Abstract
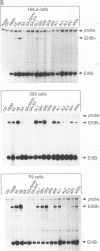
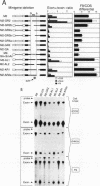
We have investigated the regulation of splicing of one of the alternatively spliced exons in the rat fibronectin gene, the EIIIB exon. This 273-nucleotide exon is excluded by some cells and included to various degrees by others. We find that EIIIB is intrinsically poorly spliced and that both its exon sequences and its splice sites contribute to its poor recognition. Therefore, cells which recognize the EIIIB exon must have mechanisms for improving its splicing. Furthermore, in order for EIIB to be regulated, a balance must exist between the EIIIB splice sites and those of its flanking exons. Although the intron upstream of EIIIB does not appear to play a role in the recognition of EIIIB for splicing, the intron downstream contains sequence elements which can promote EIIIB recognition in a cell-type-specific fashion. These elements are located an unusually long distance from the exon that they regulate, more than 518 nucleotides downstream from EIIIB, and may represent a novel mode of exon regulation.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barone M. V., Henchcliffe C., Baralle F. E., Paolella G. Cell type specific trans-acting factors are involved in alternative splicing of human fibronectin pre-mRNA. EMBO J. 1989 Apr;8(4):1079–1085. doi: 10.1002/j.1460-2075.1989.tb03476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. L. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992 May 29;69(5):795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- Black D. L. Does steric interference between splice sites block the splicing of a short c-src neuron-specific exon in non-neuronal cells? Genes Dev. 1991 Mar;5(3):389–402. doi: 10.1101/gad.5.3.389. [DOI] [PubMed] [Google Scholar]
- Buckler A. J., Chang D. D., Graw S. L., Brook J. D., Haber D. A., Sharp P. A., Housman D. E. Exon amplification: a strategy to isolate mammalian genes based on RNA splicing. Proc Natl Acad Sci U S A. 1991 May 1;88(9):4005–4009. doi: 10.1073/pnas.88.9.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis K. C., Baker B. S. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989 Mar 24;56(6):997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Sharp P. A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989 Dec 1;59(5):789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Clackson T., Winter G. 'Sticky feet'-directed mutagenesis and its application to swapping antibody domains. Nucleic Acids Res. 1989 Dec 25;17(24):10163–10170. doi: 10.1093/nar/17.24.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouet d'Orval B., d'Aubenton Carafa Y., Sirand-Pugnet P., Gallego M., Brody E., Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991 Jun 28;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. Nucleotide substitutions within the cardiac troponin T alternative exon disrupt pre-mRNA alternative splicing. Nucleic Acids Res. 1989 Oct 11;17(19):7905–7921. doi: 10.1093/nar/17.19.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z., Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991 Dec;11(12):6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emeson R. B., Hedjran F., Yeakley J. M., Guise J. W., Rosenfeld M. G. Alternative production of calcitonin and CGRP mRNA is regulated at the calcitonin-specific splice acceptor. Nature. 1989 Sep 7;341(6237):76–80. doi: 10.1038/341076a0. [DOI] [PubMed] [Google Scholar]
- Eperon L. P., Estibeiro J. P., Eperon I. C. The role of nucleotide sequences in splice site selection in eukaryotic pre-messenger RNA. Nature. 1986 Nov 20;324(6094):280–282. doi: 10.1038/324280a0. [DOI] [PubMed] [Google Scholar]
- Fu X. Y., Ge H., Manley J. L. The role of the polypyrimidine stretch at the SV40 early pre-mRNA 3' splice site in alternative splicing. EMBO J. 1988 Mar;7(3):809–817. doi: 10.1002/j.1460-2075.1988.tb02879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M. E., Nadal-Ginard B. Myosin light-chain 1/3 gene alternative splicing: cis regulation is based upon a hierarchical compatibility between splice sites. Mol Cell Biol. 1990 May;10(5):2133–2144. doi: 10.1128/mcb.10.5.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattoni R., Schmitt P., Stevenin J. In vitro splicing of adenovirus E1A transcripts: characterization of novel reactions and of multiple branch points abnormally far from the 3' splice site. Nucleic Acids Res. 1988 Mar 25;16(6):2389–2409. doi: 10.1093/nar/16.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H., Manley J. L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990 Jul 13;62(1):25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Gorman C. M., Rigby P. W., Lane D. P. Negative regulation of viral enhancers in undifferentiated embryonic stem cells. Cell. 1985 Sep;42(2):519–526. doi: 10.1016/0092-8674(85)90109-6. [DOI] [PubMed] [Google Scholar]
- Goux-Pelletan M., Libri D., d'Aubenton-Carafa Y., Fiszman M., Brody E., Marie J. In vitro splicing of mutually exclusive exons from the chicken beta-tropomyosin gene: role of the branch point location and very long pyrimidine stretch. EMBO J. 1990 Jan;9(1):241–249. doi: 10.1002/j.1460-2075.1990.tb08101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- Guo W., Mulligan G. J., Wormsley S., Helfman D. M. Alternative splicing of beta-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 1991 Nov;5(11):2096–2107. doi: 10.1101/gad.5.11.2096. [DOI] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley M. L., Maniatis T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell. 1991 May 17;65(4):579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- Helfman D. M., Ricci W. M. Branch point selection in alternative splicing of tropomyosin pre-mRNAs. Nucleic Acids Res. 1989 Jul 25;17(14):5633–5650. doi: 10.1093/nar/17.14.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Roscigno R. F., Mulligan G. J., Finn L. A., Weber K. S. Identification of two distinct intron elements involved in alternative splicing of beta-tropomyosin pre-mRNA. Genes Dev. 1990 Jan;4(1):98–110. doi: 10.1101/gad.4.1.98. [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Inoue K., Higuchi I., Sakamoto H., Shimura Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science. 1991 May 10;252(5007):833–836. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol Cell Biol. 1990 Feb;10(2):696–704. doi: 10.1128/mcb.10.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer A. R., Conway G. C., Kozak D. The essential pre-mRNA splicing factor SF2 influences 5' splice site selection by activating proximal sites. Cell. 1990 Jul 13;62(1):35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo H. C., Nasim F. H., Grabowski P. J. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991 Mar 1;251(4997):1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- Leavitt J., Gunning P., Porreca P., Ng S. Y., Lin C. S., Kedes L. Molecular cloning and characterization of mutant and wild-type human beta-actin genes. Mol Cell Biol. 1984 Oct;4(10):1961–1969. doi: 10.1128/mcb.4.10.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Goux-Pelletan M., Brody E., Fiszman M. Y. Exon as well as intron sequences are cis-regulating elements for the mutually exclusive alternative splicing of the beta tropomyosin gene. Mol Cell Biol. 1990 Oct;10(10):5036–5046. doi: 10.1128/mcb.10.10.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Piseri A., Fiszman M. Y. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science. 1991 Jun 28;252(5014):1842–1845. doi: 10.1126/science.2063196. [DOI] [PubMed] [Google Scholar]
- Maniatis T. Mechanisms of alternative pre-mRNA splicing. Science. 1991 Jan 4;251(4989):33–34. doi: 10.1126/science.1824726. [DOI] [PubMed] [Google Scholar]
- Mardon H. J., Sebastio G., Baralle F. E. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987 Oct 12;15(19):7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattox W., Ryner L., Baker B. S. Autoregulation and multifunctionality among trans-acting factors that regulate alternative pre-mRNA processing. J Biol Chem. 1992 Sep 25;267(27):19023–19026. [PubMed] [Google Scholar]
- Mayeda A., Krainer A. R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992 Jan 24;68(2):365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- McKeown M. Alternative mRNA splicing. Annu Rev Cell Biol. 1992;8:133–155. doi: 10.1146/annurev.cb.08.110192.001025. [DOI] [PubMed] [Google Scholar]
- Mullen M. P., Smith C. W., Patton J. G., Nadal-Ginard B. Alpha-tropomyosin mutually exclusive exon selection: competition between branchpoint/polypyrimidine tracts determines default exon choice. Genes Dev. 1991 Apr;5(4):642–655. doi: 10.1101/gad.5.4.642. [DOI] [PubMed] [Google Scholar]
- Nasim F. H., Spears P. A., Hoffmann H. M., Kuo H. C., Grabowski P. J. A Sequential splicing mechanism promotes selection of an optimal exon by repositioning a downstream 5' splice site in preprotachykinin pre-mRNA. Genes Dev. 1990 Jul;4(7):1172–1184. doi: 10.1101/gad.4.7.1172. [DOI] [PubMed] [Google Scholar]
- Noble J. C., Prives C., Manley J. L. Alternative splicing of SV40 early pre-mRNA is determined by branch site selection. Genes Dev. 1988 Nov;2(11):1460–1475. doi: 10.1101/gad.2.11.1460. [DOI] [PubMed] [Google Scholar]
- Norton P. A., Hynes R. O. In vitro splicing of fibronectin pre-mRNAs. Nucleic Acids Res. 1990 Jul 25;18(14):4089–4097. doi: 10.1093/nar/18.14.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Grabowski P. J., Konarska M. M., Seiler S., Sharp P. A. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- Reed R., Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986 Aug 29;46(5):681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Robberson B. L., Cote G. J., Berget S. M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990 Jan;10(1):84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryner L. C., Baker B. S. Regulation of doublesex pre-mRNA processing occurs by 3'-splice site activation. Genes Dev. 1991 Nov;5(11):2071–2085. doi: 10.1101/gad.5.11.2071. [DOI] [PubMed] [Google Scholar]
- Saga Y., Lee J. S., Saraiya C., Boyse E. A. Regulation of alternative splicing in the generation of isoforms of the mouse Ly-5 (CD45) glycoprotein. Proc Natl Acad Sci U S A. 1990 May;87(10):3728–3732. doi: 10.1073/pnas.87.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Patel R. S., Fonda D., Hynes R. O. Multiple sites of alternative splicing of the rat fibronectin gene transcript. EMBO J. 1987 Sep;6(9):2573–2580. doi: 10.1002/j.1460-2075.1987.tb02547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. W., Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989 Mar 10;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- Smith C. W., Patton J. G., Nadal-Ginard B. Alternative splicing in the control of gene expression. Annu Rev Genet. 1989;23:527–577. doi: 10.1146/annurev.ge.23.120189.002523. [DOI] [PubMed] [Google Scholar]
- Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985 Dec;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- Streuli M., Saito H. Regulation of tissue-specific alternative splicing: exon-specific cis-elements govern the splicing of leukocyte common antigen pre-mRNA. EMBO J. 1989 Mar;8(3):787–796. doi: 10.1002/j.1460-2075.1989.tb03439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland S., Smith K. K., Marotti K. R. Hormonal induction of differentiation in teratocarcinoma stem cells: generation of parietal endoderm by retinoic acid and dibutyryl cAMP. Cell. 1980 Sep;21(2):347–355. doi: 10.1016/0092-8674(80)90471-7. [DOI] [PubMed] [Google Scholar]
- Tacke R., Goridis C. Alternative splicing in the neural cell adhesion molecule pre-mRNA: regulation of exon 18 skipping depends on the 5'-splice site. Genes Dev. 1991 Aug;5(8):1416–1429. doi: 10.1101/gad.5.8.1416. [DOI] [PubMed] [Google Scholar]
- Talerico M., Berget S. M. Effect of 5' splice site mutations on splicing of the preceding intron. Mol Cell Biol. 1990 Dec;10(12):6299–6305. doi: 10.1128/mcb.10.12.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Maniatis T. Positive control of pre-mRNA splicing in vitro. Science. 1992 Apr 10;256(5054):237–240. doi: 10.1126/science.1566072. [DOI] [PubMed] [Google Scholar]
- Tsai A. Y., Streuli M., Saito H. Integrity of the exon 6 sequence is essential for tissue-specific alternative splicing of human leukocyte common antigen pre-mRNA. Mol Cell Biol. 1989 Oct;9(10):4550–4555. doi: 10.1128/mcb.9.10.4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler A. M., Lane W. S., Stolk J. A., Roth M. B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992 May;6(5):837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]