wild-type Tregs, along-with up-regulation of the Th2 cytokine, Il4 (Fig. 5). Thus, elevated miR-21 expression in Tregs can promote both Treg and Th2 gene expression. This phenotype is consistent with the gene expression pattern we observed with Bcl6−/− Tregs (Sawant et al. 2012).
Figure 5. MiR-21 promotes both Treg and Th2 gene expression in Tregs.
QPCR analysis of miR-21, FoxP3, Il10 and Il4 in wild-type Tregs transduced with miR-21 RV relative to control RV transduced Tregs. Cells were re-stimulated with anti-CD3 and anti-CD28 for 24 hrs following sorting of RV+ Tregs for cytokine measurements. Data from both control and miR-21 RV are averaged from 3–4 separate transductions and cell sorts.
MiR-21 target genes in T cells
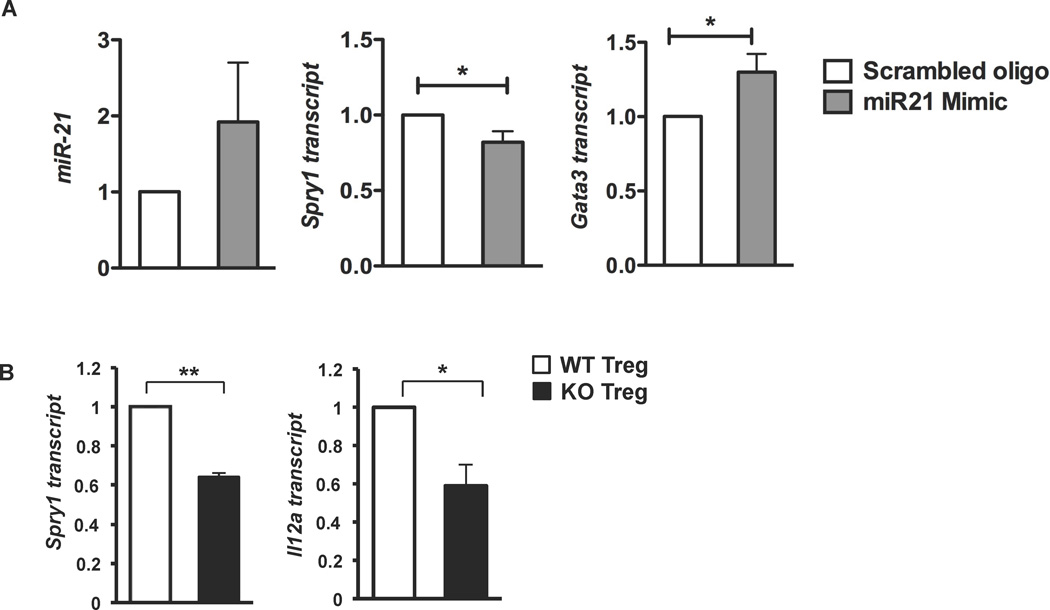
A large number of miR-21 target genes have been described (Lu et al. 2009; Jung and Calin 2010). To assess miR-21 target genes in CD4+ T cells, we treated cells with miR-21 mimic or scrambled control and analyzed six of the most well characterized target genes (Supp. Fig. 2). Of these six genes, only Sprouty1 (Spry1), a negative regulator of the MAP kinase pathway, was consistently decreased by mimic treatment (Supp. Fig. 2, Fig. 6A). Along with decreasing Spry1, miR-21 mimic augmented Gata3 mRNA (Fig. 6A). Since the MAP kinase pathway has been linked to promoting Gata3 protein stability (Yamashita et al. 2005), and Gata3 can auto-activate its own expression (Ouyang et al. 2000), miR-21 may augment Gata3 expression and thus Th2 gene expression by decreasing Spry1 and increasing MAP kinase activity. We then analyzed miR-21 gene targets in Bcl6−/− Tregs (Fig. 6B). We observed a significant decrease of Spry1 mRNA in Bcl6−/− Tregs compared to the wild-type Tregs, indicating that the regulation of Spry1 by miR-21 occurs in Tregs. The promotion of Th2 differentiation by miR-21 via Spry1, may possibly explain the elevated Th2 gene expression exhibited by Bcl6−/− Tregs and their failure to control Th2 responses in vivo (Sawant et al. 2012). Additionally, Bcl6 repression of miR-21 in Tregs may be required to limit Th2 gene expression in the Treg lineage. Using gene expression from Sawant et al (Sawant et al. 2012), we determined the relative expression of 54 potential miR-21 target genes in Bcl6−/− Tregs versus wild-type Tregs (Supplementary Table). Of these 54 genes, two were decreased more than 2-fold: Spry1 and Satb1. Satb1 is known to promote Th2 differentiation (Cai et al. 2006; Ahlfors et al. 2010; Notani et al. 2010), and lower levels of Satb1 do not explain the enhanced Th2-like phenotype of Bcl6−/− Tregs. Over 30 other potential miR-21 target genes were decreased in Bcl6−/− Tregs by more than 20% compared to wild-type (Supplementary Table), however, a clear immunological role has not been described for the majority of these genes.
Figure 6. MiR-21 targets Spry1 and Il12a.
A. QPCR analysis of miR-21, Spry1 and Gata3 following 16 hrs treatment of naïve T cells with scrambled control and miR-21 mimic (1 µ M). Data are averaged from at least 3 independent experiments.
B. QPCR analysis of Spry1 and Il12a in sorted Tregs from Bcl6−/−Foxp3-gfp (black bars) and wild-type-Foxp3-gfp mice (white bars). N = 3 mice per group.
*p<0.05, **p<0.01(Student’s t-test) (error bars, s.e.m.)
Il12a, another reported miR-21 gene target in myeloid cells is involved in Th1/Th2 polarization (Lu et al. 2011). In Tregs, Il12a is a component of the Treg suppressive cytokine IL-35 in conjunction with Ebi3 (Collison et al. 2007). We observed a strong decrease in Il12a mRNA in Bcl6−/− Tregs compared to the wild-type Tregs (Fig. 6B). The decrease in Il12a mRNA in Bcl6−/− Tregs may lead to less production of IL-35, which is important in control of Th2 responses in a dust-mite allergen challenge model (Huang et al. 2011). One interesting possibility is that reduced IL-35 secretion of Bcl6−/− Tregs factors into their inability to suppress Th2 allergic airway inflammation in vivo. Thus, one target gene of miR-21 in Tregs encodes part of an important immunosuppressive cytokine that may be critical for Treg control of Th2 immune responses.
Discussion
Cytokines such as IL-6 and IL-21 are thought to up-regulate miR-21 via Stat3 activation during inflammation, cellular differentiation or growth and survival responses (Loffler et al. 2007; Iliopoulos et al. 2010; van der Fits et al. 2011; Barnes et al. 2012). Here we have found that Bcl6 represents a counter-balance to up-regulation of miR-21 by Stat3. This pathway may be particularly important in regulatory T cells, which are critical for controlling inflammation. Intriguingly, Stat3 and Bcl6 appear to bind to a common site in the miR-21 promoter, indicating the potential for competitive regulation of miR-21 by these factors in cells in an inflammatory environment. Our results imply that T cells within an inflammatory environment can undergo Th2 differentiation due to miR-21 up-regulation, and that this is enhanced in the absence of Bcl6. The deregulation of miR-21 in Bcl6−/− T cells could augment Th2 differentiation, and lead to deregulated Th2 inflammation as is seen in Bcl6−/− mice (Dent et al. 1997; Dent et al. 1998). Since miR-21 regulates Th2 differentiation by a separate pathway than the canonical pathway of IL-4 and Stat6, our findings may explain why Th2-type inflammation occurs in Bcl6−/− mice independently of IL-4 and Stat6 (Dent et al. 1998). In fact, the Th2-type inflammation in Bcl6−/− mice is driven, in part, by IL-6 produced by myeloid cells (Mondal et al. 2010; Ohtsuka et al. 2011). Higher levels of IL-6 in Bcl6−/− mice could lead to higher miR-21 in CD4 T cells, which would pre-dispose the T cells to Th2 differentiation.
Stat3 is required for full Th2 differentiation (Stritesky et al. 2011), and the failure to up-regulate miR-21 can explain the incomplete Th2 differentiation seen with Stat3−/− T cells. The counter-regulation of miR-21 by Bcl6 and Stat3 can potentially explain the opposing roles of these transcription factors in Th2 differentiation, with Bcl6 inhibiting and Stat3 promoting Th2 differentiation, respectively. Deregulation of miR-21 in Bcl6−/− Treg cells can also explain the inability of these cells to control Th2 inflammation (Sawant et al. 2012). The regulation of miR-21 by Bcl6 may also relate to the specific development of myocarditis in Bcl6−/− mice, as increased miR-21 expression has been associated with cardiac hypertrophy (Thum et al. 2008).
Curiously, while four other groups have studied the regulation of miR-21 by Stat3, these studies have all relied on chromatin immuno-precipitation of the miR-21 promoter as an indirect test of Stat3 activity (Loffler et al. 2007; Iliopoulos et al. 2010; van der Fits et al. 2011; Barnes et al. 2012). Only one of these studies tested Stat3 function on miR-21 transcription: siRNA was used to knock-down Stat3, resulting in loss of miR-21 expression in mammary epithelial cells (Iliopoulos et al. 2010). In our current study, we show genetic ablation of Stat3 in T cells results in loss of miR-21 induction by IL-6. While none of these other studies used a luciferase assay to show Stat3 activating the miR-21 promoter, we were unable to activate our miR-21 promoter luciferase construct in our T cell system with IL-6. Thus, there may be some unusual regulation of the miR-21 promoter by Stat3, which may be worthy of further investigation.
MiR-21 has been shown to inhibit Th1 differentiation and promote Th2 differentiation by acting in myeloid cells and inhibiting the expression of IL-12 (Lu et al. 2011). Here, we show that miR-21 promotes Th2 differentiation by a novel pathway, in a T cell autonomous manner. One possible mechanism for how miR-21 can promote Th2 differentiation is by inhibiting the expression of the transcript for Spry1, a MAP kinase pathway inhibitor. Since the transcriptional activity of the master Th2 factor Gata3 can be stabilized by MAP kinase activity (Yamashita et al. 2005), we propose that increased miR-21 decreases Spry1, leading to more MAP kinase activity and increased Gata3 expression. We observed that miR-21 represses Spry1 transcript in conventional T cells about 1.25-fold, and 1.5- to 2-fold in Treg cells (Figure 6; Supplementary Table). Although these changes in expression are relatively slight, this level of transcript decrease is typical for microRNAs, which fine-tune gene expression via relatively small changes in transcript levels rather than by an on-off mechanism (O'Connell et al. 2012). Given the potential of miR-21 to affect the MAP kinase pathway and Gata3 expression, the regulation of miR-21 by Bcl6 might also explain the post-transcriptional control of Gata3 and Th2 differentiation by Bcl6 (Kusam et al. 2003). In this earlier work, we observed that high levels of Bcl6 could lead to decreased Gata3 protein without affecting Gata3 transcript. Repression of miR-21 by Bcl6 could lead to higher Sprouty1, less MAP kinase activity and decreased Gata3 protein stability.
MiR-21 is specifically increased in Tregs versus conventional T cells (Cobb et al. 2006; Rouas et al. 2009), suggesting that miR-21 may have an important role in Treg biology. MiR-21 is up-regulated in human Tregs, and can positively regulate Foxp3 expression (Rouas et al. 2009). Since many Treg genes are dependent upon Foxp3 for their expression, regulation of miR-21 by Bcl6 can control expression of Foxp3 and thus other Treg genes. IL-10 is a well-characterized BCL6 target gene in T cells (Kusam et al. 2003) and is positively regulated by miR-21 (Sheedy et al. 2009). Thus Bcl6 may regulate IL-10 indirectly in Tregs via miR-21 regulation. MiR-21 can also decrease Il12a expression in Tregs, leading to less secretion of the immune-suppressive cytokine IL-35, which may be important for Treg control of Th2 responses. The constellation of gene changes mediated by increased expression of miR-21 can potentially explain the unusual gene signature of Bcl6−/− Tregs with augmented Treg and Th2 genes resulting in potent suppression of T cell proliferation in vitro yet exacerbated Th2 inflammation in vivo (Sawant et al. 2012).
While miR-21 function has been extensively studied in cancer, the function of miR-21 in T cells and inflammation has not been well characterized. To our knowledge, this is the first study identifying miR-21 as a novel target gene for Bcl6 in Treg cells, and reporting a T cell autonomous role for miR-21 in promoting Th2 differentiation. Finally, we have identified miR-21 target genes that shed light on the role of miR-21 in conventional T cells and Tregs.
Materials and Methods
Mice
Bcl6−/− mice on a mixed C57BL/6-129Sv background (Dent et al. 1997; Dent et al. 1998) were used between 5 and 10 weeks of age, and the mice used were active and relatively healthy. Foxp3-gfp, Rag1−/− and BoyJ mice were obtained from the Jackson Laboratory (Bar Harbor, ME). CD4-specific Stat3−/− mice have been described (Stritesky et al. 2011). Mice were bred under specific pathogen-free conditions at the laboratory animal facility at IUSM and were handled according to protocols approved by the IUSM Animal Use and Care Committee.
MiR profiling and qRT-PCR assessment of miRs
RNA was extracted from FACS-sorted CD4+CD25+Foxp3+ Tregs from wild-type and Bcl6−/− Foxp3gfp mice following 16 hr activation in vitro with anti-CD3 (5 µg/ml) and anti-CD28 (10 µg/ml). The samples were hybridized to the miRCURY™ LNA array version 11.0 (Exiqon, Denmark), which tests for all miRNAs registered in the miRBASE version 13.0 at the Sanger Institute. Validation of miR expression was performed using TaqMan miR assays (Applied Biosystems). MiRs sno202, sno234 and U6 were used as controls, with U6 as the sole control for samples with limiting RNA.
Generation of Bone Marrow Chimeras
Bone marrow (BM) cells (5×106 each type) from donor wild-type BoyJ (CD45.1+) and Bcl6−/− (CD45.1−) mice (each on Foxp3gfp background) were injected i.v. into Rag−/− recipient mice, sub-lethally irradiated (350 Gy) 24 hrs prior. The lymphoid compartment in the recipients was allowed to reconstitute for 4–5 months. Mice were immunized with OVA/Alum i.p. 2 weeks prior to FACS sorting of the CD45.1+ wild-type and CD45.1− Bcl6−/− CD25+Foxp3+ Tregs for miR assessment.
Reporter assays
Full-length, SB1 and SB2 miR-21 promoter constructs (1 µg each) and control and Bcl6 expression constructs (1 µg CXN or 0.8 µg CXN plus 0.2 µg CXN-Bcl6 for the respective conditions) were transiently transfected into 1×106 Jurkat T cells with X-tremeGENE HP DNA transfection reagent (Roche), according to the manufacturer’s protocol. Luciferase measurements were performed 24 hrs after transfection following 6 hr activation of cells with PMA (10 ng/ml) and Ionomycin (0.3 µM) using Luciferase Assay System (Promega).
Cloning of mmu-miR-21 and plasmid construction for miR-21 reporter vectors
The miR-21 gene representing the primary transcript (~300 bp) was PCR amplified from mouse genomic DNA and cloned into a retroviral vector co-expressing H2Kk. Full-length mouse miR-21 promoter region was PCR amplified from mouse genomic DNA and was inserted using Mlu-I and Xho-I restriction enzyme sites into the pGL3-basic vector (Promega). The control retrovirus expresses a leader transcript plus the internal ribosomal entry site and H2Kk.
Mouse T helper cell differentiation assays and retroviral transductions
Naïve T cells (CD4+CD62L+) were purified from lymph nodes and spleen using the MACS system (Miltenyi Biotech). Naïve CD4+ T cells were activated with plate-bound anti-CD3 (5 µg/ml; 145-2C11) and anti-CD28 (10 µg/ml; 37.51) and cultured under standard Th0 and Th2 differentiation conditions (Mondal et al. 2010). Retroviral transductions of T cells were performed as described (Mondal et al. 2010).
MiR-21 mimic and inhibitor
Naïve T cells activated under Th0 conditions were treated with 1 µ M control (scrambled oligo), miR-21 mimic (double stranded RNA oligo) and antagomiR (single stranded DNA oligo) (Exiqon). The oligos were cholesterol linked that enable efficient delivery into cells, without the need of any transfection protocol. Gene expression was assessed 16 hrs following treatment. For assessment of cytokine productions, cells were treated with the oligos over a 5-day period, following which the cultures were re-stimulated with anti-CD3 (10 µg/ml) overnight to obtain supernatants for ELISA.
Gene expression and Cytokine Secretion analysis
RNA isolation, cDNA synthesis, QPCR assays, T cell stimulations and ELISAs were performed as described (Mondal et al. 2010).
Statistical analysis
p values were calculated using Students’ T-test. A p value < 0.05 was considered to show a significant difference.
Supplementary Material
Highlights.
microRNA-21 (miR-21) is a novel target gene for Bcl6 in Treg cells
Bcl6 represses miR-21 transcription through a Stat3 binding element in the promoter, indicating opposing regulation of miR-21 by Bcl6 and Stat3
Expression of miR-21 promoted Th2 differentiation in non-polarized T cells by a novel intrinsic pathway
The pro-Th2 activity of miR-21 was associated with increased Gata3 expression and decreased expression of the miR-21 target Sprouty1
ACKNOWLEDGEMENTS
We would like to thank Susan Rice and Kim Stoner of the IU Cancer Center Flow cytometry facility for help with cell sorting. We greatly appreciate the contributions of Arpita Mondal, Paulla Teuscher, Heng Zhao and Abhirami Iyer for PCR assays. We thank Janice Blum and Christopher Touloukian for helpful advice and discussions. This work was supported by NIAID grants 1R21AI079349-01A1, 1R21AI090150-01 and 1R21AI092212-01 to A.L.D., and American Heart Association pre-doctoral fellowship 10PRE4620001 to D.V.S.
Abbreviations
- Treg
Regulatory T cell
- miR
MicroRNA
- Tfh
follicular helper T cells
- RV
retrovirus
- Spry1
Sprouty1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahlfors H, Limaye A, Elo LL, Tuomela S, Burute M, Gottimukkala KV, Notani D, Rasool O, Galande S, Lahesmaa R. SATB1 dictates expression of multiple genes including IL-5 involved in human T helper cell differentiation. Blood. 2010;116:1443–1453. doi: 10.1182/blood-2009-11-252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes NA, Stephenson S, Cocco M, Tooze RM, Doody GM. BLIMP-1 and STAT3 counterregulate microRNA-21 during plasma cell differentiation. J Immunol. 2012;189:253–260. doi: 10.4049/jimmunol.1101563. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collison LW, Workman CJ, Kuo TT, Boyd K, Wang Y, Vignali KM, Cross R, Sehy D, Blumberg RS, Vignali DA. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:566–569. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- Dalla-Favera R, Migliazza A, Chang CC, Niu H, Pasqualucci L, Butler M, Shen Q, Cattoretti G. Molecular pathogenesis of B cell malignancy: the role of BCL-6. Curr Top Microbiol Immunol. 1999;246:257–263. doi: 10.1007/978-3-642-60162-0_32. [DOI] [PubMed] [Google Scholar]
- Dent AL, Hu-Li J, Paul WE, Staudt LS. T helper type 2 inflammatory disease in the absence of IL-4 and STAT6. Proc Natl Acad Sci U S A. 1998;95:13823–13828. doi: 10.1073/pnas.95.23.13823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a doublenegative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Huang B, Zhao J, Lei Z, Shen S, Li D, Shen GX, Zhang GM, Feng ZH. miR-142-3p restricts cAMP production in CD4+CD25− T cells and CD4+CD25+ TREG cells by targeting AC9 mRNA. EMBO Rep. 2009;10:180–185. doi: 10.1038/embor.2008.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Loo EX, Kuo IC, Soh GH, Goh DL, Lee BW, Chua KY. Airway Inflammation and IgE Production Induced by Dust Mite Allergen-Specific Memory/Effector Th2 Cell Line Can Be Effectively Attenuated by IL-35. J Immunol. 2011;187:462–471. doi: 10.4049/jimmunol.1100259. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeker LT, Bluestone JA. Small RNA regulators of T cell-mediated autoimmunity. J Clin Immunol. 2010;30:347–357. doi: 10.1007/s10875-010-9392-7. [DOI] [PubMed] [Google Scholar]
- Jeker LT, Zhou X, Gershberg K, de Kouchkovsky D, Morar MM, Stadthagen G, Lund AH, Bluestone JA. MicroRNA 10a Marks Regulatory T Cells. PLoS One. 2012;7:e36684. doi: 10.1371/journal.pone.0036684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung EJ, Calin GA. The Meaning of 21 in the MicroRNA world: perfection rather than destruction? Cancer Cell. 2010;18:203–205. doi: 10.1016/j.ccr.2010.08.015. [DOI] [PubMed] [Google Scholar]
- Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–2582. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- Kusam S, Toney LM, Sato H, Dent AL. Inhibition of Th2 differentiation and GATA-3 expression by BCL-6. J Immunol. 2003;170:2435–2441. doi: 10.4049/jimmunol.170.5.2435. [DOI] [PubMed] [Google Scholar]
- Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, Cvijic H, Ullmann AK, Stadler PF, Horn F. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2011;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, Orkin SH, Aronow BJ, Rothenberg ME. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol. 2011;187:3362–3373. doi: 10.4049/jimmunol.1101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu TX, Sherrill JD, Wen T, Plassard AJ, Besse JA, Abonia JP, Franciosi JP, Putnam PE, Eby M, Martin LJ, Aronow BJ, Rothenberg ME. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J Allergy Clin Immunol. 2012;129:1064–1075. e1069. doi: 10.1016/j.jaci.2012.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal A, Sawant D, Dent AL. Transcriptional repressor BCL6 controls Th17 responses by controlling gene expression in both T cells and macrophages. J Immunol. 2010;184:4123–4132. doi: 10.4049/jimmunol.0901242. [DOI] [PubMed] [Google Scholar]
- Notani D, Gottimukkala KP, Jayani RS, Limaye AS, Damle MV, Mehta S, Purbey PK, Joseph J, Galande S. Global regulator SATB1 recruits beta-catenin and regulates T(H)2 differentiation in Wnt-dependent manner. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000296. e1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- Ohtsuka H, Sakamoto A, Pan J, Inage S, Horigome S, Ichii H, Arima M, Hatano M, Okada S, Tokuhisa T. Bcl6 is required for the development of mouse CD4+ and CD8alpha+ dendritic cells. J Immunol. 2011;186:255–263. doi: 10.4049/jimmunol.0903714. [DOI] [PubMed] [Google Scholar]
- Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, Murphy KM. Stat6-Independent GATA-3 Autoactivation Directs IL-4-Independent Th2 Development and Commitment. Immunity. 2000;12:27–37. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- Rouas R, Fayyad-Kazan H, El Zein N, Lewalle P, Rothe F, Simion A, Akl H, Mourtada M, El Rifai M, Burny A, Romero P, Martiat P, Badran B. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol. 2009;39:1608–1618. doi: 10.1002/eji.200838509. [DOI] [PubMed] [Google Scholar]
- Sawant DV, Sehra S, Nguyen ET, Jadhav R, Englert K, Shinnakasu R, Hangoc G, Broxmeyer HE, Nakayama T, Perumal NB, Kaplan MH, Dent AL. Bcl6 controls the th2 inflammatory activity of regulatory T cells by repressing gata3 function. J Immunol. 2012;189:4759–4769. doi: 10.4049/jimmunol.1201794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, Johnson DS, Chen Y, O'Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2009;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritesky GL, Muthukrishnan R, Sehra S, Goswami R, Pham D, Travers J, Nguyen ET, Levy DE, Kaplan MH. The transcription factor STAT3 is required for T helper 2 cell development. Immunity. 2011;34:39–49. doi: 10.1016/j.immuni.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciume G, Muljo SA, Kuchen S, Casellas R, Wei L, Kanno Y, O'Shea JJ. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol. 2012;13:587–595. doi: 10.1038/ni.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- van der Fits L, van Kester MS, Qin Y, Out-Luiting JJ, Smit F, Zoutman WH, Willemze R, Tensen CP, Vermeer MH. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sezary syndrome. J Invest Dermatol. 2011;131:762–768. doi: 10.1038/jid.2010.349. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Shinnakasu R, Asou H, Kimura M, Hasegawa A, Hashimoto K, Hatano N, Ogata M, Nakayama T. Ras-ERK MAPK cascade regulates GATA3 stability and Th2 differentiation through ubiquitin-proteasome pathway. J Biol Chem. 2005;280:29409–29419. doi: 10.1074/jbc.M502333200. [DOI] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.